Abstract
In most mammals, sex is determined by the presence or absence of the SRY gene. SRY encodes a DNA binding HMG-box transcription factor which, during embryogenesis, is the initial trigger of testis differentiation from the bipotential gonad, yet its precise mode of function remains unclear. In ovarian development, R-spondin1 and Wnt4 act through the Wnt/β-catenin signaling pathway to regulate TCF-dependent expression of unknown target genes and repress testis development. Conversely, SRY may be necessary to prevent the development of ovaries by inhibiting the action of ovarian-determining genes. We hypothesize that SRY prevents Wnt/β-catenin signaling, thereby inhibiting ovarian development. In HEK293T cells, SRY repressed β-catenin-mediated TCF-dependent gene activation in the presence of a specific GSK3β inhibitor or an activated β-catenin mutant, suggesting that SRY inhibits Wnt signaling at the level of β-catenin. Three SRY mutant proteins with nuclear localization defects, encoded by XY male-to-female patients, failed to inhibit β-catenin; surprisingly four SRY sex reversed mutants with defective DNA binding activity showed near wild-type SRY inhibitory activity. Moreover the potent transactivator SRY-VP16 fusion protein also showed wild-type SRY inhibitory activity. Thus SRY inhibition of β-catenin involves neither DNA binding nor transactivation functions of SRY. β-catenin and SRY interact in-vitro and SRY expression triggered β-catenin localization into specific nuclear bodies in NT2/D1 and Hela cells. We conclude that SRY inhibits β-catenin-mediated Wnt signaling by a novel nuclear function of SRY that could be important in sex determination.
Keywords: SRY, β-catenin, Wnt signaling, sex determination, Rspo1, sex reversal
Introduction
Sex is chromosomally controlled in most mammals. In the presence of the Y chromosome, testes form and the male phenotype develops. Testes are required for male development, whereas ovaries are not required for female development. The pathways of development leading to either testis or ovary formation have been considered as either male-specific or female-specific cascade of gene regulation beginning with SRY acting as a genetic switch to divert gonadal development from the default ovarian determination pathway (Berta et al., 1990, Sinclair et al., 1990). However, the recent demonstration that Fgf9 and Wnt4 act as antagonistic signals to regulate mammalian sex determination, provided evidence for a crosstalk between male and female gonadal development (Kim et al., 2006b). In XY mice, testis determination is triggered by the transient expression of a single Y-chromosome gene, Sry (sex-determining region on the Y chromosome) whereas in XX females (in the absence of Sry), ovarian development proceeds (Berta et al., 1990). The role of SRY as a genetic switch for testis development is exemplified in humans by the identification of 46, XY females carrying mutations in the SRY gene and by 46, XX male patients with a translocation of Y chromosome material including the SRY gene (Andersson et al., 1986). In transgenic mice, overexpression of Sry during embryogenesis in an XX gonad leads to female-to-male sex reversal, confirming the requirement of Sry for testis determination (Koopman et al., 1991). These studies clearly identify SRY as a master gene in testis formation, yet the molecular mechanisms of SRY action remain poorly understood.
SRY is a member of the SOX family of proteins characterized by the presence of a DNA binding domain, the High Mobility Group (HMG) (Harley et al., 1992, Harley et al., 1994). The ability of SRY to bind and bend DNA in vitro was established more than fifteen years ago, however its direct target genes are yet to be identified. Unlike other SOX proteins, SRY lacks classic transcriptional activation and repression domains. One of the candidate target genes of SRY is another member of the SOX family of protein, SOX9 (Sry box-9). In XY mice Sox9 is upregulated shortly after the onset of Sry expression in Sertoli cells and ectopic expression of Sry in both XX and XY transgenic mice induces the expression of Sox9 (Kidokoro et al., 2005, Sekido et al., 2004). In cases of human XX males carrying a duplication of the SOX9 gene and in transgenic XX mice overexpressing Sox9, SOX9 itself is sufficient to drive testis determination suggesting that SRY and SOX9 act closely in a testis-forming developmental pathway (Huang et al., 1999, Qin and Bishop, 2005). SOX9 may be able to take over SRY function when SRY is absent or no longer expressed. Unlike SRY, gonadal target genes of SOX9 have been identified in vitro and in vivo, including Amh, Pgds and Vanin1 (De Santa Barbara et al., 1998, Wilhelm et al., 2007, Wilson et al., 2005).
Given that both SRY and SOX9 are sufficient to initiate a testis development pathway in an XX individual and because the majority of cases of XX male patients are due to Y material translocation resulting in the presence of a functional SRY gene, many have thought ovarian development to be the default pathway occurring only in the absence of specification of the male pathway. However, the recent discovery of mutations in the RSPO1 (R-spondin homolog 1) gene in female-to-male XX patients has challenged this view (Parma et al., 2006). Analysis of the testis structure in these patients revealed fully differentiated Sertoli and Leydig cells indicating that, in the absence of SRY, the male sex determination pathway was activated. This observation also indicates that RSPO1 may have two complementary functions, a “pro-ovary” function in initiating female gonadal development and an “anti-testis” function in inhibiting the male gonadal development pathway (Chassot et al., 2008). Several years ago, the analysis of a number of XX male patients led McElreavey et al. to propose that an unidentified female “Z” gene was involved in repressing male gonadal development and that SRY was actively involved in the repression of the “Z” gene in the XY gonad, allowing testis development to proceed (McElreavey et al., 1993). Therefore, it is possible that RSPO1 or downstream effectors of the RSPO1 signaling pathway could assume the function of the “Z” repressor gene (Bernard and Harley, 2007).
RSPO1 is a small secreted factor that is able to stimulate the canonical Wnt/β-catenin signaling pathway by increasing the levels of a Wnt co-receptor, LRP6 (low density lipoprotein receptor-related protein 6) at the membrane of the cell (Binnerts et al., 2007, Kazanskaya et al., 2004, Kim et al., 2005, Kim et al., 2006a, Nam et al., 2006). Importantly, the Wnt/β-catenin pathway is active in the early developing ovary and inactive in the early developing testis (Chassot et al., 2008). Furthermore, a number of SOX proteins, such as SOX1, SOX2, SOX3, SOX7, SOX9 and SOX17 inhibit the canonical Wnt/β-catenin signaling in different models of chondrocyte, neuronal and intestinal differentiation; however the mechanism of their action is still unclear (Akiyama et al., 2004, Bastide et al., 2007, Kan et al., 2004, Mansukhani et al., 2005, Sinner et al., 2007, Takash et al., 2001, Zorn et al., 1999). Here we investigated whether SRY could inhibit the canonical Wnt/β-catenin pathway. We report that SRY can physically interact with β-catenin and inhibit β-catenin-mediated transcription in HEK293T cells. We also show that SRY is able to function at non-transcriptional levels. Our data provides the first evidence for a repressive function of SRY, and support a model whereby SRY inhibits an ovary-determining pathway to prevent female development in an XY gonad.
Material and methods
DNA constructs
All mammalian expression plasmids were of pcDNA3 origin (Clontech) unless otherwise stated. DNA encoding wild-type FLAG-tagged human SRY and clinical mutants R75N, R76P and R133W were previously described (Harley et al., 2003). SRY clinical mutants S18N, R30I, I90M and L163X were produced using the Site-directed Quick change mutagenesis kit (Qiagen) according to the manufacturer’s instructions. pSlax-VP16 and -Engrailed shuttle vectors were a kind gift from Dr. Jonas Muhr as previously described (Sandberg et al., 2005) and were used to produce SRY fusion proteins. Briefly, the SRY open-reading frame was amplified by PCR from pcDNA3-FLAG-SRY using a forward oligonucleotide (containing BamHI, FLAG and KOZAK sequences) and a reverse oligonucleotide that replaces the stop codon with an EcoRI restriction site. The PCR product was then ligated to an EcoRI restriction site that was the beginning of the open-reading frame of either VP16 or Engrailed domains. The fusion proteins were subsequently subcloned into pcDNA3. A similar strategy was used to generate N+HMG, C+HMG and HMG deletion constructs of SRY. HA-tagged human SOX9 expression plasmid was previously described (Preiss et al., 2001). V5-tagged mouse SOX17, V5-tagged mouse SOX17ΔC and GST-β-catenin plasmids were a kind gift of Dr. Aaron Zorn (Zorn et al., 1999). Construct of HA-tagged S37A-β-catenin was previously described (Bernard et al., 2008). The TCF reporter plasmid kit with TOPFLASH and FOPFLASH plasmids was purchased from Millipore. E1b-Luc and S10-E1b-Luc were generated by replacing the CAT gene from E1b-CAT and S10-E1b-CAT previously described (Dubin and Ostrer, 1994).
Antibodies
Rabbit polyclonal antibody against HA-tag (Y-11, sc-805), mouse monoclonal antibody against PML (PG-M3, sc-966) and fluorochrome-coupled secondary antibodies were purchased from Santa Cruz Biotechnology. Mouse monoclonal antibody against stabilized β-catenin (αABC, 8E7, 05–665) was purchased from Upstate Biotechnology. Mouse monoclonal antibody against V5 and goat anti-rabbit Alexa fluor 700 secondary antibody were obtained from Invitrogen. Mouse monoclonal antibody against β-tubulin was purchased from Chemicon. IR rabbit anti-mouse secondary antibody Dye800 was purchased from Rockland Immunochemicals.
