Abstract
Background
The nucleic acid-binding protein Purα is involved at stalled DNA replication forks, in double-strand break (DSB) DNA repair and the cellular response to DNA replication stress. Purα interacts with HIV-1 Tat, which regulates homologous recombination-directed DNA repair (HRR).
Materials and Methods
We investigated Rad51 and HRR regulation in mouse embryo fibroblasts (MEFs) from PURA-/- knockout mice that lack Purα.
Results
Rad51 was induced in PURA-/- MEFs but was repressed when Purα was ectopically expressed in these cells. Similarly Rad51 inversely correlated with the level of Purα in normal postnatal mouse brain. HIV-1 Tat stimulated HRR DNA repair of I-SceI induced DNA DSBs and the nuclear appearance of Rad51 foci. In contrast, Purα suppressed HRR DNA repair, Rad51 expression, and Rad51 foci formation.
Conclusion
Tat stimulates the Rad51 promoter involving both Purα-dependent and Purα-independent mechanisms. Interaction between Purα and Tat may have opposing effects on Rad51 expression. The effects may on HRR may contribute to HIV-1 associated pathogenesis.
Keywords: Pur-alpha, HIV-1 Tat, Rad51, DNA repair
Purα is a protein that was originally purified from mouse brain based on its ability to bind to a DNA sequence derived from the promoter of the mouse myelin basic protein gene (1, 2). Human Purα was characterized by its ability to bind to a DNA sequence present upstream of the human c-Myc gene and was cloned from HeLa cells and sequenced (3, 4). The sequence of mouse Purα (5) is almost identical to human Purα (4) with only two amino acid residues differing. The DNA-binding domain of Purα is strongly conserved throughout evolution. Purα is a member of the Pur family of proteins along with Purβ (4) and Purγ which has two isoforms that arise from the usage of alternative polyadenylation sites (6), and it is expressed in virtually every metazoan tissue (7). Purα is a multifunctional protein that can bind to both DNA and RNA and functions in the initiation of DNA replication, control of transcription and mRNA translation (7, 8). A novel role has recently been described for Purα in the transport and targeting of mRNAs in neurons (9, 10).
It is also becoming clear that Purα is a major player in the regulation of the cell cycle and oncogenic transformation. Purα binds to several cellular regulatory proteins including the retinoblastoma protein (11), E2F-1 (12, 13, 14), Sp1 (15) and YB-1 (16). Levels of Purα fluctuate during the cell cycle, declining at the onset of S-phase and peaking during mitosis (17). Microinjection of Purα into NIH-3T3 cells caused cell cycle arrest at either the G1/S or G2/M checkpoints (18). Expression of Purα in Ras-transformed NIH-3T3 cells inhibited their ability to grow in soft agar (19). Ectopic overexpression of Purα suppressed the growth of several transformed and tumor cells including glioblastomas (20). Gene expression profiling in chronic myeloid leukemia patients revealed a down-regulation of Purα expression (21) and deletions of Purα have been reported in myelodysplastic syndrome, a condition that can progress to acute myelogenous leukemia consistent with a role for Purα as a tumor suppressor (22).
The generation and analysis of knockout mice with targeted inactivation of the PURA gene that encodes Purα revealed that Purα has an essential role in postnatal brain development (23). Mice with targeted disruption of the PURA gene in both alleles (PURA-/-) appear normal at birth, but at two weeks of age, develop neurological abnormalities culminating in death by four weeks. There are much fewer cells in the brain cortex, hippocampus and cerebellum of PURA-/- mice than in PURA+/+ controls due to a lack of proliferation of precursor cells in this region. This implicates Purα in the regulation of developmentally timed DNA replication in specific cell types in the brain. These knockout mice also provide a source of mouse embryo fibroblast primary cultures (MEFs) with PURA deletion that can be used in comparison with MEF cells as an experimental system to examine the cellular functions of Purα.
It has been found that Purα has a role in the life cycle of certain viruses including JC virus (JCV) and human immunodeficiency virus type-1 (HIV-1). In this regard, some viral regulatory proteins target Purα and these include the Tat transactivator protein of the HIV-1 (24) and the large T-antigen of the human neurotropic polyomavirus JC (25). HIV-1 Tat protein transactivates the JCV late promoter (26) and this transactivation is mediated by the interaction of Purα with Tat which stimulates JCV late promoter transcription (27) and JCV DNA replication (28). Purα-mediated activation of JCV by Tat may be important in the pathogenesis of the progressive multifocal leukoencephalopathy, which is caused by JCV and occurs predominantly in individuals with HIV-1/AIDS (29). In the life cycle of HIV-1 itself, Purα has recently been found to have a role in the Rev-mediated nuclear export of certain HIV-1 mRNAs (30).
Interestingly, we have found that both Purα and HIV-1 Tat affect cellular DNA repair, specifically homologous recombination-directed DNA repair (HRR) of double-strand breaks (DSBs). Purα negatively affects HRR and represses the expression of the HRR protein Rad51 (31) whereas Tat increases HRR DSB repair activity and induces the expression of Rad51 (32). In light of these findings and our earlier observations that Tat and Purα physically and functionally interact, we examined the effect of Tat and Purα on Rad51 and HRR utilizing MEFs derived from the PURA-/- mice and PURA+/+ controls.
