Abstract
Chickpea (Cicer arietinum, Leguminosae), an important grain legume, is widely used for food and fodder throughout the world. We sequenced the complete plastid genome of chickpea, which is 125,319 bp in size, and contains only one copy of the inverted repeat (IR). The genome encodes 108 genes, including 4 rRNAs, 29 tRNAs, and 75 proteins. The genes rps16, infA, and ycf4 are absent in the chickpea plastid genome, and ndhB has an internal stop codon in the 5′exon, similar to other legumes. Two genes have lost their introns, one in the 3′exon of the transpliced gene rps12, and the one between exons 1 and 2 of clpP; this represents the first documented case of the loss of introns from both of these genes in the same plastid genome. An extensive phylogenetic survey of these intron losses was performed on 302 taxa across legumes and the related family Polygalaceae. The clpP intron has been lost exclusively in taxa from the temperate “IR-lacking clade” (IRLC), whereas the rps12 intron has been lost in most members of the IRLC (with the exception of Wisteria, Callerya, Afgekia, and certain species of Millettia, which represent the earliest diverging lineages of this clade), and in the tribe Desmodieae, which is closely related to the tribes Phaseoleae and Psoraleeae. Data provided here suggest that the loss of the rps12 intron occurred after the loss of the IR. The two new genomic changes identified in the present study provide additional support of the monophyly of the IR-loss clade, and resolution of the pattern of the earliest-branching lineages in this clade. The availability of the complete chickpea plastid genome sequence also provides valuable information on intergenic spacer regions among legumes and endogenous regulatory sequences for plastid genetic engineering.
Keywords: Plastid genetic engineering, Genome evolution, Phylogeny of legumes, Leguminosae, Intron loss, Chickpea, Cicer
1. Introduction
Gene mapping and genomic sequencing have demonstrated that plastid genome organization is generally highly conserved among angiosperms (Palmer, 1991; Raubeson and Jansen, 2005). Most genomes are characterized by a quadripartite structure, with two copies of an inverted repeat (IR) separating the large (LSC) and small (SSC) single copy regions. The genomes usually include 120– 130 genes and range in size from 120 to 170 kilobases (kb). Gene content and gene order are conserved throughout angiosperms with the ancestral configuration depicted by the earliest-branching angiosperm clades Amborella and Nymphaeales (Goremykin et al., 2003; Raubeson et al., 2007; Jansen et al., 2007). In a typical angiosperm chloroplast genome, there are 18 genes containing introns, six in tRNA genes and the remaining twelve in protein-coding genes. Fifteen of the intron-containing genes have only two exons, and the remaining three have three exons.
Changes in this highly conserved organization of plastid genomes have been utilized to resolve phylogenetic relationships among major clades in a number of angiosperm families, including Asteraceae (Jansen and Palmer, 1987; Kim et al., 2005), Berberidaceae (Kim and Jansen, 1995), Cactaceae (Wallace and Cota, 1996), Campanulaceae (Cosner et al., 2004), Leguminosae (Bruneau et al., 1990; Lavin et al., 1990; Doyle et al., 1995; Doyle et al., 1996), Geraniaceae (Chumley et al., 2006), Lobeliaceae (Knox et al., 1993), Oleaceae (Lee et al., 2007), Onagraceae (Hachtel et al., 1991; Greiner et al., 2008), Poaceae (Doyle et al., 1992), and Ranunculaceae (Hoot and Palmer, 1994; Johansson, 1999). The types of changes that have been used include inversions, loss of the 22–25 kb IR which contains a duplicated set of rRNA and tRNA genes, expansion/contraction of the IR, and gene/intron loss (Downie and Palmer, 1992). Although all of these genomic changes have exhibited some homoplasy, they have served as powerful phylogenetic markers for several reasons: (1) these types of changes are generally rare, resulting in lower homoplasy relative to nucleotide substitutions; (2) assessing homology of these events is generally straightforward; (3) the direction of evolutionary change is easily discerned; and (4) once a rearrangement is detected it is relatively easy to survey numerous taxa for each event. The relative phylogenetic utility of the different types of plastid genomic rearrangements varies considerably with inversions exhibiting the least amount of homoplasy. However, even genomic changes that are homoplasious provide valuable phylogenetic characters within each major lineage in which they occur. One example of this phenomenon concerns the rpoC1 intron, which has been lost independently six times among angiosperms (Downie et al., 1996) but its absence is still a valuable phylogenetic marker for resolving relationships within each of these six lineages.
The Leguminosae (also Fabaceae; the legumes) is one angiosperm family that has experienced considerable numbers of plastid genomic rearrangements. Legumes are the third largest family of angiosperms with 730 genera and more than 19,000 species distributed throughout the world (Lewis et al., 2005). Legumes are second only to grasses in their agricultural and economic value, and include many important species grown for food, fodder, wood, ornamentals, and raw materials for industry and also for their ecologically important role in biological nitrogen fixation. A number of previous studies have examined the phylogenetic distribution of different plastid genome rearrangements among legumes, including the loss of one copy of the IR (Palmer and Thompson, 1982; Lavin et al., 1990), inversions of 50 kb (Palmer and Thompson, 1981; Doyle et al., 1996), and 78 kb (Bruneau et al., 1990), loss of the rpl22 and rps16 genes (Doyle et al., 1995), and loss of the rpl2 intron (Doyle et al., 1995). These genomic rearrangements, combined with DNA sequence data (Doyle, 1995; Käss and Wink, 1995, 1996; Doyle et al., 1997, 2000; Kajita et al., 2001; Wojciechowski et al., 2004), have provided valuable phylogenetic data for resolving relationships among several deep nodes of legumes. The first, and probably most dramatic example of the phylogenetic utility of a plastid genomic rearrangement among legumes is the loss of one copy of the IR by all members sampled from the tribes Carmichaelieae, Cicereae, Hedysareae, Trifolieae, Fabeae (Vicieae), Galegeae, and three genera of Millettieae (Lavin et al., 1990; Liston, 1995). The monophyly of this clade, known as the “IR-lacking clade” or IRLC (Wojciechowski et al., 2000), was later confirmed by phylogenetic analyses of DNA sequences of the plastid genes rbcL (Doyle et al., 1997; Käss and Wink, 1997) and matK (Wojciechowski et al., 2004), the plastid trnL intron (Pennington et al., 2001), and the internal transcribed spacer (ITS) regions of the nuclear ribosomal DNA (Hu et al., 2002).
In addition to utilization of plastid genome sequences for phylogenetic studies, they are very useful in engineering foreign genes. However, complete chloroplast genome sequence of only six species of crop plants were determined until 2004. Therefore, complete plastid genome sequences of several major crop species including fiber crops (Lee et al., 2006), tubers (Daniell et al., 2006, 2008), cereals (Saski et al., 2007), trees (Steane, 2005; Bausher et al., 2006; Ravi et al., 2006; Samson et al., 2007), vegetables (Ruhlman et al., 2006), fruits (Jansen et al., 2006; Daniell et al., 2006) and legumes (Saski et al., 2005; Guo et al., 2007) have been determined recently. Plastid genetic engineering offers a number of unique advantages including high level of transgene expression (DeCosa et al., 2001), multi-gene engineering in a single transformation event (Quesda-Vargas et al., 2005), transgene containment via maternal inheritance (Daniell, 2002; Daniell, 2007) or cytoplasmic male sterility (Ruiz and Daniell, 2005). Plastid transgenic lines also lack gene silencing (DeCosa et al., 2001; Lee et al., 2003), position effect due to site specific transgene integration (Daniell et al., 2002) and pleiotropic effects due to subcellular compartmentalization of transgene products (Lee et al., 2003; Daniell et al., 2001; Leelavathi et al., 2003); concerns of transgene silencing, position effect and pleiotropic effects are often encountered in nuclear genetic engineering. Therefore, transgenes have been integrated into plastid genomes to confer valuable agronomic traits, including herbicide resistance (Daniell et al., 1998), insect resistance (McBride et al., 1995; DeCosa et al., 2001), disease resistance (DeGray et al., 2001), drought tolerance (Lee et al., 2003), salt tolerance (Kumar et al., 2004), phytoremediation (Ruiz et al., 2003; Hussein et al., 2007) or expression of various therapeutic proteins or biomaterials (Verma and Daniell, 2007; Kamarajugadda and Daniell, 2006; Daniell et al., 2005). However, soybean is the only legume that has been transformed via the plastid genome so far (Dufoumantel et al., 2004, 2005) and more genome sequence information is needed to facilitate plastid genetic engineering in other economically important legumes.
During the past eight years plastid genome sequences have been completed for four legumes, including Lotus japonicus (Regel) K. Larson, Medicago truncatula Gaertn., Glycine max Merr., and Phaseolus vulgaris L. In this paper, we report on the complete genome sequence of Cicer arietinum L. (chickpea). Sequences of these five legume plastid genomes, all from taxa belonging to the subfamily Papilionoideae and two of which are from members of the IRLC (Cicer, Medicago), will enable more detailed comparisons of the organization and evolution of the plastid genomes of legumes. Our comparisons have identified two additional genomic rearrangements, the losses of introns in the clpP and rps12 (3′-end) genes, and we survey the phylogenetic distribution of these changes in 302 taxa of legumes and the related family Polygalaceae.
2. Materials and methods
2.1. Plant material and plastid isolation
Chickpea (C. arietinum L.) seeds were obtained from IARI (Indian Agricultural Research Institute) New Delhi, India. Fresh leaves were harvested from greenhouse grown chickpea seedlings. Prior to plastid isolation, plants were kept in the dark for 48 h to reduce the levels of starch. Plastid isolation was performed as described by Jansen et al. (2005) and Samson et al. (2007).
Purified plastids were used to amplify the entire plastid genome by rolling circular amplification (RCA) using the Repli-g RCA-KIT (Qiagen GmbH, Hilden, Germany) following the methods described in Jansen et al. (2005). The success of genome amplification and the quality of the DNA was verified by digesting with restriction enzymes BstXI, EcoRI, and HindIII, and visualization of the resulting fragments on ethidium bromide stained, 1% agarose gels.
2.2. Plastid genome sequencing and assembly
Purified RCA products were subjected to nebulization, followed by end repair, and size-fractioned by agarose gel electrophoresis to obtain fragment lengths ranging from 2.0 to 3.5 kb. Repaired products were blunt-end cloned into the 4blunt-TOPO vector, followed by transformation into Escherichia coli ElectroMax TM-DH5 α cells by electroporation (TOPO shotgun Cloning Kit; Invitrogen, Carlsbad, CA, USA). Transformed cells were selected on Luria–Bertani (LB) agar containing 100 μg/ml ampicillin and arrayed into 30 × 96-well microtitre plates. Sequencing reactions were carried out in both the forward and reverse directions using the BigDye Terminator v3.1 Cycle Sequencing Kit and separated by a 3730×L DNA Sequence Analyzer (Applied Biosystems, Foster City, CA, USA). Sequence data were assembled using Sequencher version 4.5 (Gene Codes, Ann Arbor, MI, USA) following quality and vector trimming. Gap regions were filled by sequencing PCR fragments generated using primers that flank the gaps. The assembly was considered complete when a quality score of ≥20 was obtained at every base position with at least 6× coverage.
2.3. Genome annotation
The annotation program Dual Organellar Genome Annotator (DOGMA; Wyman et al., 2004) was used to annotate the C. arietinum plastid genome, after uploading a FASTA-formatted file of complete nucleotide sequence to the program's server. BLASTX and BLASTN searches, against a custom database of previously published plastid genomes, identified putative protein-coding genes, tRNAs, and rRNAs. For genes with low sequence identity, manual annotation was performed, after identifying the position of the start and stop codons, as well as the translated amino acid sequence, using the plastid/bacterial genetic code.
2.4. Whole genome sequence alignment
MultiPipMaker (Schwartz et al., 2003; http://bio.cse.psu.edu) was used for multiple genome alignment of chickpea with four published legume plastid genomes from the subfamily Papilionoideae; Lotus japonicus (NC_002694, Kato et al., 2000), Medicago truncatula (NC_003119), Glycine max (NC_007942, Saski et al., 2005), and Phaseolus vulgaris (NC_009259, Guo et al., 2007). We generated the alignments of whole genomes using chickpea as the reference genome.