Cell Culture and transfections
HEK293T cells (ATCC, CRL-11268) were grown in Dulbecco’s medium. Medium was supplemented with 10% fetal bovine serum and L-glutamine in an atmosphere of 5% CO2. Transient transfections were conducted using Fugene6 (Roche) in accordance with the manufacturer’s instructions. HEK293T cells were seeded at a density of 2×05 cells per well in 6-well plates 24 hours prior to transfection. 48 hours post-transfection the culture media was removed, cell lysate collected and luciferase reporter activity was measured according to the manufacturer’s instructions (Promega). Reporter activity was normalized to β-galactosidase as an internal control (Promega).
Western blots
HEK293T cells were seeded on to 6-well plates at 2×105 cells per well and co-transfected with the indicated plasmids by using the Fugene6 (Roche). At 36 h after transfection, proteins were solubilized, and protein concentration was determined by Bradford assay (Bio-Rad Protein Assay kit; Bio-Rad). Cell lysates were incubated for 5 min at 95°C and resolved by SDS/PAGE. Proteins were transferred to a PVDF membrane (Immun-Blot PVDF membrane; Bio-Rad), incubated with primary antibodies, washed and incubated with either rabbit anti-mouse Dye800 secondary antibody or goat anti-rabbit Alexa fluor 700 secondary antibody. Membranes were scanned on the Odyssey Infrared Imaging System (LiCor). Densitometric analysis was performed by using the ImageJ software package (http://rsb.info.nih.gov/ij/).
In Vitro Protein Binding Assays
In vitro protein binding assay was carried out as previously described (Zorn et al., 1999) with minor modifications. Brifely, GST-β-catenin was expressed in BL-21 bacteria and purified on glutathione coupled agarose resin. 35S-labeled proteins were produced in TNT translation reactions (Promega). 5 μl of a translation reaction was incubated with 1 μg of GST-β-catenin protein coupled to agarose resin overnight at 4°C in 400 μl of binding buffer (25 mM HEPES pH 7.5, 12.5 mM MgCl, 150 mM KCl, 20% glycerol, 0.1% NP-40, 1 mM DTT, 1 mg/ml BSA). GST-agarose pellets were washed five times in NETN (20 mM Tris pH 8.0, 800 mM NaCl, 1 mM EDTA, 0.5% NP-40). Bound proteins were resolved on SDS-PAGE and visualized by autoradiaography using the Storm phosphor-imaging system and ImageQuant analysis software (Amersham).
Electromobility Shift Assay (EMSA)
Proteins were produced using an in vitro coupled transcription and translation process using a TNT Coupled Reticulocyte Lysate System (Promega) according to the manufacturer’s guidelines. EMSA analysis was performed using in vitro translated protein combined with 1 μl of 32P-labeled DNA probe (~40,000 cpm) using standard protocol (Harley et al., 1992). Oligonucleotide sequence of the SRY consensus probe was 5′-GGGTTAACTAAACAATAGAATCTGGTAGA-3′; core binding sequence is underlined. Oligonucleotide sequence of the TCF consensus probe was 5′-AGCTGGTAAGATCAAAGGCTGA-3′; core binding sequence is underlined. The gel was visualized by autoradiography using the Storm phosphor-imaging system and ImageQuant analysis software (Amersham).
Immunohistochemistry
Cells used for immunohistochemistry were seeded on coverslips placed into 6-well plates. Standard protocols were used for immunohistochemistry. The primary antibodies used include affinity-purified mouse monoclonal anti-FLAG (1:500) (Sigma), mouse Anti-PML monoclonal antibody (1:1000) (Santa Cruz Biotechnology). Coverslips were mounted onto slides with DAKO fluorescence mounting medium containing DAPI (final 0.6 mg/ml). Images were captured using an Olympus FV500 confocal laser scanning microscope. Image analysis was performed using ImageJ software package (http://rsb.info.nih.gov/ij/).
Results
SRY represses the Wnt signaling pathway in a cell-specific manner
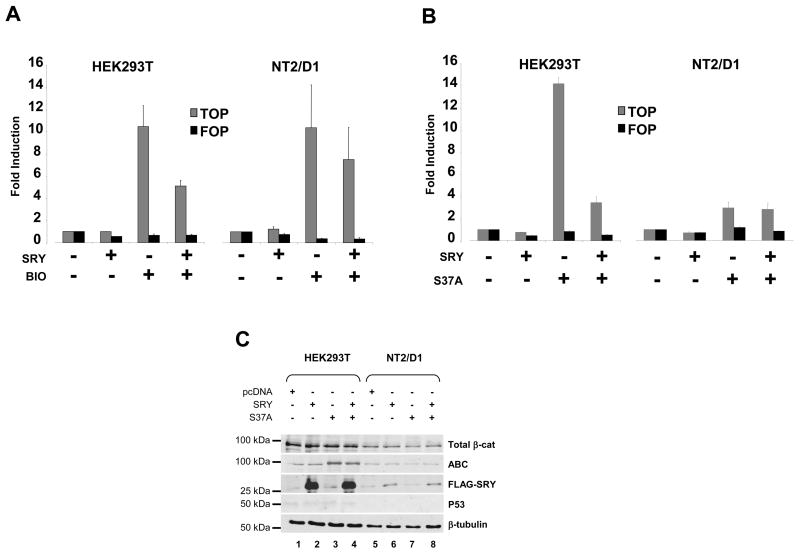
To investigate the effect of SRY on the Wnt signaling pathway we used the TOPFLASH reporter assay in three different cell types, HEK293T (Human Embryonic Kidney), NT2/D1 (Human embryonal carcinoma) and Hela (Human cervix adenocarcinoma). TOPFLASH reporter contains TCF binding sites positioned upstream of a minimal c-fos promoter driving the the luciferase reporter gene expression. FOPFLASH is an inactive control for TOPFLASH, in that it contains mutant TCF binding sites unable to bind the TCF protein. Activation of the Wnt canonical pathway in cells is measured by the transcriptional activation of TOPFLASH upon binding of TCF/β-Catenin protein complex to the TCF binding sites. To activate the TOPFLASH reporter we first used the 6-bromoindirubin-3′-oxime (BIO), a specific inhibitor of GSK3β(Ribas et al., 2006, Sato et al., 2004). Inactivation of GSK3β results in the nuclear accumulation of β-catenin, which activates the transcription of target genes in collaboration with TCF. BIO induced a strong activation of the reporter in HEK293T and NT2/D1 cells (Figure 1A) but was ineffective in Hela cells (data not shown) suggesting that Hela cells lack important factors necessary for a functional canonical Wnt pathway.
Figure 1. SRY, antagonize the canonical Wnt pathway in HEK293T cells.
(A). Cells were transfected with or without SRY together with the TCF-luciferase reporter (TOPFLASH, gray bars) or mutant TCF-luciferase reporter (FOPFLASH, black bars) and treated with 5 μM 6-bromoindirubin-3′-oxime (BIO) for 24h. BIO treatment resulted in a strong activation of the TOPFLASH reporter in HEK293T cells (left panel) or NT2/D1 cells (right panel). SRY repressed the BIO-mediated activation of TOPFLASH in HEK293T cells and to a lesser extent in NT2/D1 cells. Transfection of SRY alone did not affect the basal TOPFLASH activity. Error bars indicate standard errors of the means for triplicate samples from three independent experiments. (B). Cells were transfected with or without SRY together with the TCF-luciferase reporter (TOPFLASH, gray bars) or mutant TCF-luciferase reporter (FOPFLASH, black bars) and the expression plasmid encoding a stabilized form of β-catenin (carrying the mutation S37A). Transfection of β-catenin S37A alone resulted in a strong activation of the TOPFLASH reporter plasmid in HEK293T cells (left panel) and to a lesser extent in NT2/D1 (right panel). SRY repressed the β-catenin-mediated activation of TOPFLASH in HEK293T cells but not in NT2/D1 cells. (C) HEK293T cells and NT2/D1 cells were transfected with pcDNA, SRY alone or together with the β-catenin S37A plasmid. Immunoblotting was performed on 10 μg of whole cell extract resolved by SDS-page. β-tubulin was used as loading control.
Activation of the reporter by BIO was reduced two-fold by SRY in HEK293T cells (Figure 1A, left panel) suggesting that SRY inhibit the Wnt canonical signaling pathway downstream of GSK3β in these cells. In NT2/D1 cells, SRY showed a weaker reduction of BIO activation (Figure 1A, right panel).
