Summary
JIL-1 is the major kinase controlling the phosphorylation state of histone H3S10 at interphase in Drosophila. In this study we used three different commercially available H3S10ph antibodies as well as an acid-free polytene chromosome squash protocol that preserves the antigenicity of the H3S10 phospho-epitope to examine the role of histone H3S10 phosphorylation in transcription under both heat shock and non-heat shock conditions. We show that there is no redistribution or upregulation of JIL-1 or H3S10 phosphorylation at transcriptionally active puffs in such polytene squash preparations after heat shock treatment. Furthermore, we provide evidence that heat shock induced puffs in JIL-1 null mutant backgrounds are strongly labeled by antibody to the elongating form of RNA polymerase II (Pol IIoser2) indicating that Pol IIoser2 is actively involved in heat shock induced transcription in the absence of H3S10 phosphorylation. This was supported by the finding that there was no change in the levels of Pol IIoser2 in JIL-1 null mutant backgrounds compared to wild-type. That mRNA from the six genes that encode the major heat shock protein Hsp70 in Drosophila is transcribed at robust levels in JIL-1 null mutants was directly demonstrated by qRT-PCR. Taken together these data are inconsistent with the model that Pol II dependent transcription at active loci requires JIL-1 mediated histone H3S10 phosphorylation and instead support a model where transcriptional defects in the absence of histone H3S10 phosphorylation are a result of structural alterations of chromatin.
Keywords: Pol II, histone H3S10 phosphorylation, transcription, chromatin structure, JIL-1 kinase
Introduction
The JIL-1 tandem kinase in Drosophila localizes specifically to euchromatic interband regions of polytene chromosomes and is the predominant kinase controlling the phosphorylation state of histone H3S10 at interphase (Wang et al., 2001). JIL-1 is essential for viability and reduced levels of JIL-1 protein leads to a global disruption of chromosome structure (Jin et al., 2000; Wang et al., 2001; Zhang et al., 2003; Deng et al., 2005) as well as to extensive ectopic spreading of heterochromatic factors (Zhang et al., 2006). These findings suggested a model where maintenance of histone H3S10 phosphorylation levels at euchromatic chromatin regions is necessary to counteract heterochromatization and gene silencing (Wang et al., 2001; Ebert et al., 2004; Zhang et al., 2006; Bao et al., 2007). Recently, based on analyses of transcriptionally active regions during the heat-shock response (Nowak and Corces, 2000; Nowak et al., 2003; Ivaldi et al., 2007) an alternative model was proposed where JIL-1 is required for transcription by the RNA polymerase II (Pol II) machinery (Ivaldi et al., 2007). According to this model, rather than contributing to global chromosome structure, JIL-1 mediated histone H3S10 phosphorylation maintains a local chromatin environment that serves as a platform for the recruitment of the positive transcription elongation factor b (P-TEFb) and the consequent release of Pol II from promoter-proximal pausing (Ivaldi et al., 2007). Ivaldi et al. (2007) further suggested that histone H3S10 phosphorylation by JIL-1 is a hallmark of early transcription elongation in Drosophila and that this histone modification is required for the transcription of the majority, if not all, genes in this organism. However, this model is contradicted by the findings of Deng et al. (2007) that demonstrate that the lethality as well as some of the chromosome morphology defects associated with the null JIL-1 phenotype to a large degree can be rescued by reducing the dose of the Su(var)3-9 gene. Su(var)3-9 is a histone methyltransferase that is necessary for pericentric heterochromatin formation (Schotta et al., 2002; Elgin and Reuter, 2007) and that plays an important role in silencing of reporter genes by heterochromatic spreading (reviewed in Weiler and Wakimoto, 1995; Girton and Johansen, 2008). If JIL-1 had a critical role in promoting transcription at a majority of genes by regulating transcriptional elongation, it is difficult to envision how lethality can be rescued to near wild-type levels in the complete absence of JIL-1 and interphase histone H3S10 phosphorylation (Deng et al., 2007). Furthermore, Deng et al (2008) recently showed that JIL-1 mediated ectopic histone H3S10 phosphorylation is sufficient to induce a change in higher order chromatin structure from a condensed heterochromatin-like state to a more open euchromatic state and that these changes are not associated with enhanced transcriptional activity. Thus, these findings are incompatible with the transcriptional elongation model for JIL-1 function and we therefore attempted to repeat the experiments on which it is based using three different histone H3S10 phosphorylation (H3S10ph) antibodies as well as a newly developed acid-free polytene chromosome squash technique (DiMario et al., 2006) that preserves the antigenicity of the H3S10 phospho-epitope. We show that many of the key findings of Nowak and Corces (2000), Nowak et al. (2003), and Ivaldi et al. (2007) are likely to be artifacts caused by non-specific antibody cross-reactivity and by fixation procedures that are not suitable for reliable antibody detection of interphase phosphorylated histone H3S10. Taken together the results of Deng et al. (2007; 2008) and the findings presented here are inconsistent with the model of Ivaldi et al. (2007) that Pol II dependent transcription at active loci requires JIL-1 mediated histone H3S10 phosphorylation and instead support a model where transcriptional defects in the absence of histone H3S10 phosphorylation are a result of structural alterations of chromatin.