2.5. Survey for loss of clpP and rps12 introns
We surveyed for the presence/absence of two introns from 318 accessions of 301 legume species representing all 3 subfamilies and 198 genera, and 1 member of the related family Polygalaceae (Table 1) using primers designed that span the intron in each gene: clpPF3 and clpPR3 (5′-ATGCCMATTGGTGTTCCAAAAGTRCC and 5′-G CGTGAGGGAATGCTAGACGTTTGGT) for the clpP gene, and rps12F and rps12R (5′-CCYAAAAAACCAAACTCTGCYTTACGTAAA and 5′-TT ATTTTGGCTTTTTBGCMCCATATT) for the rps12 gene. PCR amplification products were resolved on 1% agarose gels and fragment sizes were determined by comparison to DNA size markers.
Table 1.
Survey of clpP and rps12 intron losses among 302 taxa of Fabaceae and Polygalaceae based on PCR analyses of genomic DNA samples from representative legume taxa
| TAXON | Collection/accession | clpP intron presence | rps12 intron presence |
|---|---|---|---|
| Polygalaceae | |||
| Polygala californica Nutt. ex Torr. & A. Gray | Wojciechowski & Steele 887 (ASU) | Y | Y |
| Fabaceae | |||
| Caesalpinioideae | |||
| Arcoa gonavensis Urb. | Lavin s.n. (MONT) | Y | Y |
| Bauhinia tomentosa L. | Wojciechowski 946 (ASU) (DES 199501431001) | Y | Y |
| Caesalpinia gilliesii Wall. ex Hook. | Wojciechowski 882 (ASU) | Y | Y |
| Cassia javanica L. | dePompert 48 (ASU) | Y | Y |
| Ceratonia siliqua L. | Wojciechowski 872 (ASU) | Y | Y |
| Cercis canadensis var. texensis (S. Watson) M. Hopkins | Fritsch 1465 (CAS) | Y | Y |
| Chamaecrista fasciculata (Michx.) Greene | Wojciechowski 873 (ASU) | Y | Y |
| Chamaecrista nictitans var. mensalis (Greenman) H.S. Irwin & Barneby | Wojciechowski 1465 (ASU) | Y | Y |
| Colophospermum mopane (J. Kirk ex Benth.) J. Léonard | Wojciechowski 947 (ASU) (DES 198301901003) | Y | Y |
| Conzattia multiflora Standl. | Werling 399 (ASU) | Y | Y |
| Gleditsia triacanthos L. | Wojciechowski 881 (ASU) | Y | Y |
| Gymnocladus dioicus K. Koch | Mayer & Mazzio 14545 (ASU) | Y | Y |
| Gymnocladus chinensis Baill. | P Herendeen 8-V-2003-1 (US) | Y | Y |
| Haematoxylon brasiletto H. Karst | Wojciechowski 953 (ASU) (cult., DES 197300990102) | Y | Y |
| Hoffmannseggia glauca (Ortega) Eifert | Wojciechowski 1501 (ASU) | Y | Y |
| Hymenaea courbaril L. | A Salywon 1264 (ASU) | Y | Y |
| Parkinsonia aculeata L. | A Salywon 918 (ASU) | Y | Y |
| Parkinsonia microphylla Torr. | Wojciechowski 1279 (ASU) | Y | Y |
| Petalostylis labicheoides R. Br. | Wojciechowski 945 (ASU) (cult., DES 1979971414) | Y | Y |
| Poeppigia procera C. Presl. | EJ Lott 4099 (ASU) | Y | Y |
| Senna artemisioides (Gaudich. ex DC.) Randell | Wojciechowski 1500 (ASU; cultivated) | Y | Y |
| Senna covesii (A. Gray) H.S. Irwin & Barneby | Wojciechowski 876 (ASU) | Y | Y |
| Senna lindheimeriana (Scheele) H.S. Irwin & Barneby | Wojciechowski 1275 (ASU) | Y | Y |
| Mimosoideae clade | |||
| Acacia greggii A. Gray | Salywon 915 (ASU) | Y | Y |
| Acacia willardiana Rose | Wojciechowski 952 (ASU) | Y | Y |
| Acacia hindsii Benth. | Wojciechowski 854 (ASU) | Y | Y |
| Albizia julibrissin Durazz. | Wojciechowski 908 (ASU) | Y | Y |
| Calliandra californica Benth. | Wojciechowski 950 (ASU) | Y | Y |
| Chloroleucon mangense Britton & Rose | J Rebman 5862 (ASU) | Y | Y |
| Desmanthus cooleyi (Eaton) Branner & Coville | Wojciechowski 1018 (ASU) | Y | Y |
| Desmanthus illinoensis MacMill. | Wojciechowski 1171 (ASU) | Y | Y |
| Dichrostachys cinerea (L.) Wight & Arn. | Wojciechowski 1502 (ASU; cultivated) | ||
| Havardia mexicana Britton & Rose | AL Reina 98–716 (ASU) | Y | Y |
| Inga punctata Willd. | Landrum 10430 (ASU) | N | Y |
| Leucaena retusa Benth. | Boke & Massey 419 (UC) | Y | Y |
| Lysiloma watsonii Rose | A Salywon 921 (ASU) | Y | Y |
| Mimosa dysocarpa Benth. | Damrel 107 (ASU) | Y | Y |
| Parkia nitida Miq. | UC 1589048 | Y pm? | Y ? |
| Pentaclethra macroloba Kuntze. | Landrum 10317 (ASU) | Y | Y |
| Prosopidastrum mexicanum (Dressler) Burkart | J Rebman 4021 (DES) | Y | Y |
| Prosopis glandulosa Torr. | Wojciechowski 875 (ASU) | Y | Y |
| Papilionoideae clade (early-branching lineages) | |||
| Andira inermis (Wright) DC. | SB 347 (E) | Y | Y |
| Andira parviflora Ducke | WR 11179 (INPA, K) | Y | Y |
| Bobgunnia madagascariensis (Desv.) J.H. Kirkbr. & Wiersema | Smith 1725 (K); DNA Data Bank 8220 | Y | Y |
| Bocoa viridiflora (Ducke) R.S. Cowan | Nascimento (MO); DNA Data Bank 9454 | Y | Y |
| Cladrastis platycarpa (Maxim.) Makino | P Herendeen 1-V-2003-11 (US) | Y† | Y |
| Calia arizonica (S. Watson) Yakovlev | Salywon 917 (ASU) | Y | Y |
| Calia secundiflora (Ortega) Yakovlev | Wojciechowski 951 (ASU); cult. | Y | Y |
| Hymenolobium flavum Kleinh. | RT Pennington 451 (E) | Y | Y |
| Hymenolobium nitidum Benth. | WR 11177 (K) | Y | Y |
| Myrocarpus frondosus Allem. | Tressens et al. 3443 (ASU) | Y | Y |
| Myroxylon balsamum Harms | E Martinez 4051 (ASU) | Y | Y |
| Pickeringia montana Nuttall | Wojciechowski 883 (ASU) | Y† | Y |
| Sophora stenophylla A. Gray | RK Gierisch 4997 (ASU) | Y | Y |
| Sophora nuttalliana B.L. Turner | MA Baker 11465 (ASU) | Y† | Y |
| Styphnolobium japonicum (L.) Schott | RR Halse 4523 (ASU) | Y† | Y |
| Styphnolobium conzattii (Standl.) M. Sousa & Rudd | R Torres 5231 (MEXU) | Y† | Y |
| Styphnolobium monteviridis M. Sousa & Rudd (2 access.) | E Bello 3 (MO); L Landrum 10506 (ASU) | Y, Y† | Y, Y |
| Swartzia jojori Harms | RT Pennington 938 (E) | Y | Y |
| Swartzia flaemingii Ducke | Ratter 7433 (K) | Y | Y |
| Sweetia fruticosa Spreng. | RT Pennington 897 (E) | Y | Y |
| Dalbergioid clade | |||
| Adesmia parvifolia Phil. | Lavin 8269 (MONT) | Y | Y |
| Adesmia volckmannii Phil. | Lavin 8281 (MONT) | Y | Y |
| Aeschynomene pfundii Taub. | Lavin s.n. (MONT) | Y† pm? | Y |
| Aeschynomene rudis Benth. | DE Fairbrothers et al. 82 (LSU) | Y | Y |
| Amicia glandulosa H.B. & K. | RT Pennington 654 (E) | Y | Y |
| Amorpha fruticosa L. | Wojciechowski 1378 (ASU) | Y† | Y |
| Arachis magna Krapov., W.C. Greg. & C.E. Simpson | CIAT 22248 (MONT) | Y | Y |
| Brya hirsuta Borhidi | Lavin 7110 (MONT) | Y | Y |
| Chaetocalyx nigricans Burkart | Vanni 2955 (F) | Y | Y |
| Dalea pulchra Gentry | Wojciechowski 175 (ARIZ) | Y | Y |
| Diphysa humilis Oerst. ex Benth. & Oerst. | Haber 1322 (MO) | Y | Y |
| Diphysa ormocarpoides (Rudd) M. Sousa & R. Antonio | R Torres-C. 977 (MEXU) | Y | Y |
| Eysenhardtia polystachya Sarg. | ASU 218775 | Y | Y |
| Macherium sp. | CE Hughes 4/89 (FHO) | Y | Y |
| Marina parryi (Torr. & A. Gray) Barneby | Wojciechowski 176 (ARIZ) | Y | Y |
| Ormocarpopsis itremoensis DuPuy & Labat | D DuPuy 2363 (K) | Y | Y |
| Ormocarpopsis parvifolia Dumaz-le-Grand | P Phillipson 3508 (K) | Y | Y |
| Pictetia sulcata (P. Beauv.) Beyra & Lavin | A Gentry 51037 (MO) | Y pm? | no rxn |
| Platymiscium sp. | RT Pennington 692 (E) | N pm? | Y |
| Platypodium elegans Vog. (2 access.) | RT Pennington 488, 688 (E) | Y, Y | Y, Y |
| Pterocarpus indicus Willd. | RT Pennington 718 (E) | Y | Y |
| Stylosanthes hamata Taub. | Beyra-M. s.n. (MONT) | Y pm? | Y |
| Tipuana tipu (Benth.) Kuntze | Lavin 6184 (MONT) | Y | Y |
| Vatairea macrocarpa Ducke | RT Pennington s.n. (E) | Y | Y |
| Vatairea sp. | RT Pennington 475 (E) | Y | Y |
| Genistoid clade | |||
| Anarthrophyllum desideratum Reiche var. mustersi Speg. | RH Fortunato 4971 (ARIZ) | Y | Y |
| Bowdichia virgilioides Kunth (2 access.) | AM de Carualho 3981 (US); BA Perelra 1110 (US) | Y, Y | Y, Y |
| Brongniartia peninsularis Rose | J Rebman 4214 (ASU) | Y | Y |
| Brongniartia ulbrichiana Harms | CE Hughes 2459 (K) | Y | Y |
| Crotalaria pumila Ortega | Wojciechowski 930 (ASU) | Y | Y |
| Crotalaria incana L. | N Harriman 16341 (ASU) | Y | Y |
| Crotalaria pumila Ortega | Wojciechowski 930 (ASU) | Y | Y |
| Crotalaria saltiana Andrews | CC Chaung 4723 (ASU) | Y | Y |
| Cyclolobium brasiliense Benth. (2 access.) | Klittgaard & de Lima 86 (K); JRI Wood 17607 (K) | Y, Y | Y, Y |
| Cytisus scoparius Link | Wojciechowski 1000 (ASU) | Y | Y |
| Dichilus lebeckioides DC. | A Balsinhas 3464 (K) | Y | Y |
| Dicraeopetalum mahafaliense (M. Pelt.) Yakovlev | Capuron 28529-SF (TEF) | Y | Y |
| Diplotropis martiusii Benth. | Beck et al. 166 (US) | Y | Y |
| Genista monspessulana (L.) L.A.S. Johnson | Wojciechowski 897 (ASU) | Y | Y |
| Harpalyce brasiliana Benth. | Ganer 2674 (K) | Y | Y |
| Lamprolobium fruticosum Benth. | Clarkson & Nelder 8827 (K) | Y | Y |
| Lebeckia sericea Thunb | JJM van der Meruve 215 (K) | Y | Y |
| Lupinus consentini Walp. | CE Hughes 1974 (FHO) | Y | Y |
| Lupinus sericeus Pursh | Wojciechowski 1460 (ASU) | Y | Y |