Materials and Methods
Cell lines and cell cultures
Primary human fetal astrocytes were cultured from human fetal tissues as described previously (33, 34). Primary mouse embryo fibroblasts (MEFs) are derived from homozygous PURA-/- mouse embryos, heterozygous PURA+/- mouse embryos and their partner wild-type (WT) PURA-/- mouse embryos. PURA-/-(+Purα) MEFs/were established by transfecting PURA-/- MEFs with pcDNA3-Purα containing full length Purα cDNA and Purα-/- MEFs/GFP-Purα with pTRE-GFP-Purα containing tagged GFP Purα. MEFs were cultured in DMEM medium supplemented with 10% FBS and antibiotics. Cells were maintained in a humid incubator at 37°C with 5% CO2.
Immunofluorescence
Cells were plated for immunofluorescence microscopy and grown on poly-D-lysine coated glass chamber slides. Spontaneous Rad51 foci formation was determined in exponentially growing cells. DNA damage-induced Rad51 foci were determined in cells treated with 2 mM HU for 24 hours. Cells were grown for 24 h in chamber slides and then treated with 2 mM hydroxyurea (HU) for another 24 hours. Untreated cells and treated cells were fixed for 15 minutes in 3% paraformaldehyde at room temperature. After a wash in phosphate buffered saline (PBS), cells were permeabilized for 15 min at room temperature in PBS containing 0.3% Triton X-100. Slides were washed three times in PBS for 5 min and subsequently blocked with PBS containing 10% normal goat serum for 30 min. Subsequently, anti-Rad51 polyclonal antibody was applied to the slides for 60 min at room temperature followed by a goat-anti-rabbit secondary antibody conjugated with Rhodamine (1: 400 dilution, Molecular Probes, Eugene USA). After extensive washing, slides were mounted with coverslips using antifading solution containing DAPI. For detection of RAD51 foci, fluorescence microscopy was used. For each data point at least 100 nuclei were analyzed. Nuclei containing more than 10 strongly fluorescing foci were counted as positive. Images of Rad51 foci were recorded using an inverted fluorescence microscopy with a digital deconvolution system with a 100x oil immersion objective. Thirty-two optical sections per nucleus were generated at 0.4 mm distance. The 3D staining cells were viewed and each fluorescence was counted using Slidebook software. The volume of Rad51 foci and GFP-Purα staining were quantified in the same total volume of nucleus. The colocalization volume is counted by merging the red staining for Rad51 and Green staining for Purα.
Rad51 promoter assay by transient transfection
Cells were transfected with pRad51-LUC, which contains the human Rad51 promoter (-722 to +37) driving expression of the luciferase gene (35). After 48 hours, the cells were lysed and luciferase activity was measured using a luciferase assay kit according to the manufacturer’s instructions (Promega, Madison, WI, USA).
Homologous recombination assay
pDR-GFP (obtained from Dr. Jasin’s laboratory, Sloan-Kettering Institute, New York, NY) containing a mutated GFP gene with an 18 bp I-SceI site is transfected into PURA-/- MEFs cells using Lipofectamine 2000 (Invitrogen Corporation, Carlsbad, CA). The stable transfected cell lines are selected by growing in medium containing 5 μg/ml of puromycin. Puromycin-resistant colonies are screened by Southern blot analyses for an intact DR-GFP reporter. A 714-bp GFP coding fragment obtained by PCR system 9700 (Perkin Elmer Inc., Wellesley, MA) with primers forward 5′-ATGGTGAGCAAGGGCGAGGAGCT-3′ and reverse 5′-CTTGTACAGCTCGTCCATGCCGA-3′ from template pDR-GFP labeled with 32P as the probe. Ten micrograms of genomic DNA from puromycin-resistant colonies were digested by SalI and HindIII and separated on a 0.7% agarose gel and transferred to a nylon membrane. Hybridization is carried out under standard conditions using the 32P-labeled 714-bp probe to test whether these puromycin-resistant colonies have integrated an intact DR-GFP fragment. A radiosensitive screen exposed with the hybridized membrane is analyzed using a Phosphorimager (Storm 840; Amersham) with the ImageQuant analysis software (Amersham). To evaluate HRR of DNA DSBs, MEF-DRGFP cells are prepared for transfection by pCMV3xnlsI-SceI DNA using LipofectaminePlus as described above. At 48 h post transfection, cells are collected by trypsinization and GFP expression assayed by flow cytometry (Beckman Coulter, Epics Elite ESP) using an argon ion laser emitting at 488 nm. The frequency of recombination events is calculated from the frequency of GFP-positive cells. All experiments are repeated at least three times and the results are shown as average.
Cell cycle analysis
To analyze cell cycle profiles, harvested cells were stained for flow cytometer with propidium iodide solution. Cell cycle were measured by a FACScan flow cytometry and analyzed by using CELLQUEST software.