Materials and Methods
Drosophila melanogaster stocks and heat shock induction
Fly stocks were maintained at 23°C according to standard protocols (Roberts, 1998). Canton-S was used for wild-type preparations. The JIL-1z2 null allele is described in Wang et al. (2001) as well as in Zhang et al. (2003) and the recombined JIL-1z2 Su(var)3-906 chromosome in Deng et al. (2007). The P-element insertion mutant allele tws02414 was obtained from the Bloomington Stock Center and the twsP allele (Mayer-Jaekel et al., 1993) was the generous gift of Dr. D.M. Glover (University of Cambridge, Cambridge, England). Balancer chromosomes and markers are described in Lindsley and Zimm (1992). For heat shock experiments wandering third instar larvae were subjected to 25 min of heat shock treatment at 37°C as described previously by Nowak et al. (2003).
Immunohistochemistry
Salivary gland nuclei smush preparations were made as described in Wang et al. (2001), standard polytene chromosome squash preparations were performed as in Kelley et al. (1999) using the 5 min fixation protocol, and acid-free squash preparations were done following the procedure of DiMario et al. (2006). Antibody labeling of these preparations was performed as described in Jin et al. (1999) and in Wang et al. (2001). Primary antibodies used in this study include rabbit anti-H3S10ph (Epitomics, Upstate, and Cell Signaling), mouse anti-H3S10ph (Upstate and Cell Signaling), rabbit anti-histone H3 (Cell Signaling), mouse anti-lamin Dm0 (Gruenbaum et al., 1988), rabbit anti-HSF (gift from Dr. C. Wu, National Cancer Institute, Bethesda, MD), mouse anti-Pol IIoser2 (H5, Covance), rabbit anti-JIL-1 (Jin et al., 1999), chicken anti-JIL-1 (Jin et al., 2000), and anti-JIL-1 mAb 5C9 (Jin et al., 2000). DNA was visualized by staining with Hoechst 33258 (Molecular Probes) in PBS. The appropriate species- and isotype-specific Texas Red-, TRITC-, and FITC-conjugated secondary antibodies (Cappel/ICN, Southern Biotech) were used (1:200 dilution) to visualize primary antibody labeling. The final preparations were mounted in 90% glycerol containing 0.5% n-propyl gallate. The preparations were examined using epifluorescence optics on a Zeiss Axioskop microscope and images were captured and digitized using a high resolution Spot CCD camera. Confocal microscopy was performed with a Leica confocal TCS NT microscope system. A separate series of confocal images for each fluorophor of double labeled preparations were obtained simultaneously with z-intervals of typically 0.5 μm using a PL APO 63X/1.40 oil objective. A maximum projection image for each of the image stacks was obtained using the ImageJ software (http://rsb.info.nih.gov/ij/). Images were imported into Photoshop where they were pseudocolored, image processed, and merged. In some images non-linear adjustments were made to the channel with Hoechst labeling for optimal visualization of chromosomes.
Immunoblot analysis
Protein extracts were prepared from salivary glands dissected from third instar larvae (or in some experiments from whole larvae) homogenized in a buffer containing: 20 mM Tris-HCl pH8.0, 150 mM NaCl, 10 mM EDTA, 1 mM EGTA, 0.2% Triton X-100, 0.2% NP-40, 2 mM Na3VO4, 1 mM PMSF, 1.5 μg/ml aprotinin. Proteins were separated by SDS-PAGE according to standard procedures (Laemmli, 1970). Electroblot transfer was performed as in Towbin et al. (1979) with transfer buffer containing 20% methanol and in most cases including 0.04% SDS. For these experiments we used the Bio-Rad Mini PROTEAN II system, electroblotting to 0.2 μm nitrocellulose, and using anti-mouse or anti-rabbit HRP-conjugated secondary antibody (Bio-Rad) (1:3000) for visualization of primary antibody. A buffer containing: 0.9% NaCl, 100 mM Tris, pH 7.5, 0.1% Tween-20, 5% BSA was used for blocking procedures in experiments using the Pol IIoser2 mAb H5 as per the manufacturer's instructions (Covance). Antibody labeling was visualized using chemiluminescent detection methods (SuperSignal West Pico Chemiluminescent Substrate, Pierce). The immunoblots were digitized using a flatbed scanner (Epson Expression 1680).