| Lupinus tegeticularis var. duranii (Eastw.) Barneby | Wojciechowski 729 (DAV) | Y | Y |
| Maackia amurensis Rupr. | P Herendeen 1-V-2003-5 (US) | Y | Y |
| Ormosia formosana Kaneh. | JM Hu 1095 (DAV) | Y | Y |
| Piptanthus nepalensis Sweet | W Hodgson 10787 (ASU) | Y | Y |
| Plagiocarpus axillaris Benth. | Barrit 1406 (K) | Y | Y |
| Poecilanthe falcata (Vell.) Heringer | Klittgaard & de Lima 4 (K) | Y | Y |
| Poecilanthe subcordata Benth. | Harley 21205 (K) | Y | Y |
| Spartium junceum L. | Wojciechowski 999 (ASU) | Y | Y |
| Templetonia retusa (Vent.) R. Br. | Lewis et al. 3833 (K) | Y | Y |
| Thermopsis rhombifolia Nutt. ex Pursh | Wojciechowski 807 (MONT) | Y | Y |
| Ulex europaeus L. | D Damrel 2304 (ASU) | Y | Y |
| Mirbelioid & Indigoferoid clades | |||
| Aotus ericoides G. Don | Wojciechowski 866 (ASU) (UCSCA 81.114) | N? | Y |
| Bossiaea cordigera Benth. ex Hook. (f.) | Wojciechowski 860 (ASU) (UCSCA 81.47) | Y | Y |
| Daviesia latifolia R. Br. | Wojciechowski 863 (ASU) (UCSCA 94.379) | Y† | Y |
| Gastrolobium sp. | MD Crisp 9294 (CANB) | Y | Y |
| Gompholobium minus Sm. | MD Crisp 9153 (CANB) | Y | Y |
| Hovea purpurea Sweet | Wojciechowski 869 (ASU) | Y | Y |
| Hypocalyptus coluteoides (Lam.) R. Dahlgren | C Burman 1236 (CANB) | N? | Y |
| Indigofera sphaerocarpa A. Gray | Van Devender 94–445 (ASU) | Y | Y |
| Indigofera sp. | Schrire 2556 (K) | Y? | no rxn |
| Isotropis foliosa Crisp | MD Crisp 9121 (CANB) | Y | Y |
| Jacksonia sp. | MD Crisp 9114 (CANB) | Y | Y |
| Microcharis sessilis (Thulin) Schrire | Thulin & Warfa 6263 (UPS) | Y | Y |
| Phylloxylon spinosa Du Puy, Labat & Schrire | Schrire 2531 (K) | Y | Y |
| Millettioids-Phaseoloids clade | |||
| Alysicarpus vaginalis DC. | JB Nelson 702 (ASU) | Y | N |
| Apios Americana Medik. | LC Higgins 12117 (ASU) | Y | Y |
| Austrosteenisia blackii (F. Muell.) R. Geesink | Pedley 5005 (K) | Y | Y |
| Bituminaria bituminosa (L.) C. H. Stirton | Soza & Gross1140 (RSA) | Y | Y |
| Campylotropis macrocarpa (Bunge) Rehder | Wojciechowski 864 (ASU) (UCBBG 95.0459) | Y | N |
| Clitoria mariana L. | Wojciechowski & Lavin 1198 (ASU) | Y | Y |
| Cologania angustifolia Kunth. (2 access.) | W Hodgson 5573 (ASU); Wojciechowski 1412 (ASU) | no rxn, Y | Y, Y |
| Cologania longifolia A. Gray | Wojciechowski s.n. | Y | Y |
| Dalbergiella nyasae Baker f. | Muller 2686 (K) | Y | Y |
| Derris laxiflora Benth. | JM Hu 1081 (DAV) | Y | Y |
| Desmodium angustifolium DC. | AL Reina et al. s.n. (ASU) | Y | N |
| Desmodium batocaulon A. Gray | Wojciechowski 1474 (ASU) | Y | N |
| Desmodium incanum DC. | RD Worthington 32961 (ASU) | Y | N |
| Desmodium psilocarpum A. Gray | Wojciechowski 929 (ASU) | Y | N |
| Desmodium rosei B. G. Schub. | Wojciechowski s.n. | Y | N |
| Desmodium tortuosum (Sw.) DC. | WM Bush 368 (ASU) | Y | N |
| Erythrina cristi-galli L. | Wojciechowski 894 (ASU) | Y | Y |
| Glycine max Merr. | Univ. California Davis (cult.) | Y | Y |
| Hoita macrostachya (DC.) Rydb. | S Boyd 11472 (ASU) | Y | Y |
| Kummerowia stipulacea Makino | AR Diamond 15666 (ASU) | Y | N |
| Kunstleria ridleyi Prain | NN Kat 193 (L) (JM Hu 1230) | Y | Y |
| Lablab purpureus Sweet | Wojciechowski 1514 (ASU) (USDA 639019) | Y | Y |
| Lespedeza cuneata G. Don | Wojciechowski & Lavin 1182 (ASU) | Y | N |
| Lonchocarpus eriocarinalis Micheli | Lavin s.n. (MONT) | Y | Y |
| Lonchocarpus phaseolifolius Benth. | Hughes 7/89 (FHO) | Y | Y |
| Macroptilium gibbosifolium (Ortega) A. Delgado | ASU 242502 | Y | Y |
| Millettia leptobotrys Dunn | JM Hu 1164 (DAV) | Y | Y |
| Millettia thonningii Baker | Lavin s.n. (MONT) | Y | Y |
| Ostryocarpus stulhmannii (Taub.) R. Geesink | Corby 2162 (K) | Y | Y |
| Otholobium glandulosum (L.) J.W. Grimes | D Kelch 028 (UC) | Y | Y |
| Pediomelum mephiticum (S. Wats.) Rydb. | A Salywon & Wojciechowski 1058 (ASU) | Y | Y |
| Phaseolus filiformis Benth. (2 access.) | Wojciechowski 1513 (ASU) (DLEG 890043); D Damrel V55 (ASU) | Y, Y | Y, Y |
| Phaseolus vulgaris L. | Univ. California Davis (cult.) | Y | Y |
| Philenoptera eriocalyx (Harms) Schrire (var. wankiensis) | JM Hu 1090 (DAV) | Y | Y |
| Piscidia piscipula Sarg. | Lavin & Luckow 5793a (TEX) | Y | Y |
| Platycyamus regnellii Benth. | HC de Lima 5 (RJ) | Y | Y |
| Pongamiopsis amygdalina (Baill.) R. Vig. | DuPuy 560 (K) | Y | Y |
| Psophocarpus tetragonolobus DC. | Wojciechowski 1512 (ASU) (DLEG 910202) | Y | Y |
| Psoralea argophylla Pursh | Wojciechowski 1436 (ASU) | Y | Y |
| Pueraria montana (Lour.) Merr. | Wojciechowski & Lavin 1189 (ASU) | Y | Y |
| Rhynchosia senna Gillies ex Hook. & Arn. | ASU 235516 | Y | Y |
| Rupertia physodes (Douglas ex Hook.) J.W. Grimes | Wojciechowski & Steele 889 (ASU) | Y | Y |
| Strophostyles helvola (L.) Elliott | D Cothran 7 (MONT) | Y | Y |
| Tephrosia leiocarpa A. Gray | DLEG 880028 | Y pm? | Y |
| Tephrosia tenella A. Gray | PD Jenkins s.n. (ARIZ) | Y | Y |
| Tephrosia virginiana (L.) Pers. | G. Neesom s.n. | Y pm? | Y |
| Vigna subterranea (L.) Verdc. | Lavin s.n. (MONT) (USDA 241993) | Y | Y |
| Vigna unguiculata (L.) Walp. | JC Baudet 114 (MONT) | Y | Y |
| Wajira grahamiana (Wight & Arn.) Thulin & Lavin | Lavin 1623 (MONT) | Y | Y |
| Loteae, Robinieae, and Sesbania clades | |||
| Anthyllis vulneraria L. | G Allan 45 (RSA) | Y | Y |
| Coronilla coronata L. | A Mayer 39 (M/MSB) | Y | Y |
| Coronilla minima L. | D Podlech 54633 (M/MSB) | Y | Y |
| Coronilla varia L. | SP McLaughlin & JE Bowers 6823 (ARIZ) | Y | Y |
| Dorycnium pentaphyllum Scop. | F Schuhwerk 92/111 (M/MSB) | Y | Y |
| Hammatolobium kremerianum C. Muell. | G Allan 43 (RSA) | Y | Y |
| Hippocrepis glauca Ten. | Hepper 9239 (K) | Y | Y |
| Hippocrepis unisiliquosa L. | G Allan 55 (RSA) | Y | Y |
| Lotus corniculatus L. | Wojciechowski 1545 (ASU) | Y | Y |
| Lotus creticus L. | G Allan 5 (RSA) | Y† | Y |
| Lotus grandiflorus Greene | Wojciechowski 885 (ASU) | Y | Y |
| Lotus purshianus Clem. & E.G. Clem. | Wojciechowski 707 (DAV) | Y | Y |
| Lotus rigidus Greene | Wojciechowski & Sanderson 156 (ARIZ) | Y† | Y |
| Coursetia glandulosa A. Gray | Wojciechowski 1200 (ASU) | Y | Y |
| Genistidium dumosum I.M. Johnston | Lavin 210890 (MONT) | Y | Y |
| Gliricidia maculata (H.B. & K.) Steud. | CE Hughes 675 (FHO) | Y | Y |
| Hebestigma cubense Urb. | Lavin 7144a (MONT) | Y | Y |
| Hybosema robustum M. Sousa & Lavin | CE Hughes 92/92 (FHO) | Y | Y |
| Lennea modesta Standl. & Steyerm. | Lavin & Delgado 8210a (MEXU) | Y† | Y |
| Olneya tesota A. Gray | Wojciechowski 877 (ASU) | Y? | Y |
| Peteria thompsonae S. Watson | Wojciechowski, Steele, & Morse 1531 (ASU) | Y | Y |
| Poissonia heterantha (Griseb.) Lavin | Lavin 5856 (TEX) | Y | Y |
| Poissonia hypoleuca (Speg.) Lillo | Lavin 5787 (TEX) | Y | Y |
| Poissonia weberbaueri (Harms) Lavin | Hutchinson 7259 (F) | Y | Y |
| Poitea glyciphylla (Poiret) Lavin | Lavin 8030-4 (MONT) | Y | Y |
| Poitea immarginata (C. Wright) Lavin | Lavin 7105 (MONT) | Y | Y |
| Robinia neomexicana A. Gray | Wojciechowski 717 (ARIZ) | Y† | Y |
| Robinia pseudoacacia L. | Univ. California Davis, cult. | Y† | Y |
| Sphinctospermum constrictum (S. Watson) Rose | Lavin 5120 (MONT) | Y | Y |
| Sesbania drummondii (Rydb.) Cory | Lavin s.n. (TEX) | Y | Y |
| Sesbania emerus (Aubl.) Urb. | CE Hughes 80/87 (FHO) | Y | Y |
| Sesbania punicea Benth. | Wojciechowski & Lavin 1185 (ASU) | Y† | Y |
| IR-Lacking clade (IRLC) | |||
| Afgekia filipes (Dunn) R. Geesink | JF Maxwell 90–246 (L); (Hu 1231) | N | Y |
| Afgekia sericea Craib | C Chermsirivanthana 996 (E) | N | Y |
| Alhagi maurorum Medik. | USDA 502281 | N | N |
| Astragalus americanus (Hook.) M.E. Jones | Wojciechowski 851 (DAV) | N | N |
| Astragalus canadensis L. var. brevidens (Gand.) Barneby | Wojciechowski & Sanderson 302 (ARIZ) | N | N |
| Astragalus garbancillo Cav. | CE Hughes 2041 (FHO) | N | N |
| Astragalus lonchocarpus Torr. | Wojciechowski & Sanderson 143 (ARIZ) | N | N |
| Astragalus nothoxys A. Gray | Wojciechowski & Sanderson 177 (ARIZ) | N | N |
| Astragalus pelecinus (L.) Barneby | Wojciechowski & Sanderson 294 (USDA 186284) | N | N |
| Callerya atropurpurea (Wall.) Schot (2 access.) | OSC 322, OSC 323; Liston | N, N | Y, Y |
| Callerya australis (Endl.) Schot (2 access.) | OSC 326, OSC 327; Liston | N, N | Y, Y |
| Callerya megasperma (F. Muell.) Schot (2 access.) | OSC 325, Liston; Wilson 7646 (CANB); J Trusty | N, N | Y, Y |
| Callerya pilipes (F. M. Bailey) Schot | Gray 08360 (CANB); J Trusty | N | Y |