Results
Purα regulates Rad51 expression
Rad51 plays a critical role in repairing DNA double-strand breaks by promoting homologous recombination-directed pairing and strand exchange between damaged DNA and homologous DNA duplexes. It has been shown that Rad51 is down regulated in primary cells and overexpressed in proliferating cells and tumor tissues. In contrast, our previous studies have showed that Purα expression is increased in brain tissue during development, reaching a peak stage at postnatal day 15 (23). In this study, we found that Rad51 and PCNA levels were reduced at day 15, the peak of Purα gene expression (Figure 1A). Rad51 expression was increased in PURA+/- and PURA-/- mouse embryo fibroblasts (MEFs) (Figure 1B) relative to wild-type mice. Introduction of ectopically expressed Purα reduced the level of Rad51 in PURA-/- MEFs. These results suggest that Purα downregulates Rad51 gene expression or promotes Rad51 protein degradation.
Figure 1.
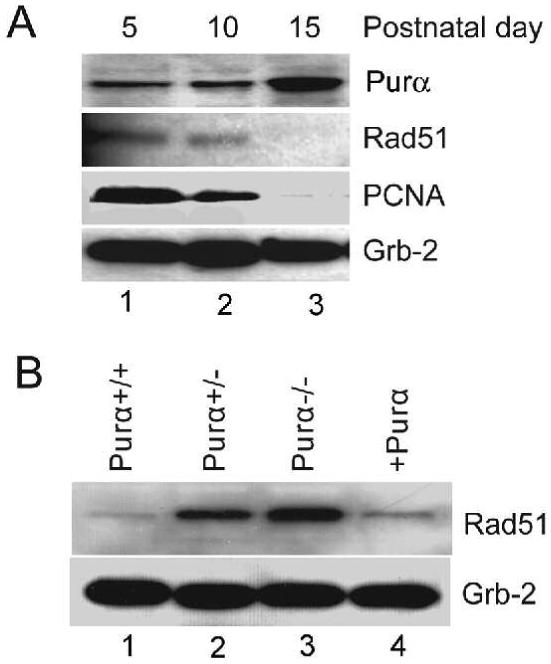
Purα suppresses Rad51 expression. A. The expression of Rad51 and Purα in the brain was measured during mouse development. Whole tissue protein extracts from mouse brains were prepared at postnatal days 5, 10 & 15 and were immunoblotted with antibodies against Purα, anti-PCNA, anti-Rad51 and PCNA. Grb-2 protein was used as a loading control. B. Expression of Rad51 was measured in MEFs isolated from PURA+/+ (wild-type), PUR+/- and PUR-/- mice. The lane labeled +Purα represents PUR-/- MEFs where Purα was ectopically expressed as described in Materials and Methods.
It is well established that locations where DNA damage has been induced and stalled DNA replication forks recruit Rad51 at sites of DSB repair, leading to the formation of foci visible by immunofluorescence (IF) using anti-Rad51 antibodies (Figure 2A). Rad51 foci seen in untreated cells are thought to reflect spontaneous DSB or DSB generated during DNA replication from other forms of damage. To investigate whether increased Rad51 expression in PURA-/- cells reflects increased DNA damage or repair activity, we examined Rad51 foci formation. The results in Figure 2B indicate that there are more Rad51 foci positive cells in untreated PURA-/- cells (Purα-) cells than wild-type cells (statistical significant in t-test p<0.05; Figure 2B left) and that ectopic expression of Purα in PURA-/- cells (+Purα) reduces the number of Rad51 foci positive cells to the levels seen in wild-type cells. Long-term exposure to the DNA replication inhibitor, hydroxyurea (HU), causes stalled replication forks to collapse into double-strand breaks. We found that the percent of cells with Rad51 foci increases in wild-type, PURA-/- and PURA-/- (+Purα) following treatment with HU for 24 hours (Figure 2B, right). A slightly higher percentage of cells containing Rad51 foci were found in PURA-/- cells than in wild-type and PURA-/- (+Purα) cells. Furthermore, we found that Purα is translocated into the nucleus in some of HU-treated PURA-/- (+Purα) cells (Figure 2C). There are significantly less Rad51 foci in Purα positive nuclei compared to Purα negative nuclei (Figure 2D). These results demonstrate that Purα significantly reduced Rad51 foci and Rad51 expression and that these events are related to both spontaneous DNA damage (-HU) and replication stress (+HU).
Figure 2.