To investigate the SRY action downstream of GSK3β, the expression plasmid encoding SRY was co-transfected with or without the expression plasmid encoding a mutant form of β-catenin (S37A, Figure 1B). The S37A mutation produces a more transcriptionally active form of β-catenin than the wild-type protein and strongly activates the TOPFLASH reporter (Bernard et al., 2008, Liu et al., 2002, Rubinfeld et al., 1997). In both HEK293T and NT2/D1 cells, in the absence of SRY, β-catenin S37A stimulated TOPFLASH with the stronger response observed in HEK293T cells (Figure 1B, left panel). In the presence of transfected SRY, activation by S37A was inhibited in HEK293T cells (Figure 1B, left panel) but not in NT2/D1 cells (Figure 1B, right panel). This indicates that SRY inhibits canonical Wnt signaling upstream of TCF/β-catenin in HEK293T cells. The weak or lack of repression observed in NT2/D1 could be due in part to the poor transfection efficiency achieved (20%) when compared to that in HEK293T cells (80%). To investigate this, we prepared whole cell extracts from cells transfected with SRY and/or S37A β-catenin and examined protein levels by immunoblotting (Figure 1C). Levels of stabilized form of β-catenin were visualized using the ABC antibody (Figure 1C) that recognize β-catenin that is not phosphorylated at residues S37 and T41, two of the sites that are phosphorylated by GSK3β (van Noort et al., 2002). Reflecting the transfection efficiency, strong expression of both SRY (Figure 1C, lanes 2 and 4) and the stabilized form of β-catenin (Figure 1C, lanes 3 and 4) were observed in HEK293T cells transfected with SRY and/or S37A β-catenin. In addition, HEK293T cells transfected with SRY showed less stabilized b-catenin than cells lacking SRY (Figure 1C, compare lanes 3 and 4). In NT2/D1 cells, weak expression of SRY (Figure 1C, lanes 6 and 8) was detected and no change in the levels of stabilized form of β-catenin were detected (Figure 1C, compare lane 5 and 6). The weak expression of SRY in NT2/D1 cells could explain partially why SRY is unable to represses TOPFLASH activity induced by either BIO treatment or S37A β-catenin transfection. Furthermore, it is possible that the Wnt canonical pathway in NT2/D1 cells is not fully functional as reported previously (Misiuta et al., 2006).
Because excess of β-catenin has been shown to induce P53 levels when co-transfected with P53 (Damalas et al., 1999), we investigated in our system if β-catenin increased P53 protein levels and if SRY could repress such effect (Figure 1E). P53 was hardly detected in HEK293T cells and not at all in NT2/D1 cells. Levels were unaffected by the overexpression of β-catenin suggesting that, the levels of β-catenin expression was not sufficient to trigger an anti-apoptotic response. This observation is consistent with previous report showing that β-catenin transfection alone is not sufficient to observe an increase in P53 protein levels (Damalas et al., 1999).
Like other SOX proteins, SRY reduces the levels of intracellular β-catenin
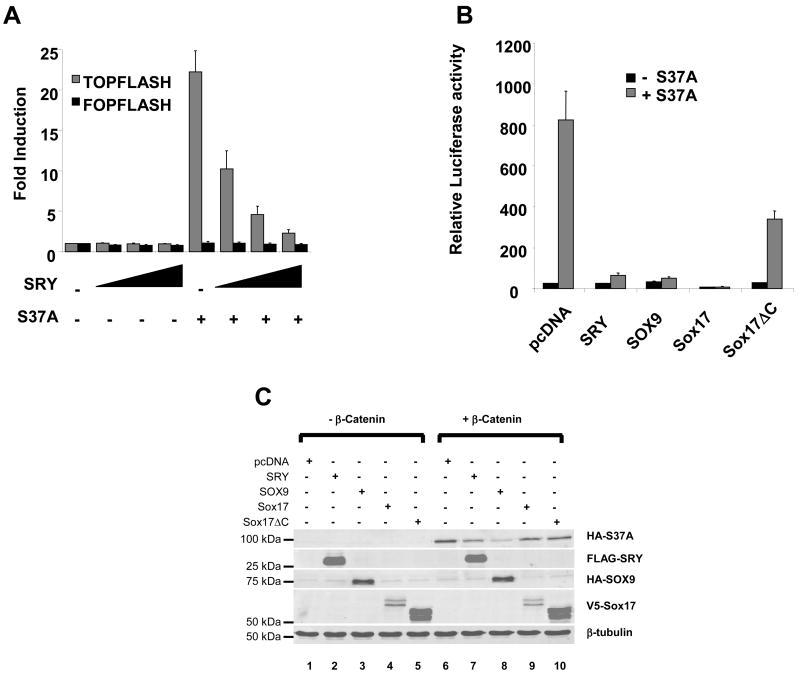
The assay of SRY inhibitory action in HEK293T cells was optimized by varying the concentration of SRY expression plasmid transfected with or without S37A β-catenin plasmid (Figure 2A). S37A β-catenin alone stimulated TOPFLASH more than 20-fold (Figure 2A, black bars). With increasing concentration of SRY, activation by S37A was dose-dependently inhibited to basal levels of TOPFLASH activity. Remarkably, SRY itself was unable to drive the expression of TOPFLASH which suggests that, despite a high sequence similarity between TCF and SRY DNA-binding sites, SRY may not bind TCF sites in the TOPFLASH construct in vivo (see below).
Figure 2. SRY, SOX9 and Sox17 antagonize the canonical Wnt pathway.
(A). HEK293T cells were transfected with increasing concentrations of SRY together with the TCF-luciferase reporter (TOPFLASH, gray bars) or mutant TCF-luciferase reporter (FOPFLASH, black bar) and the expression plasmid encoding a stabilized form of β-catenin (carrying the mutation S37A). Transfection of β-catenin S37A alone resulted in a significant activation of the TOPFLASH reporter plasmid (more than 20-fold). SRY repressed the β-catenin-mediated activation of TOPFLASH in a dose-dependent manner. Transfection of SRY alone did not affect the basal TOPFLASH activity. SRY or β-catenin S37A did not affect FOPFLASH activity. Error bars indicate standard errors of the means for triplicate samples from three independent experiments. (B). HEK293T cells were transfected with either SRY, SOX9, Sox17 or Sox17 ΔC together with the TCF-luciferase reporter (TOPFLASH, gray bars) or mutant TCF-luciferase reporter (FOPFLASH, black bar) and the expression plasmid encoding a stabilized form of β-catenin (carrying the mutation S37A). SRY, SOX9 and Sox17 repressed the β-catenin-mediated activation of TOPFLASH. The mutant Sox17 ΔC, which lacks the β-catenin interaction domain of Sox17, did not strongly inhibit the β-catenin-mediated activation of TOPFLASH suggesting that β-catenin/SOX proteins interaction is necessary for the repression of the Wnt canonical pathway. Error bars indicate standard errors of the means for triplicate samples from two independent experiments. (C) HEK293T cells were transfected with pcDNA, SRY, SOX9, Sox17 or Sox17 ΔC alone or together with the β-catenin S37A plasmid. Immunoblotting was performed on 10 μg of whole cell extract resolved by SDS-page. β-tubulin was used as loading control
In HEK293T cells, both SOX9 and SOX17 strongly repressed the β-catenin activation of TOPFLASH (Figure 2B) consistent with previous reports(Akiyama et al., 2004, Zorn et al., 1999). A mutant SOX17 plasmid, SOX17ΔC, lacking the C-terminus domain involved in β-catenin interaction, showed reduced inhibitory activity (Figure 2B). Altogether, these data indicate that SRY is a potent inhibitor of Wnt signaling at the level of TCF/β-catenin complex similar to other SOX proteins.
SOX9 inhibits Wnt signaling in chondrocytes by binding to, and inducing the degradation of β-catenin (Akiyama et al., 2004). Since SRY was able to inhibit β-catenin S37A transcriptional activity, we investigated the effect of SRY on the levels of β-catenin protein in HEK293T cells. We co-transfected the S37A β-catenin plasmid together with expression plasmids for SRY, SOX9, SOX17 or SOX17 ΔC because endogenous levels in these cells are too low to detect. Exogenous β-catenin protein levels were detected in whole cell extracts using a HA antibody recognizing the HA-tagged S37A mutant β-catenin (Figure 2C). Upon transfection of the S37A plasmid alone, β-catenin protein levels were markedly increased, as expected (Figure 2C, compare lane 1 and lane 6). Overexpression of SOX9 with β-catenin S37A strongly reduced the levels of exogenous β-catenin by 83% (Figure 2C, compare lane 6 and lane 8) confirming previous observations (Akiyama et al., 2004). Overexpression of SOX17 also reduced the levels of β-catenin by 33%, albeit to a lesser extent than SOX9 probably due to the lower level of SOX17 protein levels (Figure 2C, compare 6 and lane 9) whereas mutant SOX17 ΔC, lacking the β-catenin interacting domain, did not to reduce the levels of β-catenin (Figure 2C, compare lane 6 and lane 10). Overexpression of SRY led to a reduction of β-catenin protein levels by 50% (Figure 2C, compare lane 6 and lane 7). Similar to SOX9 action in chondrocytes (Akiyama et al., 2004), SRY may target β-catenin for degradation in HEK293T cells leading to an inhibition of β-catenin transcriptional activity.