| Callerya reticulata (Benth.) Schot (4 access.) (=Millettia reticulata Benth.) | OSC 324, Liston; “HD”, and “OTG25”, J Trusty, cult.; Wojciechowski 1278 (ASU), cult. | N, N, N, N | Y, Y, Y, Y |
| Calophaca hovenii Schrenk | ID Baitulin et al. s.n. (K) DNA Bank 22172 | N | N |
| Caragana arborescens Lam. | Wojciechowski & Sanderson 413 (ARIZ); USDA 310390 | N | N |
| Caragana pygmaea DC. | Wojciechowski & Lavin 1134 (MONT) | N | N |
| Chesneya elegans Fomin | M Nydegger 43494 (M/MSB) | N | N |
| Chesneya parviflora Jaub et. Spach | J Leonard 5840 (K) | N | N |
| Cicer arietinum L. | Wojciechowski & Sanderson 189 (ARIZ; cult.) | N | N |
| Cicer canariense A. Santos Guerra & G. P. Lewis | KP Steele 38 (USDA 557453) | N | N |
| Cicer macracanthum Popow | KP Steele 75 (USDA 599080) | N | N |
| Cicer pinnatifidum Jaub. & Spach | Wojciechowski 409 (DAV) (USDA 458555) | N | N |
| Cicer yamashitae Kitam. | KP Steele 71 (USDA 504550) | N | N |
| Clianthus puniceus Lindl. | OSC 7140; Liston | N | N |
| Colutea arborescens L. | Wojciechowski & Sanderson 406 (ARIZ) | N | N |
| Ebenus cretica L. (2 access.) | R Gadringer et al KR36-1; N Bohling & T Raus 7262 (M/MSB) | N, N | N, N |
| Ebenus longipes Boiss. & Balansa | M Nydegger 45688 (M/MSB) | N | N |
| Endosamara racemosa (Roxb.) R. Geesink | JF Maxwell 90–202 (L) | N | Y |
| Erophaca baetica Boiss. subsp. orientalis (Chater & Meikle) Podlech | JR Edmonson & MAS McClintock 2803 (K), DNA Data Bank 22170 | N | N |
| Galega officinalis L. | USDA 325341 | N | N |
| Glycyrrhiza acanthocarpa J.M. Black | LAS Johnson & Constable 47187 (UC) | N | N |
| Glycyrrhiza aspera Pall. | D Podlech 31059 (M/MSB) | N | N |
| Glycyrrhiza astragalina Gill. (2 access.) | G Seijo 1511 (K); Weigand et al. 6894 (M/MSB) | N, N | N, N |
| Glycyrrhiza glabra L. (2 access.) | A Al-Harasi s.n. (K); J. Lamond 3228 (E) | N, N | N, N |
| Glycyrrhiza lepidota Pursh | Wojciechowski 714 (DAV) | N | N |
| Glycyrrhiza pallidiflora Maxim. | (K) DNA Data Bank 22180 | N | N |
| Glycyrrhiza triphylla Fisch. & C.A. Mey. | TF Hewer H.4000 (E) | N | N |
| Gueldenstaedtia himalaica Baker | BN Starling et al. 241 (K) DNA Bank 22182 | N | N |
| Gueldenstaedtia verna (Georgi) Boriss. | Ulanova & Bassargin s.n. (K) DNA Bank 22183 | N | N |
| Halimodendron halodendron (L.) Voss | Gillis & Good (ASU) | N | N |
| Hedysarum alpinum L. | Riewe & Marsh 290 (ASU) | N | N |
| Hedysarum boreale Nutt. | Lavin s.n. (MONT) | N | N |
| Hedysarum sulphurescens Rydb. | Lavin s.n. (MONT) | N | N |
| Lathyrus aphaca L. | KP Steele 34 (USDA 286527) | N | N |
| Lathyrus jepsonii Greene | Pinkava 11905 (ASU) | N | N |
| Lathyrus latifolius L. | Wojciechowski 543 (DAV) | N | N |
| Lathyrus odoratus L. | KP Steele 46 (USDA 420254) | N | N |
| Lathyrus sativus L. | KP Steele 29 (USDA 283562) | N | N |
| Lens ervoides Grande | KP Steele 67 (USDA 572330) | N | N |
| Lessertia annularis Burch | H Merxmuller & W Giess (M/MSB) | N | N |
| Lessertia benguellensis Baker | AE vanWyk 8758 (M/MSB) | N | N |
| Lessertia herbacea Druce | Wojciechowski & Sanderson 299 (ARIZ); cult. | N | N |
| Medicago monantha Trautv. | KP Steele 177 (USDA) | N | N |
| Medicago sativa L. | Wojciechowski 561 (ARIZ) | N | N |
| Medicago truncatula Gaertn. | Wojciechowski 1014 (ASU); (cult “A17”; D Cook) | N | N |
| Melilotus indica All. | Wojciechowski 540 (DAV) | N | N |
| Melilotus officinalis Lam. | Wojciechowski 308 (DAV) | N | N |
| Millettia japonica A. Gray | Tsugaru 768 (KYO); J Trusty | N | Y |
| Ononis arvensis L. | KP Steele 32 (USDA 440578) | N | N |
| Ononis biflora Desf. | KP Steele 41 (USDA 244319) | N | N |
| Onobrychis montana DC. | CT Mason & PB Mason 3773 (ARIZ) | N | N |
| Oxytropis deflexa var. sericea Torr. & A. Gray | Wojciechowski & Sanderson 132 (ARIZ) | N | N |
| Oxytropis lambertii Pursh | Wojciechowski & Sanderson 155 (ARIZ) | N | N |
| Parochetus communis Buch.-Ham. ex D. Don | Wojciechowski 901 (ARIZ); cultivated (Liston) | N | N |
| Pisum sativum L. | Wojciechowski 1015 (ASU); cultivar “Sparkle” (N Weeden) | N | N |
| Smirnowia turkestana Bunge | KH Rechinger 51917 (K) | N | N |
| Sphaerophysa salsula DC. | RR Halse 5170 (ASU) | N | N |
| Spongiocarpella purpurea (P.C. Li) Yakovlev | BN Starling et al. 146 (K) | N | N |
| Sulla coronaria (L.) Medik. | USDA 459103 (UC Riverside Ag Garden) | N | N |
| Sutherlandia frutescens R. Br. | Wojciechowski & Sanderson 266 (ARIZ) | N | N |
| Swainsona campylantha F. Muell. | JZ Weber 776 (M/MSB) | N | N |
| Swainsona pterostylis (DC.) Bakh. f. | Wojciechowski & Sanderson 296 (ARIZ) | N | N |
| Taverniera cuneifolia Arn. | M Thulin 10821 (K) DNA Bank 22569 | N | N |
| Taverniera lappacea DC. | A Radcliffe-Smith 5474 (K) DNA Bank 22568 | N | N |
| Tibetia himalaica (Baker) H.P. Tsui | TN Ho et al. 1867 (E) | N | N |
| Trifolium dubium Sibth. | KP Steele s.n. | N | N |
| Trifolium nanum Torr. | CU Boulder course # 4520 collection (ARIZ) | N | N |
| Trifolium pratense L. | KP Steele 129 (USDA 237713) | N | N |
| Trifolium repens L. | Wojciechowski 730 (DAV); cult. | N | N |
| Trigonella foenum-graecum L. | KP Steele 16 (USDA 567879) | N | N |
| Trigonella kotschyi Fenzl ex Boiss. | KP Steele 64 (USDA 206775) | N | N |
| Vavilovia formosa (Steven) Federov | Axbepgob, Uupzoeba, & Panebapsh s. n. (K) | N | N |
| Vicia faba L. | Wojciechowski 998 (ASU) (USDA 469175) | N | N |
| Vicia ludoviciana Nutt. ex Torr. & A. Gray | S McLaughlin & JE Bowers 3185 (ARIZ) | N | N |
| Vicia narbonensis L. | KP Steele 122 (USDA 294300) | N | N |
| Wisteria brachybotrys Siebold & Zucc. | K DNA Bank 22664; J Trusty | N | Y |
| Wisteria floribunda (Willd.) DC. | P Herendeen 1-V-2003-8 (US) | N | Y |
| Wisteria frutescens (L.) Poir. (2 access.) | AL & HN Moldenke 29243 (ARIZ); USDA 2774 | N, N | Y, Y |
| Wisteria macrostachya Nutt. ex Torr. & A. Gray | W Lathrup 081, cult.; J Trusty | N | Y |
| Wisteria sinensis Sweet (2 access.) | K DNA Bank 22082; “WT 20”, J Trusty | N, N? | Y, ? |
Intron present, “Y”; intron not present (i.e., loss), “N”; †, extra minor bands present; ?, reaction weak or inconclusive; “pm?”, polymorphic for intron presence and loss. Clade descriptions follow Wojciechowski et al. (2004).
3. Results
3.1. Size, gene content, order, and organization of chickpea plastid genome
Chickpea has a circular plastid genome 125,319 bp in length with only one copy of the IR region (Fig. 1, GenBank accession number EU 835853). Gene order in chickpea is similar to the ancestral angiosperm gene order (Raubeson et al., 2007) except for the loss of one copy of the IR and by the presence of a single, large inversion of approximately 50 kb that reverses the order of the genes between rbcL and rps16. The same inversion is present in the four other completely sequenced legume plastid genomes Glycine max (Saski et al., 2005), Lotus japonicus (Kato et al., 2000), Medicago truncatula, and Phaseolus vulgaris (Guo et al., 2007), and is apparently shared by the majority of papilionoid legumes (Doyle et al., 1996). The AT content of the chickpea plastid genome is 66.1%, similar to other legumes including Glycine max (64.63%), Lotus japonicus (64.0%), Phaseolus vulgaris (64.56%), and Medicago truncatula (66.03%).
Fig. 1.
Circular gene map of Cicer arietinum plastid genome. Genes shown on the outside of the circle are transcribed in the clockwise direction, and those on the inside of the circle are transcribed in the counter-clockwise direction (GenBank accession number EU 835853).
The chickpea plastid genome has 108 total genes, including 4 rRNA genes, 29 tRNA genes, and 75 protein-coding genes. Three genes, rps16, infA, and ycf4, found in most angiosperm plastid genomes, including representatives of the early-branching lineages (Goremykin et al., 2003; Hansen et al., 2007; Raubeson et al., 2007) are not present in the chickpea plastid genome. In ndhB, there is an internal stop codon, similar to other legume plastid genomes. There is no stop codon (Met…Val) in the rps8 gene, a characteristic feature of the Medicago truncatula plastid genome.
Fifteen genes contain one or two introns, nine of which are in protein-coding genes while six are in the tRNA genes. The protein-coding gene rpl2 in chickpea, which contains a single intron of 669 bp has 16 amino acids missing at the 5′-end relative to most other legumes (Fig. 2), while Medicago has 11 amino acids missing in the same portion of the rpl2 gene. Among intron-containing genes, trnK-UUU has the largest intron (2491 bp), and it includes the matK gene. The smallest intron is in trnL-UAA (555 bp). The ycf3 gene has two introns of 733 and 737 bp.