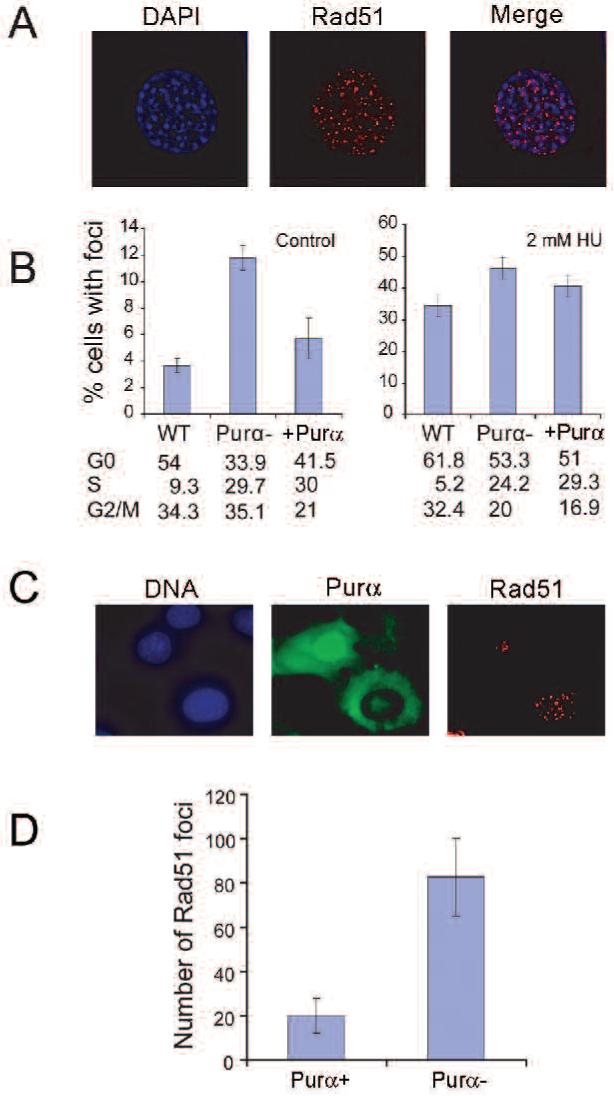
Purα suppresses Rad51 foci formation. Rad51 foci formation was visualized in MEFs isolated from PURA+/+, PUR-/- and PUR-/- MEFs expressing Purα. Cells were treated with and without 2 mM hydroxyurea for 24 hours, labeled with anti-Rad51 antibody and visualized under fluorescence microscopy. Red represents rhodamine-labeling of Rad51 foci and blue is DAPI labeling of nuclear DNA. Cells containing more than 10 Rad51 foci were counted as positive. A. A typical Rad51 foci staining from untreated PUR-/- MEFs. B. The plot of percentage of Rad51 foci positive cells for the three types of cells either untreated (left histogram) or treated with 2 mM HU for 16 hours. C. PURA-/- MEFs ectopically expressing Purα and treated with 2 mM HU for 16 hours. D. The plot of percentage of Rad51 foci in HU-treated PURA-/- (+Purα) cell nuclei that are either immunopositive for Purα (left) or immunopositive (right). The means (symbols) and standard errors (error bars) were standardized from three independent experiments.
Purα and HIV-1 Tat regulate Rad51 transcription
Promoter analysis showed that there are several Purα binding sites within the Rad51 promoter region. To test whether Purα regulates Rad51 promoter activity, we used a reporter plasmid with the Rad51 promoter driving luciferase expression in wild-type, PURA-/- and PURA-/- (+Purα-) MEFs. As shown in Figure 3A, we found that HIV-1 Tat significantly stimulated Rad51 promoter activity.
Figure 3.
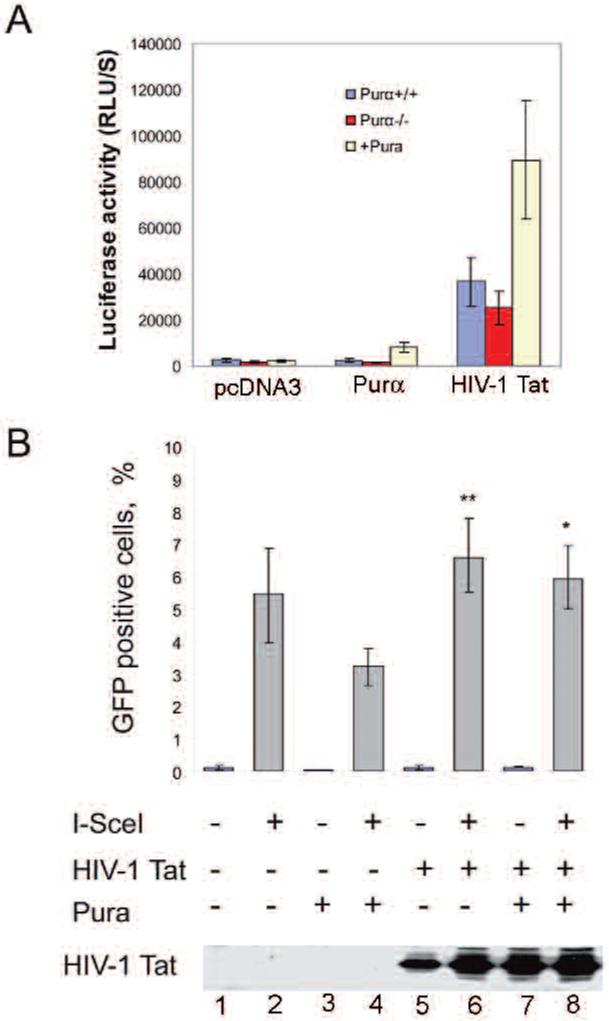
HIV-1 Tat stimulates Rad51 promoter activity and homologous recombination. A. MEFs were transfected with pGL3Rad51-luciferase plasmid along with pcDNA3-vector, pcDNA3-Purα or pcDNA-Tat expression plasmid using Lipofectamine (Invitrogen). Luciferase activity was determined after 36 h. The data represent results from three independent experiments. B. The effects of HIV-1 Tat and Pura on homologous recombination-directed DNA repair were examined. PURA-/- and PURA-/- (+Purα) MEFs were transfected with pcDNA3 or pcDNA3-TAT along with pCMV-3nls-I-SceI for 48 hours. The effect of HIV-1 TAT in PURA-/- (+Purα) cells was significantly different from control, *p<0.01, but that in PURA-/- cells was not, **p>0.05.