SRY and β-catenin proteins interact in vitro
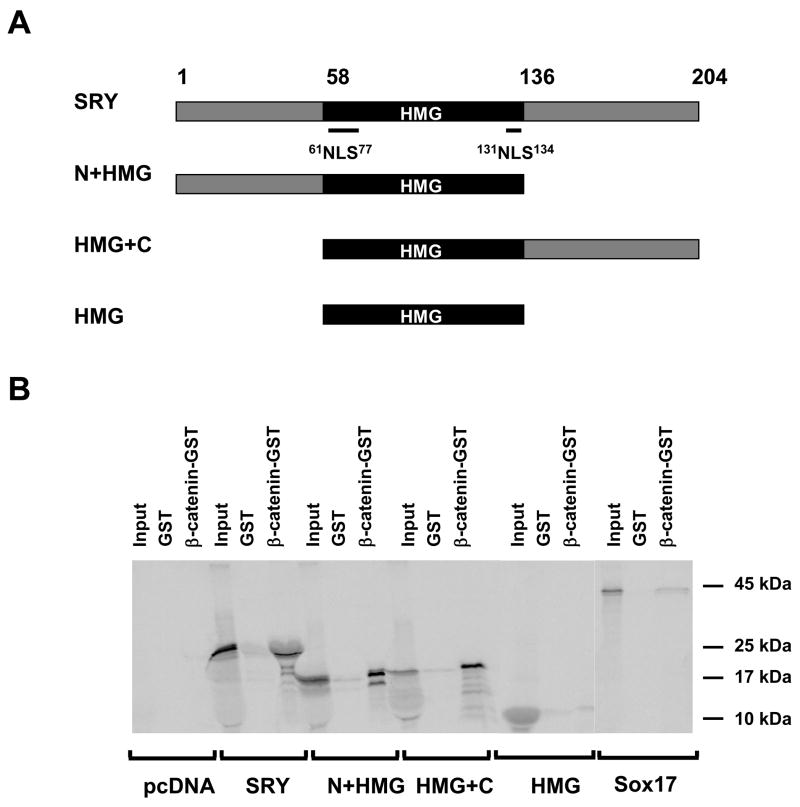
Previous reports suggest that a direct interaction between different non-HMG box regions of SOX9 and SOX17 and the armadillo repeat region of β-catenin is essential for the inhibitory effect of both SOX9 and SOX17 on β-catenin signaling (Akiyama et al., 2004, Zorn et al., 1999). To test the possibility that SRY interacts with β-catenin we used the full length β-catenin as bait and full-length or truncated version of SRY as prey (Figure 3A). We observed that full-length SRY protein is able to interact withβ-catenin (Figure 3B). The affinity of this interaction is as strong, if not stronger, than that of the SOX17/β-catenin interaction. When the truncated SRY constructs were tested, the HMG box alone failed to interact with β-catenin, whereas both the HMG+C-terminus and N+HMG-terminus constructs were able to interact with β-catenin to a similar degree as the full-length protein. This indicates that the SRY interaction with β-catenin requires either the N- or the C-terminus part of the SRY protein, at least in vitro.
Figure 3. SRY interact with β-catenin in vitro.
(A). Schematic representation of SRY deletion constructs used. The HMG box is in black (Amino acids 58–136), N-NLS and C-NLS within the HMG domain are indicated by a line. (B). Interaction of in vitro translated 35S SRY deletions constructs with GST-β-catenin. Input correspond to 10% of the total reaction used for binding assay.
SRY induces β-catenin to localize into nuclear speckles in NT2/D1 and Hela cells but not HEK293T cells
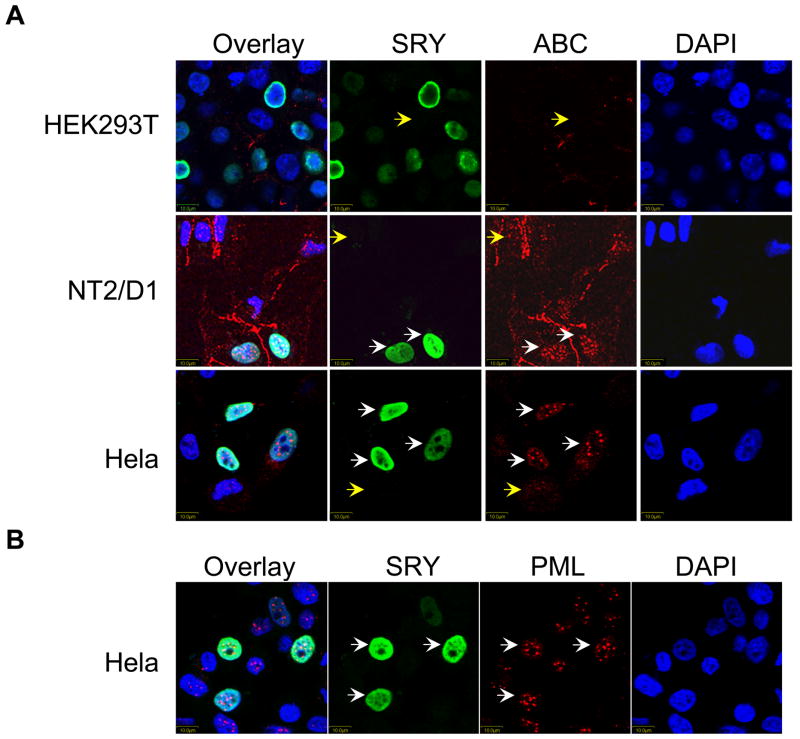
Since SRY reduced the levels of β-catenin protein, we investigated this phenomenon using immunohistochemistry assessing endogenous stabilized form of β-catenin (Figure 4). We anticipated that β-catenin immunoreactivity would be reduced in SRY-transfected cells. We transfected SRY in HEK293T, NT2/D1 and Hela cell lines and used a specific antibody to detect endogenous stabilized β-catenin (ABC antibody). In all three cell lines, in the absence of SRY, the stabilized form of β-catenin is only fainlty detected with the exception of membrane staining in NT2/D1 cells (Yellow arrows, Figure 4A). In SRY-transfected HEK293T cells, very weak staining of β-catenin was observed, whereas in NT2/D1 and Hela cellsβ-catenin is apparent and localized in specific nuclear speckles (Figure 4A). Remarkably, in NT2/D1 and Hela cells, a specific staining of the stabilized form of β-catenin was detected only in SRY positive cells, which could indicate that SRY induces specific reorganization of the stabilized form of β-catenin within the cell.
Figure 4. SRY induces β-catenin accumulation in nuclear bodies in NT2/D1 and Hela cells but not in HEK293T cells.
Immunofluorescence assay of β-catenin localization in HEK293T, NT2/D1 and Hela cells transfected with SRY. (A). SRY and β-catenin double immunofluorescence showing SRY in green and β-catenin in red.β-catenin is undetectable in HEK293T cells (Top panel) and localized in nuclear bodies in NT2/D1 cells (middle panel) and Hela cells (bottom panel). (B). SRY and PML double immunofluorescence showing SRY in the nucleus (green) and PML in PML bodies (red) in Hela cells. DAPI, nuclear stain. White arrows indicate SRY-positive cells and Yellow arrows indicate SRY-negative cells.
To identify the nature of the nuclear speckles observed in NT2/D1 and Hela cells, we performed immunohistochemistry in Hela cells using a specific antibody recognizing the PML protein, a known marker of specific nuclear speckles (Figure 4B). PML staining was evident in nuclear bodies of Hela cells, whether these cells were positive or negative for SRY. Both β-catenin and PML antibodies were raised in mouse precluding a co-immunohistochemistry study. Given the similarity of the speckle size and number with β-catenin staining in SRY positive cells in NT2/D1 and Hela cells, and PML staining in Hela cells, we concluded that SRY induces the localization of β-catenin in nuclear speckles resembling PML bodies in a cell-specific manner. Interestingly, the localization of β-catenin in nuclear speckles was only observed in NT2/D1 and Hela cells in which SRY does not represses the Wnt canonical signaling.
SRY does not require a strong transcriptional activation fucntion to inhibit the Wnt canonical signaling
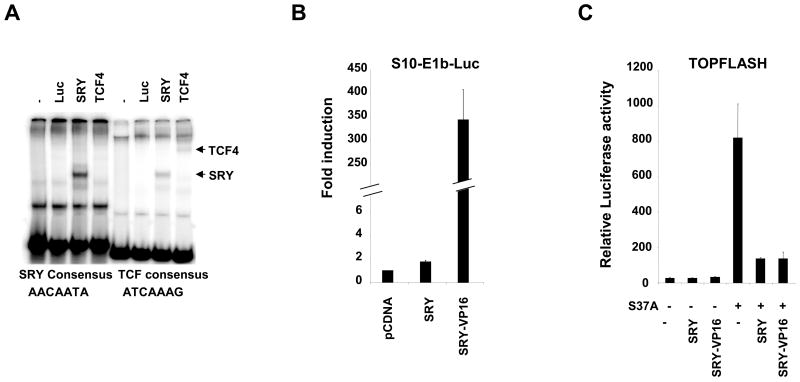
Because the core TCF binding sites on the TOPFLASH reporter (ATCAAAG) are similar to the core consensus binding site for SRY (AACAATA), we first investigated if SRY protein was able to bind TCF sites to explore the possibility that the inhibitory effect of SRY was due to competitive binding between SRY and TCF on TOPFLASH. By electromobility shift assay, SRY bind with lower affinity to the TCF consensus binding site when compared to its consensus binding site (Figure 5A, SRY, compare left and right panels). Conversely, TCF4 bind to its TCF consensus binding site but not to the SRY consensus binding site. These data suggest that a competitive binding between TCF and SRY on TOPFLASH reporter is not a likely mechanism of crosstalk between SRY and Wnt signaling.
Figure 5. Transcriptional function SRY.