Fig. 2.
Amino acid alignment of rps12 protein sequences from chickpea (Cicer arietinum) and other representative legumes; alfalfa, (Medicago sativa), soybean (Glycine max), frenchbean (Phaseolus vulgaris), and lotus (Lotus japonicus).
3.2. Phylogenetic distribution of intron losses in clpP and rps12 genes among legumes
In the Cicer arietenum chloroplast genome we observed the absence of the clpP and rps12 3′-end introns. Observation of loss of both of these introns represents the first documented case of such loss within the same plastid genome. Therefore, 301 legume taxa from 198 genera and one member of the related family Polygalaceae (Table 1) were subjected to a PCR-based survey for the presence/absence of the clpP and rps12-3′-end introns. These taxa represent all major groups of legumes, with 23 caesalpinioid species (18 genera), 18 mimosoid species (15 genera), 260 species of papilionoids (165 genera), and the genus Polygala L. in the family Polygalaceae (potential sister group to Leguminosae in Fabales; sensu APG II, 2003). For clpP, expected fragments sizes should be 1100–1300 bp if the intron is present and 300–350 bp if the intron is lost (Fig. 3). For rps12, expected fragments sizes should be 800– 850 bp if the intron is present and 250–280 bp if the intron is lost (Fig. 4). Comparison of the sizes of both the clpP and rps12 PCR products from a diversity of legumes, represented in Figs. 3 and 4, reveals fragments very similar in length, a result consistent with a process by which plastid introns are excised precisely and entirely, as observed earlier by Doyle et al. (1995) and in other taxa by Downie et al. (1991). Indeed, sequence analysis of ten taxa selected from our survey that included those with or without the clpP and rps12 introns confirms that intron excision has occurred at precisely the same points in the gene sequence in each taxon lacking the intron (MFW, unpublished data). Furthermore, the minor variation in length of the clpP PCR products we observed (Fig. 3A) was due to the length of the intron in each taxon, which ranged from 702 bp (Lespedeza cuneata) to 799 bp (Lotus corniculatus) in the taxa we sequenced, whereas the length of the rps12-3′ intron was 529–532 bp in the taxa analyzed (Glycine max, Phaseolus vulgaris, Callerya reticulata; MFW, unpublished data).
Fig. 3.

Ethidium bromide stained 1.0% agarose gel showing PCR amplified products for selected legume taxa screened for the presence/absence of the clpP intron. (A) Lanes 1–10: (1) Lupinus tegeticulatus, present; (2) Genista monspessulana, present; (3) Glycine max, present; (4) Phaseolus vulgaris, present; (5) Millettia thonningii, present; (6) Desmodium psilocarpum, present; (7) Lespedeza cuneata, present; (8) Wajira grahamiana, present; (9) Lotus purshianus, present; (10) Robinia neomexicana, present. (B) Lanes 1–10: (1) Sesbania punicea, present; (2) Callerya atropurpurea, absent; (3) Millettia japonica, absent; (4) Wisteria floribunda, absent; (5) Glycyrrhiza lepidota, absent; (6) Cicer arietinum, absent; (7) Medicago sativa, absent; (8) Pisum sativum, absent; (9) Caragana arborescens, absent; (10) Astragalus canadensis, absent. M = 1 kb + DNA marker ladder.
Fig. 4.

Ethidium bromide stained 1.0% agarose gel showing PCR amplified products for selected legume taxa screened for the presence/absence of the rps12 intron. (A) Lanes 1–10: (1) Lupinus tegeticulatus, present; (2) Genista monspessulana, present; (3) Glycine max, present; (4) Phaseolus vulgaris, present; (5) Millettia thonningii, present; (6) Desmodium psilocarpum, absent; (7) Lespedeza cuneata, absent; (8) Wajira grahamiana, present; (9) Lotus purshianus, present; (10) Robinia neomexicana, present. (B) Lanes 1–10: (1) Sesbania punicea, present; (2) Callerya atropurpurea, present; (3) Millettia japonica, present; (4) Wisteria floribunda, present; (5) Glycyrrhiza lepidota, absent; (6) Cicer arietinum, absent; (7) Medicago sativa, absent; (8) Pisum sativum, absent; (9) Caragana arborescens, absent; (10) Astragalus canadensis, absent. M = 1 kb + DNA marker ladder.
The presence of extra minor PCR products, reactions that were weak or inconclusive, and polymorphic for intron presence/absence have been identified in our survey for these two introns (Table 1). Virtually all of these examples were found in reactions surveying for the first (of two) clpP intron in taxa that presumably have the intron present. Possible explanations include partial excision reactions, poor quality DNA (many taxa have been sampled from herbarium specimens), sequence variation in primer binding sites or rearrangements in/near the gene containing the intron.
Our survey (Table 1) reveals that the loss of the clpP intron is, with a few exceptions, limited to the large IR-lacking clade (Fig. 5). Loss of the clpP intron was detected in individual accessions of the mimosoid Inga Mill. and the papilionoids Aotus Sm., and Hypocalyptus Thunb., Platymiscium Vogel, although the latter three results are ambiguous and need to be confirmed with sampling of additional specimens. The rps12 intron is also lost in all members of the IRLC surveyed, with notable exceptions (all accessions of Callerya Endl., Wisteria Nutt., Afgekia Craib, and Millettia japonica A. Gray). This intron is also lost independently in the papilionoid tribe Desmodieae (Fig. 5), a monophyletic group nested in the “Millettioids-Phaseoloids” clade (Kajita et al., 2001; Wojciechowski et al., 2004). Desmodieae, which consists of 30 genera and ca. 530 species distributed in tropical to warm temperate regions of the world (Lewis et al., 2005), is represented in this analysis by the genera Alysicarpus Desv., Campylotropis Bunge, Desmodium Desv., Kummerowia Schindl., and Lespedeza Michx. (Table 1).
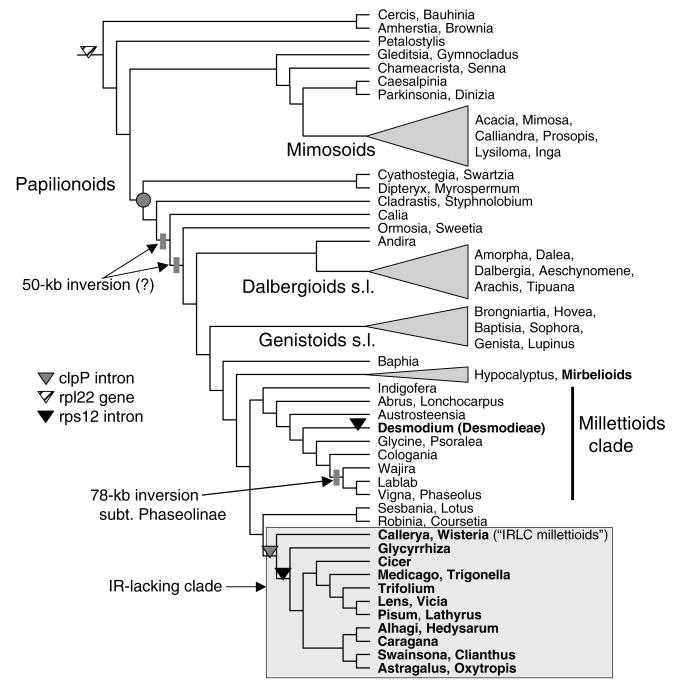
Fig. 5.
Simplified representation of phylogenetic relationships among major groups of legumes, summarized from Wojciechowski et al. (2004). Names of informal groups and clades are shown. The phylogenetic distribution of six of nine plastid genomic rearrangements (from Table 2) is depicted on the tree.
4. Discussion
4.1. Comparison of gene/intron content and genome organization among legumes
A number of previous studies indicated that legume plastid genomes have experienced substantial numbers of rearrangements (Palmer and Thompson, 1981, 1982; Palmer et al., 1988; Lavin et al., 1990; Bruneau et al., 1990; Doyle et al., 1995, 1996). Complete sequencing of plastid genomes of five legumes (Cicer, Glycine, Lotus, Medicago, and Phaseolus), combined with earlier gene mapping studies, have revealed nine genomic rearrangements, including two large inversions, the loss of the IR, three gene losses and three intron losses, two of which are reported here (Table 2). Thus, the legumes represent one of only a few angiosperm families that have experienced multiple, plastid genomic rearrangements and gene/intron losses (Jansen et al., 2007), and serve as an excellent choice in which to investigate contrasting patterns of plastid DNA evolution. Others families known to have comparable plastid genomic rearrangements include Asteraceae (two inversions; Jansen and Palmer, 1987; Kim et al., 2005), Campanulaceae (up to 42 inversions, two gene losses, 8 putative transpositions; Cosner et al., 1997, 2004; Haberle et al., 2008), Geraniaceae (12 inversions, 8 IR boundary changes; Chumley et al., 2006), Lobeliaceae (11 inversions; Knox et al., 1993), Oleaceae (4 inversions, 1 gene loss, 1 intron loss, 5 gene duplications; Lee et al., 2007), Poaceae (3 inversions, 3 gene losses, 2 intron losses; Doyle et al., 1992), and Ranunculaceae (9 inversions, 1 intron loss; Hoot and Palmer, 1994; Johansson, 1999).
Table 2.
Plastid genomic rearrangements among legumes
| Rearrangement | Phylogenetic distribution | Reference |
|---|---|---|
| IR-loss | IRLC clade | Lavin et al. (1990), Liston (1995), and Wojciechowski et al. (2000) |
| 50 kb inversion | All papilionoid tribes except Swartzieae, Sophoreae, and Dipterygeae | Doyle et al. (1996) and Pennington et al. (2001) |
| 78 kb inversion | Subtribe Phaseolinae of the papilionoid tribe Phaseoleae | Bruneau et al. (1990) |
| rpl22 gene loss | All legumes | Doyle et al. (1995) |
| rps16 gene loss | 15 of 28 papilionoid tribes | Doyle et al. (1995) |
| ycf4 gene loss | 15 of 28 papilionoid tribes | Doyle et al. (1995) |
| clpP intron loss | IRLC papilionoids | Current study |
| srpl2 intron loss | Bauhinia (tribe Cercideae), Soemmeringia (tribe Dalbergieae), Mucuna (tribe Phaseoleae), tribe Desmodieae | Doyle et al., (1995), Bailey et al. (1997) and Lai et al. (1997) |
| rps12 intron loss | IRLC papilionoids, tribe Desmodieae | Current study |
The causes for the propensity of plastid genomic changes in these lineages are unknown but several explanations have been proposed. For legumes, it was suggested that the loss of the IR has a destabilizing effect on genome organization (Palmer, 1991; Palmer and Thompson, 1982). However, given that most of the angiosperms with highly rearranged plastid genomes still retain two copies of the IR (i.e., Campanulaceae, Lobeliaceae, Oleaceae, Poaceae, Ranunculaceae, and most Geraniaceae) IR-loss does not provide a general explanation for the extensive rearrangements in plastid genomes. Moreover, the fact that the majority of the known plastid genomic rearrangements in legumes are also found in papilionoid taxa with two copies of the IR in their genomes (Table 2; Fig. 5) argues against this explanation as well. Another possible explanation for the higher incidence of rearrangements in some lineages is the presence of dispersed sequence repeats, which could facilitate rearrangements by intramolecular recombination (Palmer, 1991). In wheat (Ogihara et al., 1988) and Oenothera (Hupfer et al., 2000), such repeats have been directly implicated in inversions, and the strong correlation detected between the number of dispersed repeats and the extent of genomic rearrangements in several lineages is consistent with this explanation (Pombert et al., 2005, 2006; Haberle et al., 2008). Recent comparisons of the number and distribution of repeated sequences in completely sequenced legume plastid genomes have demonstrated the presence of numerous dispersed repeats, many more than in related rosid genomes that are not rearranged (Saski et al., 2005).