Purα and HIV-1 Tat interfere with homologous recombination-directed DNA Repair (HRR)
To analyze the effect of Purα on HRR, PURA-/- and PURA-/- (+Purα) cells were transfected with a recombination substrate DR-GFP that carries a tandem repeat of the GFP gene in which one copy is inactivated by the I-SceI sequence (scGFP) and the other by truncation of the C-and N-termini (iGFP). HRR of the I-SceI induced DSB restores the GFP gene. Cell lines that have stably integrated the construct were obtained by puromycin selection. To measure the efficiency of HRR, the pCMV3nls-I-SceI plasmid was transfected into these cells and the fraction of GFP expressing cells was measured by flow cytometry 48h later. Purα expression reduces by half the number of GFP positive cells observed after I-SceI transfection. These results suggest that Purα inhibits HRR.
Previous studies have demonstrated that HIV-1 Tat increases Rad51 expression and enhances homologous recombination (32). Further HIV-1 Tat has been shown to interact with Purα and this interaction is involved in the stimulation of transcription of the JC virus promoter by Tat (27). To further investigate the effect of HIV-1 Tat on HRR and the role of Purα, we expressed HIV-1 Tat in PURA-/- cells in the presence or absence of co-expressed Purα. We found that HIV-1 Tat expression significantly increased HRR in control PURA-/- cells and that in the presence of co-expressed Purα, Tat enhanced HRR significantly over cells expressing Purα alone (Figure 3B). These results suggest that Purα and HIV-1 Tat have opposite effects on HRR.
HIV-1 Tat induces DNA replication and Rad51 expression in primary human fetal astrocytes
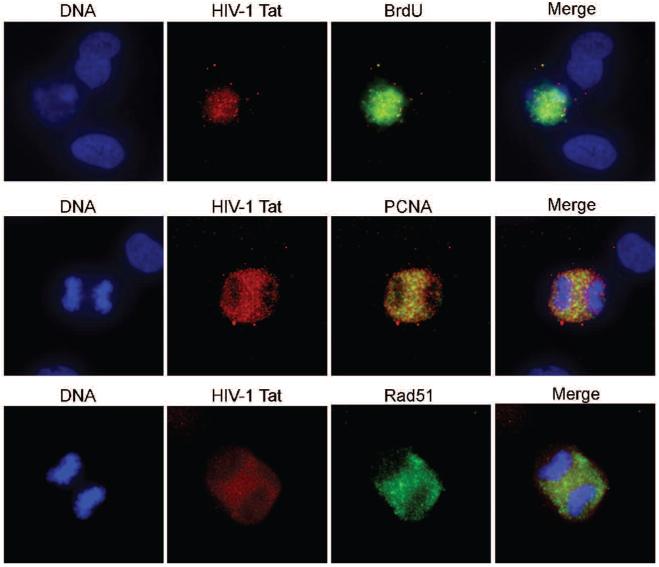
Our previous studies have shown that Rad51 expression is activated in PML tissue from patients infected with HIV-1 and JCV (36). To determine whether HIV-1 Tat affects Rad51 expression in primary cells, we transfected primary human fetal astrocytes with pCMV-Tat construct. Figure 4 shows that DNA replication and Rad51 expression are activated in HIV-1 Tat expressing cells. Furthermore, HIV-1 Tat positive cells entered mitosis and are particularly arrested at anaphase. These indicate that HIV-1 Tat may induce unexpected DNA replication and mitosis, possibly leading to chromosomal instability.
Figure 4.
HIV-1 Tat activates primary human fetal astrocytes. Cells were grown for one week before transfected with pcDNA3 HIV-1 Tat plasmid DNA. 48 hours after transfection, cells were labeled with rabbit anti-HIV-1 Tat and with mouse ant-BrdU, anti-PCNA or anti-Rad51 antibodies. Stained cells were analyzed by immunofluorescence microscopy. Top row, astrocytes expressing Tat actively replicating DNA (BrdU-positive, top panels). In Tat-positive mitotic cells, PCNA (middle panels) and Rad51 (lower panels) are expressed between the chromosomes.
Discussion
Our results indicate that Purα inhibits the level of expression of Rad51 protein and the formation of Rad51 foci. Thus, the Purα protein levels in the developing mouse brain and in cultured MEFs are inversely correlated with the level of Rad51. The decreased levels of Rad51 level may be caused by either the inhibition of Rad51 transcription or an increase in the rate of Rad51 protein degradation by Purα. There are a number of possible reasons for the increased incidence of Rad51 foci formation in the absence of Purα. During DNA replication, DNA replication arrest can induce DNA damage and unrepaired single-strand breaks can transit into double-strand breaks (DSBs). It is possible that Purα acts as a caretaker protein that is involved in the stabilization and repair of stalled replication forks and/or DSBs. In the absence of Purα, this would lead to an increase in DSBs and enhanced Rad51 foci formation. Our earlier studies on the involvement of Purα in the cellular response to DNA replication stress support this hypothesis (31).