(A). Electrophoretic mobility shift assay of synthesised SRY protein or TCF4 in the presence of 32P-radiolabelled SRY consensus binding site (left panel) or TCF consensus binding site (right panel). SRY fusion proteins bind weakly to the TCF consensus binding site, and TCF4 does not bind to SRY consensus binding site. (B). HEK293T cells were transfected with SRY fusion construct together with the S10-E1b-Luciferase reporter. Transfection of SRY alone resulted in a modest activation of the S10-E1b-luciferase reporter (1.7-fold) whereas transfection of SRY-VP16 resulted in a strong activation of the reporter (more than 340-fold). Error bars indicate standard errors of the means for triplicate samples from three independent experiments. (C). HEK293T cells were transfected with SRY fusion constructs together with the TCF-luciferase reporter (TOPFLASH, gray bars) and the expression plasmid encoding a stabilized form of β-catenin (carrying the mutation S37A). Transfection of β-catenin S37A alone resulted in a significant activation of the TOPFLASH reporter plasmid. Both SRY and SRY-VP16 repressed the β-catenin-mediated activation of TOPFLASH to the same extent. Transfection of SRY fusion constructs alone did not affect the basal TOPFLASH activity. Error bars indicate standard errors of the means for triplicate samples from two independent experiments.
In vitro assays have reported that SRY can act as a transcriptional activator (Dubin and Ostrer, 1994) or as a transcriptional repressor (Desclozeaux et al., 1998) depending on the promoter context. Furthermore, SRY acts as a weak transcriptional activator on a specific Sox9 gonad enhancer (Sekido and Lovell-Badge, 2008), K. K., P. B., and V. H., unpublished observations). To test if SRY acts as a transcriptional activator when inhibiting Wnt signaling, we generated a fusion protein of SRY harboring a strong activation domain (VP16) to amplify the signal (Sandberg et al., 2005). The SRY-VP16 fusion protein was stably translated in vitro, and its DNA binding activity retained native SRY DNA binding properties (data not shown). Next we studied the effect of SRY, or SRY-VP16 constructs on S10-E1b-Luc, a reporter containing 10 SOX binding sites in front of a minimal E1b viral promoter (Dubin and Ostrer, 1994). SRY induced a very modest 1.7-fold activation of the reporter whereas SRY-VP16 activated the reporter more than 340-fold (Figure 5B) and did not activate the control reporter indicating that SRY-VP16 is highly active. We then tested the transcriptional effect of SRY-VP16 constructs on the β-catenin-induced TOPFLASH activity (Figure 5C). Both SRY and SRY-VP16 were able to repress β-catenin activation of TOPFLASH to the same extent, which demonstrates that the addition of a strong transcriptional activator to the SRY protein did not change its wild-type activity. This suggests that SRY does not require a transcriptional activation function to inhibit Wnt signaling in HEK293T cells.
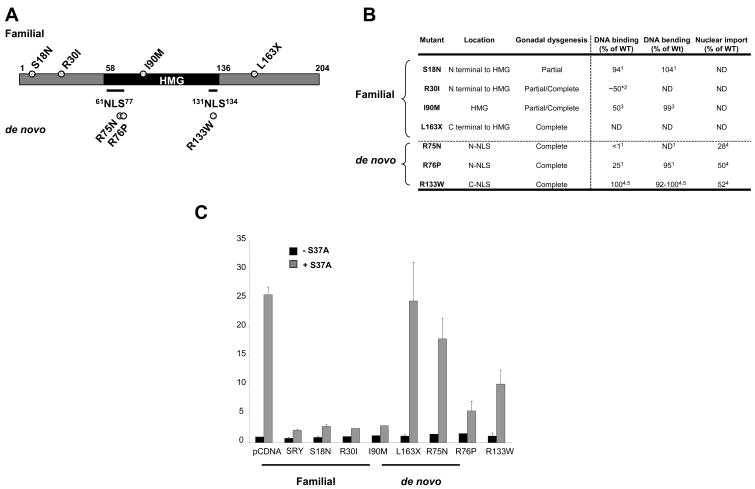
DNA binding activity of SRY is not required for SRY-mediated inhibition of β-catenin activity
To assess the biological significance of SRY inhibition of β-catenin signaling we studied the function of seven clinical mutants of SRY known to cause XY male-to-female sex reversal in human patients (Figure 6A). These mutants were classified into two categories: familial mutations, carried by non-syndromic fathers and causing milder symptoms in the proband (partial to complete gonadal dysgenesis), and de novo mutations which are highly penetrant and cause complete gonadal dysgenesis (Figure 6B). Using the TOPFLASH assay described above, we compared mutant SRY activities with the wild-type SRY inhibitory effect on β-catenin-induced activation (Figure 6C). Strikingly, three out of four familial mutants retained activity comparable to wild-type SRY -that is, all were able to inhibit TOPFLASH activation- in keeping with the mild phenotypic effect of these mutations. Among these three mutants are R30I and I90M which have similarly impaired DNA-binding activities (50% of wild-type DNA-binding activity, Figure 6B), suggesting that the DNA-binding activity of SRY may not be essential for its inhibition of β-catenin signaling. The fourth familial mutant tested, L163X, lacks 42 amino acids of the SRY C-terminus (Tajima et al., 1994). Both the HMG domain and nuclear localization sequences of the SRY protein remain intact in the L163X mutant and it is expected to display wild-type DNA-binding and nuclear import. In the TOPFLASH assay, L163X displayed a complete loss of wild-type activity suggesting that the C-terminal 164–203 part of SRY is essential for the inhibitory function.
Figure 6. SRY does not need a DNA binding activity but requires normal nuclear import to inhibit β-catenin-mediated transcription.
(A). Schematic representation of the familial and de novo mutants used. (B). Biochemical defects of the seven clinical mutants of SRY causing XY male-to-female sex reversal. ND, Not determined. *, percentage of wild-type activity had to be broadly estimated due to the non-numerical reporting of DNA-binding activity of the R30I SRY variant. Biochemical defect obtained from respective references (1(Mitchell and Harley, 2002); 2(Assumpcao et al., 2002); 3(Pontiggia et al., 1994); 4(Harley et al., 2003); 5(Li et al., 2001)). (C). HEK293T cells were transfected with wild-type or clinical mutant SRY plasmids together with the TCF-luciferase reporter (TOPFLASH, gray bars) or mutant TCF-luciferase reporter (FOPFLASH, black bar) and the expression plasmid encoding a stabilized form of β-catenin (carrying the mutation S37A). S18N, R30I, I90M mutants repressed the β-catenin-mediated activation of TOPFLASH to the same extend as the wild-type SRY. All other mutants showed various degree of ineffectiveness in repressing β-catenin-mediated TOPFLASH activation. Error bars indicate standard errors of the means for triplicate samples from three independent experiments.
All three de novo mutants showed impaired inhibitory function when compared to wild-type SRY. Interestingly, R133W and R76P, which have similar nuclear import defects, 52% and 50% of wild-type activity respectively (Harley et al., 2003, Sim et al., 2005), displayed differing abilities to inhibit TOPFLASH activity induced by β-catenin. R133W, which retains almost wild-type DNA binding and bending activities (Harley et al., 2003, Li et al., 2001), was less potent in inhibiting β-catenin-induced TOPFLASH activity than R76P which retains only 33% of DNA binding activity (Harley et al., 2003). We conclude that SRY does not seem to require DNA-binding properties to inhibit β-catenin signaling. Furthermore, the finding that de novo SRY mutants tested, all of which have known nuclear import defects, do not inhibit β-catenin function as efficiently as wild-type protein implying that the nuclear localization of SRY is important for inhibition of β-catenin signaling. The loss of ability of SRY clinical mutants to inhibit β-catenin signaling implies that this repressive function of SRY is likely to be a requisite for male sex determination in humans.
Discussion
Vertebrate SOX proteins are potent effectors of β-catenin signaling required for essential functions such as chondrogenesis, intestinal epithelium differentiation and neuronal differentiation. Here we extend these observations by linking the master sex-determining gene SRY, the founding member of the SOX family, with the Wnt/β-catenin pathway by showing that SRY can inhibit β-catenin signaling in HEK293T cells. While the inhibitory effect of several SOX proteins on Wnt/β-catenin pathway is known in these cells, the molecular mechanism of inhibition is still unclear. Perhaps this confusion resides in our inability to clearly discriminate between the transcriptional and non-transcriptional function of SOX proteins.
A non-transcriptional function of SRY
We have demonstrated that SRY is able to interact directly with β-catenin in vitro and strongly reduce the β-catenin protein levels within HEK293T cells. These observations are consistent with activities of SOX17 and SOX9 which can both interact with, and reduce β-catenin levels (Akiyama et al., 2004, Sinner et al., 2007). Since the reduction of β-catenin levels due to SOX17 or SOX9 was attributed to an increased proteasomal degradation of β-catenin, it is also possible that SRY acted in a similar fashion and this warrants further investigation. Interestingly, while both the C-terminus and N-terminus of SRY interact with β-catenin in vitro, only the C-terminus of the protein was essential in mediating its inhibitory activity as revealed by L163X sex reversed mutant. This is analogous to the β-catenin interaction domains of SOX17 and SOX9 proteins which also lie within the C-terminus of each protein, although these domains share little similarity in amino acid sequence (Akiyama et al., 2004, Zorn et al., 1999). These data suggest that SRY antagonizes the Wnt/β-catenin pathway at the non-genomic level via a direct protein-protein interaction between SRY and β-catenin rather than through conventional DNA binding and transactivation functions. This is supported by the observation that the DNA binding activity of SRY is not required for inhibition of the Wnt/β-catenin pathway in HEK293T cells and by the fact that an SRY fusion protein harboring a strong transcriptional activation domain did not change SRY activity.