4.2. Phylogenetic distribution of chloroplast genomic rearrangements among legumes
We have plotted the distribution of several of the less ambiguous of the nine legume plastid genomic rearrangements (Table 2) on a phylogenetic tree based on cladistic analyses of complete nucleotide sequences of the plastid matK gene to assess the phylogenetic implications of these rare genomic changes (Fig. 5; tree summarized from Wojciechowski et al., 2004). Clearly, most plastid genomic rearrangements among legumes are restricted to the papilionoids with the exception of the loss of the rpl22 gene (Downie and Palmer, 1992; Doyle et al., 1995), which characterizes all taxa sampled from all three subfamilies of legumes, and the loss of the rpl2 intron in numerous species of the caesalpinioid genus Bauhinia (Lai et al., 1997). Interestingly, the rpl22 gene has not been lost from any other land plants (Downie and Palmer, 1992), and a functional copy has been isolated from the nuclear genome in Pisum sativum (Gantt et al., 1991).
The loss of one copy of the IR, as originally suggested by Lavin et al. (1990), has occurred only once among legumes and is restricted to a large clade of papilionoid legumes that includes the traditional tribes Carmichaelieae, Cicereae (chickpea), Galegeae, Hedysareae, Trifolieae, and Fabeae (Vicieae) and several genera formerly treated in the tribe Millettieae (so-called “IRLC millettioids”), including Callerya, Wisteria, Afgekia, Endosamara R. Geesink, and probably Antheroporum Gagnep. and Sarcodum Lour. (Hu et al., 2000, 2002; Hu and Chang, 2003). Taxa lacking the IR have been shown to form a monophyletic group informally known as the IRLC, which is well supported in phylogenetic trees based plastid matK and nuclear rDNA sequence analyses (Hu et al., 2000, 2002; Wojciechowski et al. 2000, 2004). From its taxonomic distribution, this mutational event in the plastid genome occurred relatively later in the evolution and diversification of the Papilionoideae, a molecular synapomorphy for a large (ca. 4400 species), derived group of primarily herbaceous taxa with a temperate distribution (Wojciechowski et al., 2000, 2004) and an estimated age of 39 Ma (Lavin et al., 2005). Like the now established monophyly of the taxa marked by loss of the IR (Palmer et al., 1988), results from molecular phylogenetic studies have provided both greater resolution and corroborating evidence for the relationships of many of the temperate and tropical groups long considered “derived” within papilionoids based upon the presence of morphological and/or chemical characters that have served as important taxonomic markers (Polhill, 1994). For example, the hypothesis for a single origin of canavanine biosynthesis (production of a non-protein amino acids such as l-canavanine, a close analog of arginine) in Papilionoideae (Bell, 1981), which occurs in all the tribes that comprise the IRLC plus the related tropical tribes that comprise their sister group (i.e., Millettieae, Phaseoleae, and allies), has been supported by recent analyses of plastid rbcL and matK gene sequences in legumes (Kajita et al., 2001; Wojciechowski et al., 2004). Distribution of the two legume plastid genome inversions provided additional support for clades identified in phylogenetic trees based on analyses of gene sequences. The 50-kb inversion defines an early evolutionary split in the diversification of the papilionoid clade with all members of this clade having the inversion except for taxa from the tribes Sophoreae, Swartzieae, and Dipterygeae (Doyle et al., 1996; Pennington et al., 2001; Wojciechowski et al., 2004), although the exact membership of this clade remains unresolved (due to lack of sampling all relevant taxa). The 78-kb inversion is much more limited in its distribution, being restricted to a majority of the genera that traditionally comprise subtribe Phaseolinae of the tribe Phaseoleae (Bruneau et al., 1990), which is also supported as a monophyletic group in trees based on plastid gene sequences (Thulin et al., 2004; Wojciechowski et al., 2004). Recent evidence indicates this inversion may be a synapomorphy for this clade, defined by the most recent common ancestor of the genera Wajira Thulin and Phaseolus L. (M. Moore, M.F. Wojciechowski, A. Delgado, and P.S. Soltis, unpublished data).
Two other gene losses in legumes have been detected in at least one genus in 15 (rps16 and ycf4) of the 28 papilionoid tribes (sensu Lewis et al., 2005). The taxonomic distribution of the rps16 loss based on filter hybridizations (Doyle et al., 1995) suggested multiple, independent losses within papilionoids but more rigorous PCR and sequencing strategies are needed to confirm these events. Among 64 completely sequenced seed plant plastid genomes there have been four independent losses of rps16: in Pinus, legumes, two members of the Malphigiales (Passiflora and Populus) and the monocot Dioscorea (Jansen et al., 2007). This gene has also been lost in the genus Adonis in the Ranunculaceae based on filter hybridization data (Johansson, 1999). The loss of ycf4 (formerly called ORF184) has been documented in Pisum (Nagano et al., 1991; Smith et al., 1991) and it is lacking in three (Cicer, Glycine, and Medicago) of the five completely sequenced legume plastid genomes. An earlier survey of the phylogenetic distribution of this loss among 392 legume genera based on filter hybridization screens with ycf4 gene-specific probes indicated at least 15 independent losses within tribe Phaseoleae alone (Doyle et al., 1995). Both the genome sequences and the filter hybridization data suggest that considerable homoplasy will limit the phylogenetic utility of this gene loss within legumes.
The two plastid genomic changes identified by sequencing the chickpea genome provide valuable information for resolving relationships among the IRLC papilionoid legumes (Table 2 and Fig. 5). Intron losses for both the clpP and rps12 genes have been identified in other angiosperm lineages as well. For example, both of the clpP gene introns have been lost in Poaceae, Onagraceae, Oleaceae, and Pinus (reviewed in Jansen et al., 2007), and the intron in the 3′-end of rps12 has been lost independently twice in the monocot order Asparagales (McPherson et al., 2004). However, the losses in Cicer represent the first documented case of the loss of introns from both of these genes in the same plastid genome. The clpP intron loss, which appears to have occurred only once within Leguminosae, provides additional support for the monophyly of the IRLC.
While the data suggest that loss of the rps12 intron generally coincides phylogenetically with the loss of the IR (Table 1), the distribution of the rps12 intron loss is more informative because it marks a slightly less-inclusive clade within the IRLC that provides additional data to resolve relationships among the early-branching lineages of the IRLC. That Callerya, Wisteria, Afgekia, and Millettia japonica unambiguously possess the intron in the rps12 gene is interesting because trees based on nucleotide sequences (Wojciechowski et al., 2000, 2004; Hu et al., 2002) have not been able to resolve the relationships of these and other lineages at the base of this clade. Indeed, results from molecular phylogenetic analyses are not in agreement on this point, with some data suggesting Glycyrrhiza L. is the sister group to the rest of the IRLC or Glycyrrhiza + Callerya and/or Wisteria s.l. (i.e., including other IRLC millettioids) are sister to the rest of the IRLC. While the consensus seems to be that Callerya + Wisteria s.l. form a clade (e.g., Hu and Chang, 2003), the presence of the rps12 intron in Callerya, Wisteria, and other members of the IRLC millettioids suggests they comprise the earliest-branching lineages that form the sister group to the rest of the IRLC, which are characterized by the loss of the intron from rps12. Furthermore, this result indicates that the loss of this intron occurred subsequent to the loss of one copy of the IR in these taxa. A second, independent loss of the rps12 intron has occurred in the more distantly related tribe Desmodieae (Fig. 5), which retains both copies of the IR, a group that is also marked by loss of the rpl2 intron (Doyle et al. 1995; Bailey et al. 1997).
5. Conclusions
Legume plastid genomes have undergone considerable diversification in gene/intron content and gene order, and these changes provide valuable information for resolving phylogenetic relationships among and within some major clades identified on the basis of analyses of DNA sequences. The two new genomic changes identified in the present study provide additional support of the monophyly of the IR-loss clade, and resolution of the early-branching pattern in this clade. In addition to providing insight into plastid genome evolution and phylogenetic relationships of legumes, the availability of complete plastid genome sequences facilitates plastid genetic engineering for improvement of agronomic traits and production of vaccines, biopharmaceuticals, biomaterials and industrial enzymes. Complete plastid genome sequences provide valuable information on spacer regions for integration of transgenes at optimal sites via homologous recombination, as well as endogenous regulatory sequences for optimal expression of transgenes, and should help in expanding plastid technology to other economically important crops.
Acknowledgments
Investigations reported in this article were supported in part by Grants from USDA 3611-21000-017-00D and NIH R01 GM 63879 to H.D. Research by R.K.J. was supported, in part, by NSF ATOL Grant DEB0120709. M.F.W. was supported, in part, by NSF Grant DEB0542958. The authors thank Matt Lavin, Alfonso Delgado-Salinas, Jennifer Trusty, Kelly Steele, Jer-Ming Hu, R. Toby Pennington, Mats Thulin, Aaron Listen, herbaria of The Royal Botanic Gardens at Kew (K), Edinburgh (E), Munich (M/MSB), and the Arizona State University Vascular Plant Herbarium (ASU) for tissue or DNA samples, and Dr. Michael Bausher (USDA) for help in early stages of genome sequencing. We also thank Anne Bruneau and an anonymous reviewer for their helpful comments on the manuscript.