Rad51 becomes recruited to sites of DNA damage and binds to single strand DNA to form bright foci. Rad51 is involved in homologous recombination-directed DNA repair (HRR), which provides an efficient and faithful pathway of repairing DNA damage, especially during DNA replication. However, as discussed in the introduction, Purα has the properties of a tumor suppressor protein and ablation of its expression in certain cancers is associated with genetic instability. Thus it could be possible that loss of the DSB caretaker function of Purα is responsible for this observation. Alternatively, Purα is involved in cell cycle arrest (18) and in the absence of Purα, it is possible that cell cycle progression may occur inappropriately before DNA repair has occurred. A third possibility is that the induction of Rad51 in the absence of Purα is responsible. Paradoxically when Rad51 is overexpressed it reduces DSB-induced HRR DNA repair perhaps because excess Rad51 blocks DNA strand exchange (37) and indeed the aberrant increase in Rad51 expression that is found in tumor cells may contribute to genomic instability by promoting abnormal recombination (38, 39, 40). In another recent study, we reported that JC virus infection promotes genomic instability that is associated with a large induction of Rad51 protein level (36).
Another protein that can modulate the expression of Rad51 and HRR is HIV-1 Tat as we have recently reported (32). Unlike Purα, Tat has a positive effect, i.e., it increases the level of Rad51 and the rate of HRR. Purα and Tat are able to physically and functionally interact and this is involved in regulating the transcription of certain genes, e.g., the viral late promoter of JCV (27) and the cellular TGFβ-1 promoter (41). In the light of these findings, we have now examined the effect of Purα and Tat interaction on Rad51 and HRR. Interestingly, Tat stimulated the Rad51 promoter in the absence of Purα (Figure 3A, column 8) but this induction was significantly enhanced in the presence of Purα (Figure 3A, column 9). Similarly, HRR was stimulated by Tat both in the presence and in the absence of Purα (Figure 3B). This suggests that the regulation of Rad51 and HRR by Tat involves both Purα-dependent and Purα-independent mechanisms. With regard to the Purα-independent mechanism of regulation, we have recently found that wild-type p53 negatively regulates the Rad51 promoter but the T-antigen of JCV, which antagonizes p53 function, reverses this inhibition (35). Since reciprocal modulations also occur between p53 and Tat (42), it is possible that the Purα-independent mechanism of Rad51 promoter regulation involves p53.
In conclusion, events involving the effects of Purα and HIV-1 Tat on Rad51 expression and HRR may be important in understanding HIV-1 associated pathogenesis.
Acknowledgements
We thank past and present members of the Center for Neurovirology for their insightful discussion and sharing of ideas and reagents. We also wish to thank C. Schriver for editorial assistance. This work was supported by grants awarded by the NIH to KK.
References
- 1.Haas S, Gordon J, Khalili K. A developmentally regulated DNA-binding protein from mouse brain stimulates myelin basic protein gene expression. Mol Cell Biol. 1993;13:3103–3112. doi: 10.1128/mcb.13.5.3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haas S, Thatikunta P, Steplewski A, Johnson EM, Khalili K, Amini S. A 39-kD DNA-binding protein from mouse brain stimulates transcription of myelin basic protein gene in oligodendrocytic cells. J Cell Biol. 1995;130:1171–1179. doi: 10.1083/jcb.130.5.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergemann AD, Johnson EM. The HeLa Pur factor binds single-stranded DNA at a specific element conserved in gene flanking regions and origins of DNA replication. Mol Cell Biol. 1992;12:1257–1265. doi: 10.1128/mcb.12.3.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bergemann AD, Ma ZW, Johnson EM. Sequence of cDNA comprising the human pur gene and sequence-specific single-stranded DNA-binding properties of the encoded protein. Mol Cel Biol. 1992;12:5673–5682. doi: 10.1128/mcb.12.12.5673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma ZW, Bergemann AD, Johnson EM. Conservation in human and mouse Pur alpha of a motif common to several proteins involved in initiation of DNA replication. Gene. 1994;149:311–314. doi: 10.1016/0378-1119(94)90167-8. [DOI] [PubMed] [Google Scholar]
- 6.Liu H, Johnson EM. Distinct proteins encoded by alternative transcripts of the PURG gene, located contrapodal to WRN on chromosome 8, determined by differential termination/polyadenylation. Nucleic Acids Res. 2002;30:2417–2426. doi: 10.1093/nar/30.11.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson EM. The Pur protein family: clues to function from recent studies on cancer and AIDS. Anticancer Res. 2003;23:2093–2100. [PubMed] [Google Scholar]