Analysis of gene expression by qRT-PCR
Total RNA was extracted from 12 pooled whole third instar larvae of each genotype (wild type, JIL-1z2/JIL-1z2, and JIL-1z2/JIL-1z2, Su(var)3-906) with or without heat shock using the MicroPoly(A)Purist™ Small Scale mRNA Purification Kit (Ambion) following the manufacturer's instructions. cDNA derived from this RNA using SuperScript™ II Reverse Transcriptase (Invitrogen) was used as template for quantitative real-time PCR performed with the Stratagene M×4000 real-time cycler. In addition, the PCR mixture contained Brilliant II SYBR Green QPCR Master Mix (Stratagene) as well as the corresponding primers: rp49, 5′-AACGTTTACAAATGTGTATTCCGACC-3′ and 5′-ATGACCATCCGCCCAGCATACAGG-3′; hsp70, 5′-GTCATCACAGTTCCAGCCTACTTCAAC-3′ and 5′-CTGGGTTGATGGATAGGTTGAGGTTC-3′. Cycling parameters were 10 min at 95°C, followed by 40 cycles of 30 sec at 95°C, 60 sec at 59°C, and 40 sec at 72°C. Fluorescence intensities were plotted against the number of cycles using an algorithm provided by the manufacturer. mRNA levels were quantified using a calibration curve based on dilution of concentrated cDNA. mRNA values from the larvae were normalized to that of ribosomal protein 49 (Rp49).
Results
Histone H3S10ph antibody characterization and acid-free polytene squash labeling
The aim of this study was to examine the role of histone H3S10 phosphorylation in transcriptional regulation in Drosophila. However, many of the commercially available antibodies to this histone modification have been mostly used as a marker for mitotic chromosomes (Hendzel et al., 1997; Wei et al., 1998) and as a result they are poorly characterized with regards to detection of interphase histone H3S10 phosphorylation levels and distribution. Consequently, it is important to verify the specificity and suitability of these antibodies for experimental use at interphase to avoid artifacts. In this study we have therefore adapted the “smush” preparation of Drosophila third instar salivary gland nuclei as a rapid and sensitive screening procedure for such antibodies. The smush preparation is a modified whole-mount staining technique where nuclei from dissected salivary glands are gently compressed beneath a coverslip to flatten them before fixation in a standard paraformaldehyde/PBS solution with a physiological pH (Wang et al., 2001). The procedure furthermore takes advantage of the finding that JIL-1 kinase is the kinase responsible for interphase histone H3S10 phosphorylation (Wang et al., 2001) and that both JIL-1 and H3S10 phosphorylation are upregulated on the male X chromosome (Wang et al., 2001). Consequently, reliable histone H3S10ph antibodies would be expected to show co-localization with JIL-1, show upregulation on the male X chromosome, and all labeling should be absent in homozygous JIL-1 null nuclei as illustrated in Fig. 1. The upregulation of H3S10ph labeling on the male X chromosome is a particularly useful indicator of proper antibody recognition. Furthermore, on immunoblots of salivary gland protein extracts in JIL-1 null mutant backgrounds histone H3S10 labeling should be absent or greatly reduced as well. It should be noted that whole larval extracts are not suitable for such immunoblots because of the presence of a significant population of mitotic nuclei (>5%) where the H3S10 residue is phosphorylated by the Aurora B kinase (Giet and Glover, 2001). Using these criteria we screened various commercially available mono- and polyclonal antibodies from three different manufacturers. The results are summarized in Table 1. We found that several of the antibodies failed one or more of the above criteria and that different lots of the same antibody in some cases had different properties. Of the suitable antibodies the rabbit mAb from Epitomics (epi), the rabbit pAb (lot 2 and 3) from Cell Signaling (cs), and the rabbit pAb (lot 32219) from Upstate (up) were selected for further use in the present studies.
Fig. 1. JIL-1 and histone H3S10ph antibody labeling of salivary gland nuclei smush preparations.