While inhibition of the Wnt canonical pathway by SRY was unambiguous in HEK293T cells (Human Embryonic Kidney cells), it was not evident in NT2/D1 cells (a Human Neuroblastoma) or Hela cells (derived from Human Müllerian duct cells). NT2/D1 cells are considered as good model for studying gonadogenesis because they express a number of male sex determining genes (Knower et al., 2007), however they may not be as informative when investigating the Wnt canonical pathway. The weak activation of TOPFLASH by S37A β-catenin we observed is consistent with studies showing that sustained treatment by LiCl is unable to inhibit GSK3β in NT2/D1 cells (Misiuta et al., 2006). In Hela cells, despite the non-specific action of SRY on TOPFLASH/FOPFLASH reporters, SRY had no effect on Wnt canonical signaling. Consistent with these observations, β-catenin localization in nuclear speckles is observed only in NT2/D1 and Hela cells, in which SRY did not inhibit β-catenin action. β-catenin post-transcriptional modification changes its subcellular localization (Brembeck et al., 2004) and the localization of β-catenin in specific nuclear speckles induced by SRY in NT2/D1 and Hela cells suggests that SRY may be able to modify the β-catenin protein post-transcriptionally to target it to these nuclear speckles. The presence of β-catenin in nuclear speckles such as PML bodies has already been observed and has been linked to active transcription of specific β-catenin target genes involved in growth inhibition and senescence independently of P53 activation. Canonical Wnt target genes, such as Cylin D1, were not induced suggesting β-catenin was compartmentalized and inactive (Shtutman et al., 2002). While, it is unlikely that SRY inhibition of β-catenin in HEK293T cells lead to cell death, the effect of SRY in NT2/D1 and Hela cells, may lead to β-catenin-mediated growth inhibition.
SRY and human sex-reversal
Our data on sex reversing clinical mutant implies that inhibition of β-catenin is an essential function of SRY in sex determination. Penetrant de novo mutants which cause complete sex-reversal show loss of inhibition of β-catenin which would presumably lead to a failure to repress the ovarian pathway in these patients. In contrast, patients with familial mutations of SRY showed residual β-catenin inhibitory activity. The fact that nuclear import defects affect these activities while DNA binding defects seem to play a lesser role, point to a novel nuclear function of SRY in β-catenin interaction and inhibition. The localization of β-catenin into nuclear speckles resembling PML bodies unveils part of the inhibitory mechanism given that PML bodies are known to regulate transcription by modulating the availability or the activity of transcription factors (Bernardi and Pandolfi, 2007).
Is Wnt/β-catenin the proposed “Z” factor?
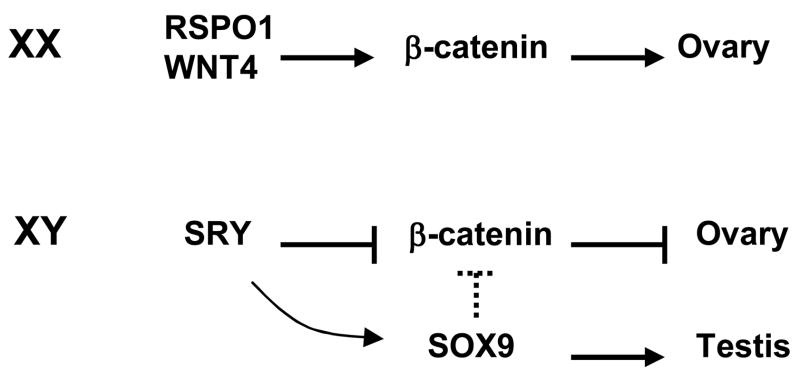
As demonstrated in vivo using reporter mouse lines, the canonical Wnt/β-catenin pathway is a female specific event in the developing ovary (Chassot et al., 2008). One of the main activators of this pathway is RSPO1 which, when mutated, gives rise to 46, XX male individuals with differentiated testes demonstrating that RSPO1 is a female sex determining gene (Parma et al., 2006). Furthermore, the lack of Rspo1 in XX mice mutant embryo leads to the loss of nuclear β-catenin indicating that the transcriptional function of β-catenin was impaired in these animals (Chassot et al., 2008). A second effector of the Wnt/β-catenin signaling pathway in the ovary is Wnt4. The lack of Wnt4 in the XX female mouse embryo leads to masculinization, with the absence of Müllerian duct and the presence of the Wolffian duct (Vainio et al., 1999). In addition, Wnt4 prevents the migration of endothelial cells into the developing ovary, an event that is male-specific (Jeays-Ward et al., 2003). We have shown previously that Wnt4 relocates β-catenin to the cell membrane, inhibiting β-catenin transcriptional activity and strengthening cell-cell contact (Bernard et al., 2008), which could explain how Wnt4 inhibits endothelial cell migration in the ovary. Wnt4 is downstream of Rspo1 in mouse ovarian differentiation (Chassot et al., 2008). Thus we speculate that activation of the nuclear β-catenin function can be triggered by the expression of Rspo1 gene, while the subsequent expression of Wnt4 leads to relocation of some cytoplasmic β-catenin to the cell membrane while the nuclear β-catenin will remain active in the developing ovary. These observations suggest that ovarian differentiation requires both membrane and nuclear forms of β-catenin whose subcellular localization is controlled by at least two different mechanisms.
The studies on RSPO1 function demonstrated for the first time that, in human females, the ovarian developmental pathway is “anti-testis” with the Wnt/β-catenin signaling pathway antagonizing male gonadal development in a normal XX individual (Parma et al., 2006). Conversely, one can assume that in the developing testis, regulatory measures are in place to prevent activation of the ovarian pathway while normal testis development proceeds. One such measure could be the function of SRY as an “anti-ovary” gene to antagonize Wnt/β-catenin signaling allowing normal testis differentiation in the XY gonad. A similar but more general hypothesis was put forward by McElreavey et al. (McElreavey et al., 1993), who suggested that SRY may repress a negative regulator, termed “Z” of male development. Our data identifies a repressive “anti-ovary” function of SRY: the inhibition of the female-specific Wnt/β-catenin signaling pathway, where the Wnt/β-catenin signaling pathway is the negative regulator “Z” (Figure 7). Another candidate that could be involved in repressing the Wnt/β-catenin pathway during testis differentiation is SOX9. SOX9 can antagonize β-catenin function in chondrocytes (Akiyama et al., 2004) and intestinal epithelium (Bastide et al., 2007), however its action on Wnt/β-catenin pathway in the developing gonad has not been thoroughly investigated so far. It is interesting to note that, in XY Sox9 -/- mice, in the presence of Sry, two ovarian genes, Wnt4 and Foxl2 are expressed (Barrionuevo et al., 2006), indicating that Sox9 alone maybe responsible for the downregulation of Wnt4 or Foxl2 in the XY gonad.
Figure 7. SRY inhibition of Wnt/β-catenin pathway during sex determination.
Model depicting proposed mechanism of SRY action on β-catenin function during sex determination. Broken lines indicate putative link.
While Wnt/β-catenin signaling is an important pathway in female ovarian determination, growing evidence suggests that other ovarian genes, such as Foxl2 are also important for ovarian development (Ottolenghi et al., 2007). Foxl2 has been a candidate gene responsible, when deleted, for XX sex-reversal in goats (Pailhoux et al., 2001) and it is expressed in ovarian tissue present in sex-reversed patients (Hersmus et al., 2008). Interestingly, in mice lacking genes relevant to the Wnt/β-catenin pathway, namely Rspo1 and Wnt4, Foxl2 expression is not changed which suggests that Foxl2 is acting independently of the Wnt/β-catenin pathway (Chassot et al., 2008, Ottolenghi et al., 2007). This suggests that multiple independent signals are involved in ovarian development as part of an “ovary organizer” (Ottolenghi et al., 2007). It is possible that SRY inhibition of Wnt/β-catenin pathway will not be sufficient to prevent ovarian development, and that SRY perhaps via SOX9 also need to repress other ovarian genes such as Foxl2.
Acknowledgments
This work was supported by NIH RO1 HD 44513 to EV and VH and NHMRC (Australia) Program Grant 334314 to VH. We thank Louisa Ludbrook and Stefan Bagheri-Fam for critical reading of the manuscript.