References
- Angiosperm Phylogeny Group. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Bot J Linn Soc. 2003;141:399–436. [Google Scholar]
- Bailey CD, Doyle JJ, Kajita T, Nemoto T, Ohashi H. The chloroplast rpl2 intron and ORF184 as phylogenetic markers in the legume tribe Desmodieae. Syst Bot. 1997;22:13–138. [Google Scholar]
- Bausher MG, Singh ND, Lee SB, Jansen RK, Daniell H. The complete chloroplast genome sequence of Citrus sinensis (L.) Osbeck var ‘Ridge Pineapple’: organization and phylogenetic relationships to other angiosperms. BMC Plt Biol. 2006;6:21. doi: 10.1186/1471-2229-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell EA. Non-protein amino acids in the Leguminosae. In: Polhill RM, Raven PH, editors. Advances in Legume Systematics, Part 2. Royal Botanic Gardens; Kew, UK: 1981. pp. 489–499. [Google Scholar]
- Bruneau A, Doyle JJ, Palmer JD. A chloroplast DNA inversion as a subtribal character in the Phaseoleae (Leguminosae) Syst Bot. 1990;15:378–386. [Google Scholar]
- Chumley TW, Palmer JD, Mower JP, Fourcade HM, Calie PJ, Boore JL, Jansen RK. The complete chloroplast genome sequence of Pelargonium × hortorum: organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol Biol Evol. 2006;23:2175–2190. doi: 10.1093/molbev/msl089. [DOI] [PubMed] [Google Scholar]
- Cosner ME, Jansen RK, Palmer JD, Downie SR. The highly rearranged chloroplast genome of Trachelium caeruleum Campanulaceae: multiple inversions, inverted repeat expansion and contraction, transposition, insertions/deletions, and several repeat families. Curr Genet. 1997;31:419–429. doi: 10.1007/s002940050225. [DOI] [PubMed] [Google Scholar]
- Cosner ME, Raubeson LA, Jansen RK. Chloroplast DNA rearrangements in Campanulaceae: phylogenetic utility of highly rearranged genomes. BMC Evol Biol. 2004;4:27. doi: 10.1186/1471-2148-4-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H. Molecular strategies for gene containment in transgenic crops. Nat Biotechnol. 2002;20:581–586. doi: 10.1038/nbt0602-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H. Transgene containment by maternal inheritance: Effective or elusive? Proc Natl Acad Sci USA. 2007;104:6879–6880. doi: 10.1073/pnas.0702219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Datta R, Varma S, Gray S, Lee SB. Containment of herbicide resistance through genetic engineering of the chloroplast genome. Nat Biotechnol. 1998;16:345–348. doi: 10.1038/nbt0498-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Lee SB, Panchal T, Wiebe PO. Expression of the native cholera toxin B subunit gene and assembly as functional oligomers in transgenic tobacco chloroplasts. J Mol Biol. 2001;311:1001–1009. doi: 10.1006/jmbi.2001.4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Khan MS, Allison L. Milestones in chloroplast genetic engineering: an environmentally friendly era in biotechnology. Trends Plant Sci. 2002;7:84–91. doi: 10.1016/s1360-1385(01)02193-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Kumar S, Dufourmantel N. Breakthrough in chloroplast genetic engineering of agronomically important crops. Trends Biotechnol. 2005;23:238–245. doi: 10.1016/j.tibtech.2005.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Lee SB, Grevich J, Saksi C, Quesada-Vargas T, Guda C, Tomkins J, Jansen RK. Complete chloroplast genome sequences of Solanum bulbocastanum, Solanum lycopersicum and comparative analyses with other Solanaceae genomes. Theor Appl Genet. 2006;112:1503–1518. doi: 10.1007/s00122-006-0254-x. [DOI] [PubMed] [Google Scholar]
- Daniell H, Wurdack KJ, Kanagaraj A, Lee SB, Saski C, Jansen RK. The complete nucleotide sequence of the cassava (Manihot esculenta) chloroplast genome and multiple losses of the atpF intron in Malpighiales. Theor Appl Genet. 2008;116:723–737. doi: 10.1007/s00122-007-0706-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCosa B, Moar W, Lee SB, Milller M, Daniell H. Overexpression of the Bt Cry2Aa2 operon in chloroplasts leads to formation of insecticidal crystals. Nat Biotechnol. 2001;19:71–74. doi: 10.1038/83559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGray G, Rajasekaran K, Smith F, Sanford J, Daniell H. Expression of an antimicrobial peptide via the chloroplast genome to control phytopathogenic bacteria and fungi. Plant Physiol. 2001;127:852–862. [PMC free article] [PubMed] [Google Scholar]
- Downie SR, Palmer JD. Use of chloroplast DNA rearrangements in reconstructing plant phylogeny. In: Soltis PS, Soltis DE, Doyle JJ, editors. Molecular Systematics of Plants. Chapman and Hall; New York: 1992. pp. 14–35. [Google Scholar]
- Downie SR, Olmstead RG, Zurawski G, Soltis DE, Soltis PS, Watson JC, Palmer JD. Six independent losses of the chloroplast DNA rpl2 intron in dicotyledons: molecular and phylogenetic implications. Evolution. 1991;45:1245–1259. doi: 10.1111/j.1558-5646.1991.tb04390.x. [DOI] [PubMed] [Google Scholar]
- Downie SR, Llanas E, Katz-Downie DS. Multiple independent losses of the rpoC1 intron in angiosperm chloroplast DNAs. Syst Bot. 1996;21:135–151. [Google Scholar]
- Doyle JJ. DNA data and legume phylogeny: a progress report. In: Crisp MD, Doyle JJ, editors. Advances in Legume Systematics, Part 7: Phylogeny. Royal Botanic Gardens; Kew, UK: 1995. pp. 11–30. [Google Scholar]
- Doyle JJ, Davis JI, Soreng RJ, Garvin D, Anderson MJ. Chloroplast DNA inversions and the origin of the grass family (Poaceae) Proc Natl Acad Sci USA. 1992;89:7723–7726. doi: 10.1073/pnas.89.16.7722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL, Palmer JD. Multiple independent losses of two genes and one intron from legume chloroplast genomes. Syst Bot. 1995;20:272–294. [Google Scholar]
- Doyle JJ, Doyle JL, Palmer JD. The distribution and phylogenetic significance of a 50-kb chloroplast DNA inversion in the flowering plant family Leguminosae. Mol Phylogenet Evol. 1996;5:429–438. doi: 10.1006/mpev.1996.0038. [DOI] [PubMed] [Google Scholar]
- Doyle JJ, Doyle JL, Ballenger JA, Dickson EE, Kajita T, Ohashi H. A phylogeny of the chloroplast gene rbcL in the Leguminosae: taxonomic correlations and insights into the evolution of nodulation. Am J Bot. 1997;84:541–554. [PubMed] [Google Scholar]
- Doyle JJ, Chappill JA, Bailey CD, Kajita T. Towards a comprehensive phylogeny of legumes: evidence from rbcL sequences and non-molecular data. In: Herendeen PS, Bruneau A, editors. Advances in Legume Systematics, Part 9. Royal Botanic Gardens; Kew, UK: 2000. pp. 1–20. [Google Scholar]
- Dufoumantel N, Pelissier B, Gracon F, Peltier G, Ferullo JM, Tissot G. Generation of fertile transplastomic soybean. Plant Mol Biol. 2004;55:479–489. doi: 10.1007/s11103-004-0192-4. [DOI] [PubMed] [Google Scholar]
- Dufourmantel N, Tissot G, Goutorbe F, Garcon F, Muhr C, Jansens S, Pelissier B, Peltier G, Dubald M. Generation and analysis of soybean plastid transformants expressing Bacillus thuringiensis Cry1Ab protoxin. Plant Mol Biol. 2005;58:658–659. doi: 10.1007/s11103-005-7405-3. [DOI] [PubMed] [Google Scholar]
- Gantt JS, Baldauf SL, Calie PJ, Weeden NF, Palmer JD. Transfer of rpl22 to the nucleus greatly preceded its loss from the chloroplast and involved the gain of an intron. EMBO J. 1991;10:3073–3078. doi: 10.1002/j.1460-2075.1991.tb07859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goremykin VV, Hirsch-Ernst KI, Wolfl S, Hellwig FH. Analysis of the Amborella trichopoda chloroplast genome sequence suggests that Amborella is not a basal angiosperm. Mol Biol Evol. 2003;20:1499–1505. doi: 10.1093/molbev/msg159. [DOI] [PubMed] [Google Scholar]
- Greiner S, Wang X, Rauwolf U, Silber MV, Mayer K, Meurer J, Haberer G, Herrmann RG. The complete nucleotide sequences of the five genetically distinct plastid genomes of Oenothera, subsection Oenothera: I. Sequence evaluation and plastome evolution. Nucl Acids Res. 2008 doi: 10.1093/nar/gkn081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X, Castillo-Ramírez S, González V, Bustos P, Luís Fernández-Vázquez J, Santamaría RI, Arellano J, Cevallos MA, Dávila G. Rapid evolutionary change of common bean (Phaseolus vulgaris L.) plastome, and the genomic diversification of legume chloroplasts. BMC Genomics. 2007;8:228. doi: 10.1186/1471-2164-8-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haberle RC, Fourcade HM, Boore JL, Jansen RK. Extensive rearrangements in the chloroplast genome of Trachelium caeruleum are associated with repeats and tRNA genes. J Mol Evol. 2008 doi: 10.1007/s00239-008-9086-4. [DOI] [PubMed] [Google Scholar]
- Hachtel W, Neuss A, vom Steim J. A chloroplast DNA inversion marks an evolutionary split in the genus Oenothera. Evolution. 1991;45:1050–1052. doi: 10.1111/j.1558-5646.1991.tb04370.x. [DOI] [PubMed] [Google Scholar]
- Hansen DR, Dastidar SG, Cai Z, Penaflor C, Kuehl JV, Boore JL, Jansen RK. Phylogenetic and evolutionary implications of complete chloroplast genome sequences of four early diverging angiosperms: Buxus (Buxaceae), Chloranthus (Chloranthaceae), Dioscorea (Dioscoreaceae), and Illicium (Schisandraceae) Mol Phylogenet Evol. 2007;45:547–563. doi: 10.1016/j.ympev.2007.06.004. [DOI] [PubMed] [Google Scholar]
- Hoot SB, Palmer JD. Structural rearrangements, including parallel inversions, within the chloroplast genome of Anemone and related genera. J Mol Evol. 1994;38:274–281. doi: 10.1007/BF00176089. [DOI] [PubMed] [Google Scholar]
- Hu JM, Chang SP. Two new members of the Callerya group (Fabaceae) based on phylogenetic analysis of rbcL sequences: Endosamara racemosa (Roxb.) Geesink and Callerya vasta (Kosterm.) Schot Taiwania. 2003;48:118–128. [Google Scholar]
- Hu JM, Lavin M, Wojciechowski MF, Sanderson MJ. Phylogenetic systematics of the tribe Millettieae (Leguminosae) based on matK sequences, and implications for evolutionary patterns in Papilionoideae. Am J Bot. 2000;87:418–430. [PubMed] [Google Scholar]
- Hu JM, Lavin M, Wojciechowski MF, Sanderson MJ. Phylogenetic analysis of nuclear ribosomal ITS/5.8 S sequences in the tribe Millettieae (Fabaceae): Poecilanthe-Cyclolobium, the core Millettieae, and the Callerya group. Syst Bot. 2002;27:722–733. [Google Scholar]
- Hupfer H, Swiatek M, Hornung S, Herrmann RG, Maier RM, Chiu WL, Sears B. Complete nucleotide sequence of the Oenothera elata plastid chromosome, representing plastome I of the five distinguishable Euoenothera plastomes. Mol Gen Genet. 2000;263:581–585. doi: 10.1007/pl00008686. [DOI] [PubMed] [Google Scholar]
- Hussein H, Ruiz ON, Terry N, Daniell H. Phytoremediation of mercury and organomercurials in chloroplast transgenic plants: enhanced root uptake, translocation to shoots and volatilization. Environ Sci Technol. 2007;41:8439–8446. doi: 10.1021/es070908q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, Palmer JD. A chloroplast DNA inversion marks an ancient evolutionary split in the sunflower family (Asteraceae) Proc Natl Acad Sci USA. 1987;84:5818–5822. doi: 10.1073/pnas.84.16.5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, Raubeson LA, Boore JL, de Pamphilis CW, Chumley TW, Haberle RC, Wyman SK, Alverson AJ, Peery R, Herman SJ, Fourcade HM, Kuehl JV, McNeal JR, Leebens-Mack J, Cui L. Methods for obtaining and analyzing whole chloroplast genome sequences. Methods Enzymol. 2005;395:348–384. doi: 10.1016/S0076-6879(05)95020-9. [DOI] [PubMed] [Google Scholar]
- Jansen RK, Kaittanis C, Saski C, Lee SB, Tomkins J, Alverson AJ, Daniell H. Phylogenetic analyses of Vitis (Vitaceae) based on complete chloroplast genome sequences: effects of taxon sampling and phylogenetic methods on resolving relationships among rosids. BMC Evol Biol. 2006;6:32. doi: 10.1186/1471-2148-6-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RK, Cai Z, Raubeson LA, Daniell H, dePamphilis CW, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, Chumley TW, Lee SB, Peery R, McNeal J, Kuehl JV, Boore JL. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA. 2007;104:19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansson JT. Three large inversions in the chloroplast genomes and one loss of the chloroplast gene rps16 suggest an early evolutionary split in the genus Adonis (Ranunculaceae) Plant Syst Evol. 1999;218:133–143. [Google Scholar]