- 8.Gallia GL, Johnson EM, Khalili K. Puralpha: a multifunctional single-stranded DNA- and RNA-binding protein. Nucleic Acids Res. 2000;28:3197–3205. doi: 10.1093/nar/28.17.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Johnson EM, Kinoshita Y, Weinreb DB, Wortman MJ, Simon R, Khalili K, Winckler B, Gordon J. Role of Pur alpha in targeting mRNA to sites of translation in hippocampal neuronal dendrites. J Neurosci Res. 2006;83:929–943. doi: 10.1002/jnr.20806. [DOI] [PubMed] [Google Scholar]
- 10.Kanai Y, Dohmae N, Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 11.Johnson EM, Chen PL, Krachmarov CP, Barr SM, Kanovsky M, Ma ZW, Lee WH. Association of human Pur alpha with the retinoblastoma protein, Rb, regulates binding to the single-stranded DNA Pur alpha recognition element. J Biol Chem. 1995;270:24352–24360. doi: 10.1074/jbc.270.41.24352. [DOI] [PubMed] [Google Scholar]
- 12.Darbinian N, Gallia GL, Kundu M, Shcherbik N, Tretiakova A, Giordano A, Khalili K. Association of Pur alpha and E2F-1 suppresses transcriptional activity of E2F-1. Oncogene. 1999;18:6398–6402. doi: 10.1038/sj.onc.1203011. [DOI] [PubMed] [Google Scholar]
- 13.Darbinian N, White MK, Gallia GL, Amini S, Rappaport J, Khalili K. Interaction between the Pura and E2F-1 transcription factors. Anticancer Res. 2004;24:2585–2594. [PubMed] [Google Scholar]
- 14.Darbinian N, White MK, Khalili K. Regulation of the Puralpha promoter by E2F-1. J Cell Biochem. 2006;99:1052–1063. doi: 10.1002/jcb.20872. [DOI] [PubMed] [Google Scholar]
- 15.Tretiakova A, Steplewski A, Johnson EM, Khalili K, Amini S. Regulation of myelin basic protein gene transcription by Sp1 and Puralpha: evidence for association of Sp1 and Puralpha in brain. J Cell Physiol. 1999;181:160–168. doi: 10.1002/(SICI)1097-4652(199910)181:1<160::AID-JCP17>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 16.Safak M, Gallia GL, Khalili K. Reciprocal interaction between two cellular proteins, Puralpha and YB-1, modulates transcriptional activity of JCVCY in glial cells. Mol Cell Biol. 1999;19:2712–2723. doi: 10.1128/mcb.19.4.2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Itoh H, Wortman MJ, Kanovsky M, Uson RR, Gordon RE, Alfano N, Johnson EM. Alterations in Pur(alpha) levels and intracellular localization in the CV-1 cell cycle. Cell Growth Differ. 1998;9:651–665. [PubMed] [Google Scholar]
- 18.Stacey DW, Hitomi M, Kanovsky M, Gan L, Johnson EM. Cell cycle arrest and morphological alterations following microinjection of NIH3T3 cells with Pur alpha. Oncogene. 1999;18:4254–4261. doi: 10.1038/sj.onc.1202795. [DOI] [PubMed] [Google Scholar]
- 19.Barr SM, Johnson EM. Ras-induced colony formation and anchorage-independent growth inhibited by elevated expression of Puralpha in NIH3T3 cells. J Cell Biochem. 2001;81:621–638. doi: 10.1002/jcb.1099. [DOI] [PubMed] [Google Scholar]
- 20.Darbinian N, Gallia GL, King J, Del Valle L, Johnson EM, Khalili K. Growth inhibition of glioblastoma cells by human Pur(alpha) J Cell Physiol. 2001;189:334–340. doi: 10.1002/jcp.10029. [DOI] [PubMed] [Google Scholar]
- 21.Bruchova H, Borovanova T, Klamova H, Brdicka R. Gene expression profiling in chronic myeloid leukemia patients treated with hydroxyurea. Leuk Lymphoma. 2002;43:1289–1295. doi: 10.1080/10428190290026358. [DOI] [PubMed] [Google Scholar]
- 22.Lezon-Geyda K, Najfeld V, Johnson EM. Deletions of PURA, at 5q31, and PURB, at 7p13, in myelodysplastic syndrome and progression to acute myelogenous leukemia. Leukemia. 2001;15:954–962. doi: 10.1038/sj.leu.2402108. [DOI] [PubMed] [Google Scholar]
- 23.Khalili K, Del Valle L, Muralidharan V, Gault WJ, Darbinian N, Otte J, Meier E, Johnson EM, Daniel DC, Kinoshita Y, Amini S, Gordon J. Puralpha is essential for postnatal brain development and developmentally coupled cellular proliferation as revealed by genetic inactivation in the mouse. Mol Cell Biol. 2003;23:6857–6875. doi: 10.1128/MCB.23.19.6857-6875.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gallia GL, Darbinian N, Tretiakova A, Ansari SA, Rappaport J, Brady J, Wortman MJ, Johnson EM, Khalili K. Association of HIV-1 Tat with the cellular protein, Puralpha, is mediated by RNA. Proc Natl Acad Sci USA. 1999;96:11572–11577. doi: 10.1073/pnas.96.20.11572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gallia GL, Safak M, Khalili K. Interaction of the single-stranded DNA-binding protein Puralpha with the human polyomavirus JC virus early protein T-antigen. J Biol Chem. 1998;273:32662–32669. doi: 10.1074/jbc.273.49.32662. [DOI] [PubMed] [Google Scholar]