Abbreviations
- SRY
Sex determining region of the Y chromosome
- RSPO1
R-spondin 1
- Wnt4
wingless-related MMTV integration site 4
- SOX9
SRY type high mobility group box 9
- Fgf9
fibroblast growth factor 9
- TCF
T-cell specific HMG-box factor
- AMH
anti-Müllerian hormone
- Pgds
prostaglandin D synthase
- LRP6
low density lipoprotein receptor-related protein receptor 6
- VP16
Virion protein 16
- PML
promyelocytic leukemia
- Foxl2
forkhead box L2
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Akiyama H, Lyons JP, Mori-Akiyama Y, Yang X, Zhang R, Zhang Z, Deng JM, Taketo MM, Nakamura T, Behringer RR, McCrea PD, de Crombrugghe B. Interactions between Sox9 and beta-catenin control chondrocyte differentiation. Genes Dev. 2004;18:1072–1087. doi: 10.1101/gad.1171104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson M, Page DC, de la Chapelle A. Chromosome Y-specific DNA is transferred to the short arm of X chromosome in human XX males. Science. 1986;233:786–788. doi: 10.1126/science.3738510. [DOI] [PubMed] [Google Scholar]
- Assumpcao JG, Benedetti CE, Maciel-Guerra AT, Guerra G, Jr, Baptista MT, Scolfaro MR, de Mello MP. Novel mutations affecting SRY DNA-binding activity: the HMG box N65H associated with 46, XY pure gonadal dysgenesis and the familial non-HMG box R30I associated with variable phenotypes. J Mol Med. 2002;80:782–790. doi: 10.1007/s00109-002-0376-9. [DOI] [PubMed] [Google Scholar]
- Barrionuevo F, Bagheri-Fam S, Klattig J, Kist R, Taketo MM, Englert C, Scherer G. Homozygous inactivation of Sox9 causes complete XY sex reversal in mice. Biol Reprod. 2006;74:195–201. doi: 10.1095/biolreprod.105.045930. [DOI] [PubMed] [Google Scholar]
- Bastide P, Darido C, Pannequin J, Kist R, Robine S, Marty-Double C, Bibeau F, Scherer G, Joubert D, Hollande F, Blache P, Jay P. Sox9 regulates cell proliferation and is required for Paneth cell differentiation in the intestinal epithelium. J Cell Biol. 2007;178:635–648. doi: 10.1083/jcb.200704152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P, Fleming A, Lacombe A, Harley VR, Vilain E. Wnt4 inhibits beta-catenin/TCF signalling by redirecting beta-catenin to the cell membrane. Biol Cell. 2008;100:167–177. doi: 10.1042/BC20070072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard P, Harley VR. Wnt4 action in gonadal development and sex determination. Int J Biochem Cell Biol. 2007;39:31–43. doi: 10.1016/j.biocel.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- Berta P, Hawkins JR, Sinclair AH, Taylor A, Griffiths BL, Goodfellow PN, Fellous M. Genetic evidence equating SRY and the testis-determining factor. Nature. 1990;348:448–450. doi: 10.1038/348448A0. [DOI] [PubMed] [Google Scholar]
- Binnerts ME, Kim KA, Bright JM, Patel SM, Tran K, Zhou M, Leung JM, Liu Y, Lomas WE, 3rd, Dixon M, Hazell SA, Wagle M, Nie WS, Tomasevic N, Williams J, Zhan X, Levy MD, Funk WD, Abo A. R-Spondin1 regulates Wnt signaling by inhibiting internalization of LRP6. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:14700–14705. doi: 10.1073/pnas.0702305104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brembeck FH, Schwarz-Romond T, Bakkers J, Wilhelm S, Hammerschmidt M, Birchmeier W. Essential role of BCL9-2 in the switch between beta-catenin’s adhesive and transcriptional functions. Genes Dev. 2004;18:2225–2230. doi: 10.1101/gad.317604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chassot AA, Ranc F, Gregoire EP, Roepers-Gajadien HL, Taketo MM, Camerino G, de Rooij DG, Schedl A, Chaboissier MC. Activation of {beta}-catenin signalling by Rspo1 controls differentiation of the mammalian ovary. Human Molecular Genetics. 2008 doi: 10.1093/hmg/ddn016. [DOI] [PubMed] [Google Scholar]
- Damalas A, Ben-Ze’ev A, Simcha I, Shtutman M, Leal JF, Zhurinsky J, Geiger B, Oren M. Excess beta-catenin promotes accumulation of transcriptionally active p53. EMBO J. 1999;18:3054–3063. doi: 10.1093/emboj/18.11.3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Santa Barbara P, Bonneaud N, Boizet B, Desclozeaux M, Moniot B, Sudbeck P, Scherer G, Poulat F, Berta P. Direct interaction of SRY-related protein SOX9 and steroidogenic factor 1 regulates transcription of the human anti-Mullerian hormone gene. Molecular & Cellular Biology. 1998;18:6653–6665. doi: 10.1128/mcb.18.11.6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desclozeaux M, Poulat F, de Santa Barbara P, Capony JP, Turowski P, Jay P, Mejean C, Moniot B, Boizet B, Berta P. Phosphorylation of an N-terminal motif enhances DNA-binding activity of the human SRY protein. Journal of Biological Chemistry. 1998;273:7988–7995. doi: 10.1074/jbc.273.14.7988. [DOI] [PubMed] [Google Scholar]
- Dubin RA, Ostrer H. Sry is a transcriptional activator. Mol Endocrinol. 1994;8:1182–1192. doi: 10.1210/mend.8.9.7838151. [DOI] [PubMed] [Google Scholar]
- Harley VR, Jackson DI, Hextall PJ, Hawkins JR, Berkovitz GD, Sockanathan S, Lovell-Badge R, Goodfellow PN. DNA binding activity of recombinant SRY from normal males and XY females. Science. 1992;255:453–456. doi: 10.1126/science.1734522. [DOI] [PubMed] [Google Scholar]
- Harley VR, Layfield S, Mitchell CL, Forwood JK, John AP, Briggs LJ, McDowall SG, Jans DA. Defective importin beta recognition and nuclear import of the sex-determining factor SRY are associated with XY sex-reversing mutations. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:7045–7050. doi: 10.1073/pnas.1137864100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley VR, Lovell-Badge R, Goodfellow PN. Definition of a consensus DNA binding site for SRY. Nucleic Acids Research. 1994;22:1500–1501. doi: 10.1093/nar/22.8.1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersmus R, Kalfa N, de Leeuw B, Stoop H, Oosterhuis JW, de Krijger R, Wolffenbuttel KP, Drop SL, Veitia RA, Fellous M, Jaubert F, Looijenga LH. FOXL2 and SOX9 as parameters of female and male gonadal differentiation in patients with various forms of disorders of sex development (DSD) J Pathol. 2008;215:31–38. doi: 10.1002/path.2335. [DOI] [PubMed] [Google Scholar]
- Huang B, Wang S, Ning Y, Lamb AN, Bartley J. Autosomal XX sex reversal caused by duplication of SOX9. American Journal of Medical Genetics. 1999;87:349–353. doi: 10.1002/(sici)1096-8628(19991203)87:4<349::aid-ajmg13>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- Jeays-Ward K, Hoyle C, Brennan J, Dandonneau M, Alldus G, Capel B, Swain A. Endothelial and steroidogenic cell migration are regulated by WNT4 in the developing mammalian gonad. Development. 2003;130:3663–3670. doi: 10.1242/dev.00591. [DOI] [PubMed] [Google Scholar]
- Kan L, Israsena N, Zhang Z, Hu M, Zhao LR, Jalali A, Sahni V, Kessler JA. Sox1 acts through multiple independent pathways to promote neurogenesis. Developmental Biology. 2004;269:580–594. doi: 10.1016/j.ydbio.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Kazanskaya O, Glinka A, del Barco Barrantes I, Stannek P, Niehrs C, Wu W. R-Spondin2 is a secreted activator of Wnt/beta-catenin signaling and is required for Xenopus myogenesis. Dev Cell. 2004;7:525–534. doi: 10.1016/j.devcel.2004.07.019. [DOI] [PubMed] [Google Scholar]
- Kidokoro T, Matoba S, Hiramatsu R, Fujisawa M, Kanai-Azuma M, Taya C, Kurohmaru M, Kawakami H, Hayashi Y, Kanai Y, Yonekawa H. Influence on spatiotemporal patterns of a male-specific Sox9 activation by ectopic Sry expression during early phases of testis differentiation in mice. Developmental Biology. 2005;278:511–525. doi: 10.1016/j.ydbio.2004.11.006. [DOI] [PubMed] [Google Scholar]
- Kim KA, Kakitani M, Zhao J, Oshima T, Tang T, Binnerts M, Liu Y, Boyle B, Park E, Emtage P, Funk WD, Tomizuka K. Mitogenic influence of human R-spondin1 on the intestinal epithelium. Science. 2005;309:1256–1259. doi: 10.1126/science.1112521. [DOI] [PubMed] [Google Scholar]
- Kim KA, Zhao J, Andarmani S, Kakitani M, Oshima T, Binnerts ME, Abo A, Tomizuka K, Funk WD. R-Spondin proteins: a novel link to beta-catenin activation. Cell Cycle. 2006a;5:23–26. doi: 10.4161/cc.5.1.2305. [DOI] [PubMed] [Google Scholar]
- Kim Y, Kobayashi A, Sekido R, DiNapoli L, Brennan J, Chaboissier MC, Poulat F, Behringer RR, Lovell-Badge R, Capel B. Fgf9 and Wnt4 act as antagonistic signals to regulate mammalian sex determination. Plos Biology. 2006b;4:e187. doi: 10.1371/journal.pbio.0040187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knower KC, Sim H, McClive PJ, Bowles J, Koopman P, Sinclair AH, Harley VR. Characterisation of urogenital ridge gene expression in the human embryonal carcinoma cell line NT2/D1. Sex Dev. 2007;1:114–126. doi: 10.1159/000100033. [DOI] [PubMed] [Google Scholar]