- Kajita TH, Ohashi Y, Tateishi Y, Bailey CD, Doyle JJ. rbcL and legume phylogeny, with particular reference to Phaseoleae, Millettieae, and Allies. Syst Bot. 2001;26:515–536. [Google Scholar]
- Kamarajugadda S, Daniell H. Chloroplast-derived anthrax vaccine and other vaccine antigens: their immunogenic and immunoprotective properties. Expert Rev Vaccines. 2006;5:839–849. doi: 10.1586/14760584.5.6.839. [DOI] [PubMed] [Google Scholar]
- Käss E, Wink M. Molecular phylogeny of the Papilionoideae (Family Leguminosae): rbcL gene sequences versus chemical taxonomy. Bot Acta. 1995;108:149–162. [Google Scholar]
- Käss E, Wink M. Molecular evolution of the Leguminosae: phylogeny of the three subfamilies based on rbcL sequences. Biochem Syst Ecol. 1996;24:365–378. [Google Scholar]
- Käss E, Wink M. Phylogenetic relationships in the Papilionoideae (Family Leguminosae) based on nucleotide sequences of cpDNA (rbcL) and ncDNA (ITS1 and 2) Mol Phylogenet Evol. 1997;8:65–88. doi: 10.1006/mpev.1997.0410. [DOI] [PubMed] [Google Scholar]
- Kato T, Kaneko T, Sato S, Nakamura Y, Tabata S. Complete structure of the chloroplast genome of a legume, Lotus japonicus. DNA Res. 2000;7:323–330. doi: 10.1093/dnares/7.6.323. [DOI] [PubMed] [Google Scholar]
- Kim YD, Jansen RK. Phylogenetic relationships of chloroplast DNA variation in the Berberidaceae. Plt Syst Evol. 1995;9:341–349. [Google Scholar]
- Kim HG, Choi KS, Jansen RK. Two chloroplast DNA inversions originated simultaneously during the early evolution of the sunflower family (Asteraceae) Mol Biol Evol. 2005;22:1–10. doi: 10.1093/molbev/msi174. [DOI] [PubMed] [Google Scholar]
- Knox EB, Downie SR, Palmer JD. Chloroplast genome rearrangements and the evolution of giant lobelias from herbaceous ancestors. Mol Biol Evol. 1993;10:414–430. [Google Scholar]
- Kumar S, Dhingra A, Daniell H. Chloroplast-expressed betaine aldehyde dehydrogenase gene in carrot cultured cells, roots and leaves confers enhanced salt tolerance. Plant Physiol. 2004;136:2843–2854. doi: 10.1104/pp.104.045187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai M, Sceppa J, Ballenger JA, Doyle JJ, Wunderlin RP. Polymorphism for the presence of the rpl2 intron in chloroplast genomes of Bauhinia (Leguminosae) Syst Bot. 1997;22:519–528. [Google Scholar]
- Lavin M, Doyle JJ, Palmer JD. Evolutionary significance of the loss of the chloroplast–DNA inverted repeat in the Leguminosae subfamily Papilionoidae. Evolution. 1990;44:390–402. doi: 10.1111/j.1558-5646.1990.tb05207.x. [DOI] [PubMed] [Google Scholar]
- Lavin M, Herendeen PS, Wojciechowski MF. Evolutionary rates analysis of Leguminosae implicates a rapid diversification of lineages during the Tertiary. Syst Biol. 2005;54:530–549. doi: 10.1080/10635150590947131. [DOI] [PubMed] [Google Scholar]
- Lee SB, Kwon H, Kwon S, Park S, Jeong M, Han S, Daniell H. Accumulation of trehalose within transgenic chloroplast confers drought tolerance. Mol Breed. 2003;11:1–13. [Google Scholar]
- Lee SB, Kaittanis C, Jansen RK, Hostetler JB, Tallon LJ, Town CD, Daniell H. The complete chloroplast genome sequence of Gossypium hirsutum: organization and phylogenetic relationships to other angiosperms. BMC Genomics. 2006;7:61. doi: 10.1186/1471-2164-7-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HL, Jansen RK, Chumley TW, Kim KJ. Gene relocations within chloroplast genomes of Jasminum and Menodora (Oleaceae) are due to multiple, overlapping inversions. Mol Biol Evol. 2007;24:1161–1180. doi: 10.1093/molbev/msm036. [DOI] [PubMed] [Google Scholar]
- Leelavathi S, Gupta N, Maiti S, Ghosh A, Reddy VS. Overproduction of an alkali-and thermo-stable xylanase in tobacco chloroplasts and efficient recovery of the enzyme. Mol Breed. 2003;11:59–67. [Google Scholar]
- Lewis G, Schrire B, Mackinder B, Lock M, editors. Legumes of the World. The Royal Botanic Gardens; Kew, UK: 2005. [Google Scholar]
- Liston A. Use of the polymerase chain reaction to survey for the loss of the inverted repeat in the legume chloroplast genome. In: Crisp M, Doyle JJ, editors. Advances in Legume Systematics 7: Phylogeny. Royal Botanic Gardens; Kew: 1995. pp. 31–40. [Google Scholar]
- McBride KE, Svab Z, Schaaf DJ, Hogan PS, Stalker DM, Maliga P. Amplification of a chimeric Bacillus gene in chloroplasts leads to an extraordinary level of an insecticidal protein in tobacco. BioTechnology. 1995;13:362–365. doi: 10.1038/nbt0495-362. [DOI] [PubMed] [Google Scholar]
- McPherson MA, Fay MF, Chase MW, Graham SW. Parallel loss of a slowly evolving intron from two closely related families in Asparagales. Syst Bot. 2004;29:296–307. [Google Scholar]
- Nagano Y, Matsuno R, Sasaki Y. Sequence and transcriptional analysis of the gene cluster trnQ-zfpA-psaI-ORF231-petA in pea chloroplasts. Curr Genet. 1991;20:431–436. doi: 10.1007/BF00317074. [DOI] [PubMed] [Google Scholar]
- Ogihara Y, Terachi T, Sasakuma T. Intramolecular recombination of chloroplast genome mediated by short direct-repeat sequences in wheat species. Proc Natl Acad Sci USA. 1988;85:8573–8577. doi: 10.1073/pnas.85.22.8573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JD. Plastid chromosomes: structure and evolution. In: Bogorad L, Vasil I, editors. Cell Culture and Somatic Cell Genetics of Plants. Academic Press; San Diego, California, USA: 1991. pp. 5–53. [Google Scholar]
- Palmer JD, Thompson WF. Rearrangements in the chloroplast genomes of mung bean and pea. Proc Natl Acad Sci USA. 1981;78:5533–5537. doi: 10.1073/pnas.78.9.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer JD, Thompson WF. Chloroplast DNA rearrangements are more frequent when a large inverted repeat sequence is lost. Cell. 1982;29:537–550. doi: 10.1016/0092-8674(82)90170-2. [DOI] [PubMed] [Google Scholar]
- Palmer JD, Osorio B, Thompson W. Evolutionary significance of inversions in legume chloroplast DNAs. Curr Genet. 1988;14:65–74. [Google Scholar]
- Pennington RT, Lavin M, Ireland H, Klitgaard BB, Preston J, Hu JM. Phylogenetic relationships of basal papilionoid legumes based upon sequences of the chloroplast trnL intron. Syst Bot. 2001;26:537–556. [Google Scholar]
- Polhill RM. Classification of the Leguminosae. In: Bisby FA, Buckingham J, Harborne JB, editors. Phytochemical Dictionary of the Leguminosae. Chapman and Hall; New York, USA: 1994. pp. xxxv–xlviii. [Google Scholar]
- Pombert JF, Otis C, Lemieux C, Turmel M. The chloroplast genome sequence of the green alga Pseudendoclonium akinetum Ulvophyceae reveals unusual structural features and new insights into the branching order of chlorophyte lineages. Mol Biol Evol. 2005;22:1903–1918. doi: 10.1093/molbev/msi182. [DOI] [PubMed] [Google Scholar]
- Pombert JF, Lemieux C, Turmel M. The complete chloroplast DNA sequence of the green alga Oltmannsiellopsisviridis reveals a distinctive quadripartite architecture in the chloroplast genome of early diverging ulvophytes. BMC Biol. 2006;4:3. doi: 10.1186/1741-7007-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesda-Vargas T, Ruiz ON, Daniell H. Over expression of the Bt cry2Aa2 operon in transgenic chloroplasts: transcription, processing, translation. Plant Physiol. 2005;138:1746–1762. doi: 10.1104/pp.105.063040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raubeson LA, Jansen RK. Chloroplast genomes of plants. In: Henry H, editor. Diversity and Evolution of Plants—Genotypic and Phenotypic Variation in Higher Plants. CABI Publishing; UK: 2005. pp. 45–68. [Google Scholar]
- Raubeson LA, Peery R, Chumley TW, Dziubek C, Fourcade HM, Boore JL, Jansen RK. Comparative chloroplast genomics: analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genomics. 2007;8:174. doi: 10.1186/1471-2164-8-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravi V, Khurana JP, Tyagi AK, Khurana P. The chloroplast genome of mulberry: complete nucleotide sequence, gene organization and comparative analysis. Tree Genet Genom. 2006;3:49–59. [Google Scholar]
- Ruhlman T, Lee SB, Jansen RK, Hostetler JB, Tallon LJ, Town CD, Daniell H. Complete chloroplast genome sequence of Daucus carota: implications for biotechnology and phylogeny of angiosperms. BMC Genomics. 2006;7:222. doi: 10.1186/1471-2164-7-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz ON, Daniell H. Engineering cytoplasmic male sterility via chloroplast genome by expression of beta-ketothiolase. Plant Physiol. 2005;138:1232–1246. doi: 10.1104/pp.104.057729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz ON, Hussein H, Terry N, Daniell H. Phytoremediation of organomercurial compounds via chloroplast genetic engineering. Plant Physiol. 2003;132:1344–1352. doi: 10.1104/pp.103.020958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson N, Bausher MG, Lee SB, Jansen RK, Daniell H. The complete nucleotide sequence of the coffee (Coffea arabica L.) chloroplast genome: organization and implications for biotechnology and phylogenetic relationships amongst angiosperms. Plt Biotechnol J. 2007;5:339–353. doi: 10.1111/j.1467-7652.2007.00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saski C, Lee SB, Daniell H, Wood T, Tomkins J, Kim HG, Jansen RK. Complete chloroplast genome sequence of Glycine Max and comparative analyses with other legume genomics. Plant Mol Biol. 2005;59:309–322. doi: 10.1007/s11103-005-8882-0. [DOI] [PubMed] [Google Scholar]
- Saski C, Lee SB, Fjellheim S, Guda C, Jansen RK, Luo H, Tomkins J, Rognli OA, Daniell H, LiuClarke J. Complete chloroplast genome sequences of Hordeum vulgare, Sorghum bicolor and Agrostis stolonifera, and comparative analyses with other grass genomes. Theor Appl Genet. 2007;115:571–590. doi: 10.1007/s00122-007-0567-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Elnitski L, Li M, Weirauch M, Riemer C, Smit A, Program NCS. Green ED, Hardison RC, Miller W. MultiPipMaker and supporting tools: alignments and analysis of multiple genomic DNA sequences. Nucleic Acids Res. 2003;31:3518–3524. doi: 10.1093/nar/gkg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AG, Wilson RM, Kaethner TM, Willey DL, Gray JC. Pea chloroplast genes encoding a 4 kDA polynucleotide of photosystem I and a putative enzyme of C1 metabolism. Curr Genet. 1991;19:403–410. doi: 10.1007/BF00309603. [DOI] [PubMed] [Google Scholar]
- Steane DA. Complete nucleotide sequence of the chloroplast genome from the Tasmanian blue gum, Eucalyptus globulus (Myrtaceae) DNA Res. 2005;12:215–220. doi: 10.1093/dnares/dsi006. [DOI] [PubMed] [Google Scholar]
- Thulin M, Lavin M, Pasquet R, Delgado-Salinas A. Phylogeny and biogeography of Wajira (Leguminosae): a monophyletic segregate of Vigna centered in the Horn of Africa region. Syst Bot. 2004;29:903–920. [Google Scholar]
- Verma D, Daniell H. Chloroplast vector systems for biotechnology applications. Plant Physiol. 2007;145:1129–1143. doi: 10.1104/pp.107.106690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace RS, Cota JH. An intron loss in the chloroplast gene rpoC1 supports a monophyletic origin for the subfamily Cactoideae of the Cactaceae. Curr Genet. 1996;29:275–281. doi: 10.1007/BF02221558. [DOI] [PubMed] [Google Scholar]
- Wojciechowski MF, Sanderson MJ, Steele KP, Liston A. Molecular phylogeny of the “temperate herbaceous tribes” of papilionoid legumes: a supertree approach. In: Herendeen P, Bruneau A, editors. Advances in Legume Systematics, Part 9. Royal Botanic Garden; Kew: 2000. pp. 277–298. [Google Scholar]
- Wojciechowski MF, Lavin M, Sanderson MJ. A phylogeny of legumes (Leguminosae) based on analysis of the plastid matK gene resolves many well-supported subclades within the family. Am J Bot. 2004;91:1846–1862. doi: 10.3732/ajb.91.11.1846. [DOI] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]