- 26.Tada H, Rappaport J, Lashgari M, Amini S, Wong-Staal F, Khalili K. Trans-activation of the JC virus late promoter by the tat protein of type 1 human immunodeficiency virus in glial cells. Proc Natl Acad Sci USA. 1990;87:3479–3483. doi: 10.1073/pnas.87.9.3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krachmarov CP, Chepenik LG, Barr-Vagell S, Khalili K, Johnson EM. Activation of the JC virus Tat-responsive transcriptional control element by association of the Tat protein of human immunodeficiency virus 1 with cellular protein Pur alpha. Proc Natl Acad Sci USA. 1996;93:14112–14117. doi: 10.1073/pnas.93.24.14112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Daniel DC, Kinoshita Y, Khan MA, Valle LD, Khalili K, Rappaport J, Johnson EM. Internalization of exogenous human immunodeficiency virus-1 protein, Tat, by KG-1 oligodendroglioma cells followed by stimulation of DNA replication initiated at the JC virus origin. DNA Cell Biol. 2004;23:858–867. doi: 10.1089/dna.2004.23.858. [DOI] [PubMed] [Google Scholar]
- 29.Khalili K, Gordon J, White MK. The polyomavirus, JCV, and its involvement in human disease. Adv Exp Med Biol. 2006;577:274–287. doi: 10.1007/0-387-32957-9_20. [DOI] [PubMed] [Google Scholar]
- 30.Kaminski R, Darbinian N, Sawaya BE, Slonina D, Amini S, Johnson EM, Rappaport J, Khalili K, Darbinyan A. Puralpha as a cellular co-factor of Rev/RRE-mediated expression of HIV-1 intron-containing mRNA. J Cell Biochem. 2007 doi: 10.1002/jcb.21503. in Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang H, Wang M, Reiss K, Darbinian-Sarkissian N, Johnson EM, Iliakis G, Amini S, Khalili K, Rappaport J. Evidence for the involvement of Puralpha in response to DNA replication stress. Cancer Biol Ther. 2007;6:596–602. doi: 10.4161/cbt.6.4.3889. [DOI] [PubMed] [Google Scholar]
- 32.Chipitsyna G, Slonina D, Siddiqui K, Peruzzi F, Skorski T, Reiss K, Sawaya BE, Khalili K, Amini S. HIV-1 Tat increases cell survival in response to cisplatin by stimulating Rad51 gene expression. Oncogene. 2004;23:2664–2671. doi: 10.1038/sj.onc.1207417. [DOI] [PubMed] [Google Scholar]
- 33.Radhakrishnan S, Otte J, Enam S, Del Valle L, Khalili K, Gordon J. JC virus-induced changes in cellular gene expression in primary human astrocytes. J Virol. 2003;77:10638–10644. doi: 10.1128/JVI.77.19.10638-10644.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Radhakrishnan S, Gordon J, Del Valle L, Cui J, Khalili K. Intracellular approach for blocking JC virus gene expression by using RNA interference during viral infection. J Virol. 2004;78:7264–7269. doi: 10.1128/JVI.78.13.7264-7269.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.White MK, Skowronska A, Gordon J, Del Valle L, Deshmane SL, Giordano A, Croul S, Khalili K. Analysis of a mutant p53 protein arising in a medulloblastoma from a mouse transgenic for the JC virus early region. Anticancer Res. 2006;26:4079–4092. [PubMed] [Google Scholar]
- 36.Darbinyan A, White MK, Akan S, Radhakrishnan S, Del Valle L, Amini S, Khalili K. Alterations of DNA damage repair pathways resulting from JCV infection. Virology. 2007;364:73–86. doi: 10.1016/j.virol.2007.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim PM, Allen C, Wagener BM, Shen Z, Nickoloff JA. Overexpression of human RAD51 and RAD52 reduces double-strand break-induced homologous recombination in mammalian cells. Nucl Acids Res. 2001;29:4352–4360. doi: 10.1093/nar/29.21.4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Raderschall E, Stout K, Freier S, Suckow V, Schweiger S, Haaf T. Elevated levels of Rad51 recombination protein in tumor cells. Cancer Res. 2002;62:219–225. [PubMed] [Google Scholar]
- 39.Richardson C, Stark JM, Ommundsen M, Jasin M. Rad51 overexpression promotes alternative double-strand break repair pathways and genome instability. Oncogene. 2004;23:546–553. doi: 10.1038/sj.onc.1207098. [DOI] [PubMed] [Google Scholar]
- 40.Xia SJ, Shammas MA, Shmookler Reis RJ. Elevated recombination in immortal human cells is mediated by HsRAD51 recombinase. Mol Cell Biol. 1997;17:7151–7158. doi: 10.1128/mcb.17.12.7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thatikunta P, Sawaya BE, Denisova L, Cole C, Yusibova G, Johnson EM, Khalili K, Amini S. Identification of a cellular protein that binds to Tat-responsive element of TGF beta-1 promoter in glial cells. J Cell Biochem. 1997;67:466–477. [PubMed] [Google Scholar]
- 42.Li CJ, Wang C, Friedman DJ, Pardee AB. Reciprocal modulations between p53 and Tat of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1995;92:5461–5464. doi: 10.1073/pnas.92.12.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]