- Koopman P, Gubbay J, Vivian N, Goodfellow P, Lovell-Badge R. Male development of chromosomally female mice transgenic for Sry. Nature. 1991;351:117–121. doi: 10.1038/351117a0. [see comment] [DOI] [PubMed] [Google Scholar]
- Li B, Zhang W, Chan G, Jancso-Radek A, Liu S, Weiss MA. Human sex reversal due to impaired nuclear localization of SRY. A clinical correlation. Journal of Biological Chemistry. 2001;276:46480–46484. doi: 10.1074/jbc.C100388200. [DOI] [PubMed] [Google Scholar]
- Liu C, Li Y, Semenov M, Han C, Baeg GH, Tan Y, Zhang Z, Lin X, He X. Control of beta-catenin phosphorylation/degradation by a dual-kinase mechanism. Cell. 2002;108:837–847. doi: 10.1016/s0092-8674(02)00685-2. [DOI] [PubMed] [Google Scholar]
- Mansukhani A, Ambrosetti D, Holmes G, Cornivelli L, Basilico C. Sox2 induction by FGF and FGFR2 activating mutations inhibits Wnt signaling and osteoblast differentiation. J Cell Biol. 2005;168:1065–1076. doi: 10.1083/jcb.200409182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElreavey K, Vilain E, Abbas N, Herskowitz I, Fellous M. A regulatory cascade hypothesis for mammalian sex determination: SRY represses a negative regulator of male development. Proceedings of the National Academy of Sciences of the United States of America. 1993;90:3368–3372. doi: 10.1073/pnas.90.8.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misiuta IE, Saporta S, Sanberg PR, Zigova T, Willing AE. Influence of retinoic acid and lithium on proliferation and dopaminergic potential of human NT2 cells. J Neurosci Res. 2006;83:668–679. doi: 10.1002/jnr.20718. [DOI] [PubMed] [Google Scholar]
- Mitchell CL, Harley VR. Biochemical defects in eight SRY missense mutations causing XY gonadal dysgenesis. Mol Genet Metab. 2002;77:217–225. doi: 10.1016/s1096-7192(02)00165-8. [DOI] [PubMed] [Google Scholar]
- Nam JS, Turcotte TJ, Smith PF, Choi S, Yoon JK. Mouse cristin/R-spondin family proteins are novel ligands for the Frizzled 8 and LRP6 receptors and activate beta-catenin-dependent gene expression. Journal of Biological Chemistry. 2006;281:13247–13257. doi: 10.1074/jbc.M508324200. [DOI] [PubMed] [Google Scholar]
- Ottolenghi C, Pelosi E, Tran J, Colombino M, Douglass E, Nedorezov T, Cao A, Forabosco A, Schlessinger D. Loss of Wnt4 and Foxl2 leads to female-to-male sex reversal extending to germ cells. Human Molecular Genetics. 2007;16:2795–2804. doi: 10.1093/hmg/ddm235. [DOI] [PubMed] [Google Scholar]
- Pailhoux E, Vigier B, Chaffaux S, Servel N, Taourit S, Furet JP, Fellous M, Grosclaude F, Cribiu EP, Cotinot C, Vaiman D. A 11.7-kb deletion triggers intersexuality and polledness in goats. Nature Genetics. 2001;29:453–458. doi: 10.1038/ng769. [DOI] [PubMed] [Google Scholar]
- Parma P, Radi O, Vidal V, Chaboissier MC, Dellambra E, Valentini S, Guerra L, Schedl A, Camerino G. R-spondin1 is essential in sex determination, skin differentiation and malignancy. Nature Genetics. 2006;38:1304–1309. doi: 10.1038/ng1907. [see comment] [DOI] [PubMed] [Google Scholar]
- Pontiggia A, Rimini R, Harley VR, Goodfellow PN, Lovell-Badge R, Bianchi ME. Sex-reversing mutations affect the architecture of SRY-DNA complexes. EMBO J. 1994;13:6115–6124. doi: 10.1002/j.1460-2075.1994.tb06958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preiss S, Argentaro A, Clayton A, John A, Jans DA, Ogata T, Nagai T, Barroso I, Schafer AJ, Harley VR. Compound effects of point mutations causing campomelic dysplasia/autosomal sex reversal upon SOX9 structure, nuclear transport, DNA binding, and transcriptional activation. Journal of Biological Chemistry. 2001;276:27864–27872. doi: 10.1074/jbc.M101278200. [DOI] [PubMed] [Google Scholar]
- Qin Y, Bishop CE. Sox9 is sufficient for functional testis development producing fertile male mice in the absence of Sry. Human Molecular Genetics. 2005;14:1221–1229. doi: 10.1093/hmg/ddi133. [DOI] [PubMed] [Google Scholar]
- Ribas J, Bettayeb K, Ferandin Y, Knockaert M, Garrofe-Ochoa X, Totzke F, Schachtele C, Mester J, Polychronopoulos P, Magiatis P, Skaltsounis AL, Boix J, Meijer L. 7-Bromoindirubin-3′-oxime induces caspase-independent cell death. Oncogene. 2006;25:6304–6318. doi: 10.1038/sj.onc.1209648. [DOI] [PubMed] [Google Scholar]
- Rubinfeld B, Robbins P, El-Gamil M, Albert I, Porfiri E, Polakis P. Stabilization of beta-catenin by genetic defects in melanoma cell lines. Science. 1997;275:1790–1792. doi: 10.1126/science.275.5307.1790. [DOI] [PubMed] [Google Scholar]
- Sandberg M, Kallstrom M, Muhr J. Sox21 promotes the progression of vertebrate neurogenesis. Nat Neurosci. 2005;8:995–1001. doi: 10.1038/nn1493. [DOI] [PubMed] [Google Scholar]
- Sato N, Meijer L, Skaltsounis L, Greengard P, Brivanlou AH. Maintenance of pluripotency in human and mouse embryonic stem cells through activation of Wnt signaling by a pharmacological GSK-3-specific inhibitor. Nat Med. 2004;10:55–63. doi: 10.1038/nm979. [DOI] [PubMed] [Google Scholar]
- Sekido R, Bar I, Narvaez V, Penny G, Lovell-Badge R. SOX9 is up-regulated by the transient expression of SRY specifically in Sertoli cell precursors. Developmental Biology. 2004;274:271–279. doi: 10.1016/j.ydbio.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Sekido R, Lovell-Badge R. Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature. 2008;453:930–934. doi: 10.1038/nature06944. [DOI] [PubMed] [Google Scholar]
- Shtutman M, Zhurinsky J, Oren M, Levina E, Ben-Ze’ev A. PML is a target gene of beta-catenin and plakoglobin, and coactivates beta-catenin-mediated transcription. Cancer Res. 2002;62:5947–5954. [PubMed] [Google Scholar]
- Sim H, Rimmer K, Kelly S, Ludbrook LM, Clayton AH, Harley VR. Defective calmodulin-mediated nuclear transport of the sex-determining region of the Y chromosome (SRY) in XY sex reversal. Mol Endocrinol. 2005;19:1884–1892. doi: 10.1210/me.2004-0334. [DOI] [PubMed] [Google Scholar]
- Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, Frischauf AM, Lovell-Badge R, Goodfellow PN. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–244. doi: 10.1038/346240a0. [see comment] [DOI] [PubMed] [Google Scholar]
- Sinner D, Kordich JJ, Spence JR, Opoka R, Rankin S, Lin SC, Jonatan D, Zorn AM, Wells JM. Sox17 and Sox4 differentially regulate beta-catenin/T-cell factor activity and proliferation of colon carcinoma cells. Molecular & Cellular Biology. 2007;27:7802–7815. doi: 10.1128/MCB.02179-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima T, Nakae J, Shinohara N, Fujieda K. A novel mutation localized in the 3′ non-HMG box region of the SRY gene in 46, XY gonadal dysgenesis. Human Molecular Genetics. 1994;3:1187–1189. doi: 10.1093/hmg/3.7.1187. [DOI] [PubMed] [Google Scholar]
- Takash W, Canizares J, Bonneaud N, Poulat F, Mattei MG, Jay P, Berta P. SOX7 transcription factor: sequence, chromosomal localisation, expression, transactivation and interference with Wnt signalling. Nucleic Acids Research. 2001;29:4274–4283. doi: 10.1093/nar/29.21.4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vainio S, Heikkila M, Kispert A, Chin N, McMahon AP. Female development in mammals is regulated by Wnt-4 signalling. Nature. 1999;397:405–409. doi: 10.1038/17068. [DOI] [PubMed] [Google Scholar]
- van Noort M, Meeldijk J, van der Zee R, Destree O, Clevers H. Wnt signaling controls the phosphorylation status of beta-catenin. Journal of Biological Chemistry. 2002;277:17901–17905. doi: 10.1074/jbc.M111635200. [DOI] [PubMed] [Google Scholar]
- Wilhelm D, Hiramatsu R, Mizusaki H, Widjaja L, Combes AN, Kanai Y, Koopman P. SOX9 regulates prostaglandin D synthase gene transcription in vivo to ensure testis development. Journal of Biological Chemistry. 2007;282:10553–10560. doi: 10.1074/jbc.M609578200. [DOI] [PubMed] [Google Scholar]
- Wilson MJ, Jeyasuria P, Parker KL, Koopman P. The transcription factors steroidogenic factor-1 and SOX9 regulate expression of Vanin-1 during mouse testis development. Journal of Biological Chemistry. 2005;280:5917–5923. doi: 10.1074/jbc.M412806200. [DOI] [PubMed] [Google Scholar]
- Zorn AM, Barish GD, Williams BO, Lavender P, Klymkowsky MW, Varmus HE. Regulation of Wnt signaling by Sox proteins: XSox17 alpha/beta and XSox3 physically interact with beta-catenin. Mol Cell. 1999;4:487–498. doi: 10.1016/s1097-2765(00)80200-2. [DOI] [PubMed] [Google Scholar]