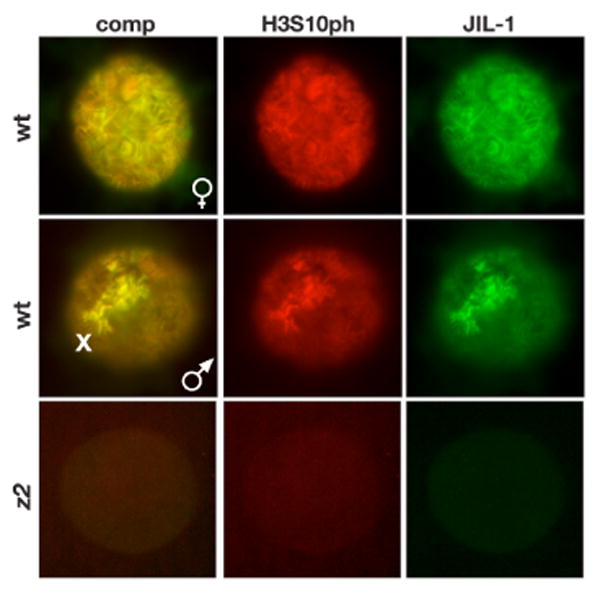
JIL-1 (in green) and H3S10ph (in red) co-localizes on chromosomes from both female (upper panel) and male (middle panel) wild type nuclei. The middle panel illustrates the characteristic upregulation of JIL-1 and H3S10ph labeling on the male X chromosome (X). Labeling of both JIL-1 and H3S10ph is absent in JIL-1z2/JIL-1z2 (z2) null mutant nuclei (lower panel). The H3S10ph antibody used was from Cell Signaling.
Table 1. Properties of histone H3S10ph antibodies.
| antibody | lot No. | immunoblot | smush | squash | note |
|---|---|---|---|---|---|
| Epitomics Rb mAb | C03143 | positive | positive | positive | centromere labeling |
| Epitomics Rb mAb | C022510 | positive | positive | positive | centromere labeling |
| Upstate Rb pAb | 23136 | positive | positive | positive | labels heat shock puffs |
| Upstate Rb pAb | 32219 | positive | positive | positive | labels heat shock puffs |
| Upstate Mo mAb | 26436 | negative | negative | ND | |
| Cell Signaling Rb pAb | lot 1 | negative | weakly positive | ND | |
| Cell Signaling Rb pAb | lot 2 | positive | positive | positive | |
| Cell Signaling Rb pAb | lot 3 | positive | positive | positive | |
| Cell Signaling Mo mAb | lot 5 | negative | weakly positive | ND |
A limitation of the smush procedure is that the visualization of chromatin structure and bands is inferior to the normal squash technique. However, as previously reported (Wang et al., 2001) the highly acidic fixation conditions of conventional squash protocols (Zink and Paro, 1989; Kelley et al., 1999) prevents reliable antibody labeling of the histone H3S10 phospho-epitope. In such preparations H3S10ph antibody labeling is extremely weak and except for rare cases the upregulation of H3S10 phosphorylation on the male X chromosome (Wang et al., 2001; data not shown) cannot be detected indicating incomplete or defective antibody recognition. In this study, to overcome these difficulties we have adopted the acid-free squash technique of DiMario et al. (2006) originally developed to preserve the fluorescence of GFP-tagged proteins in fixed preparations. As illustrated in Fig. 2A-C this technique also preserves the antigenicity of the H3S10 phospho-epitope as indicated by the robust antibody labeling in both male and female squash preparations by three different H3S10ph antibodies that includes the upregulation on the male X chromosome. The extensive co-localization of H3S10ph with JIL-1 is particularly evident in the confocal images in Fig. 2A. Furthermore, on immunoblots of protein extracts from third instar larval salivary glands H3S10ph labeling was greatly reduced in JIL-1 null mutant backgrounds confirming that the antibodies recognized the H3S10ph epitope. However, it should be noted that the Epitomics H3S10ph antibody in contrast to the other two antibodies showed strong labeling of the chromocenter (Fig. 2B, asterisks). Although, it was more difficult to properly spread the chromosomes and the chromatin structure as labeled by Hoechst was slightly less well-preserved in acid-free squashes compared to conventional squash preparations our data strongly suggest that the acid-free squash procedure is the method of choice in all antibody labeling studies of histone H3S10 phosphorylation in polytene squash preparations.
Fig. 2. Immunocytochemistry and immunoblot characterization of three different H3S10ph antibodies.
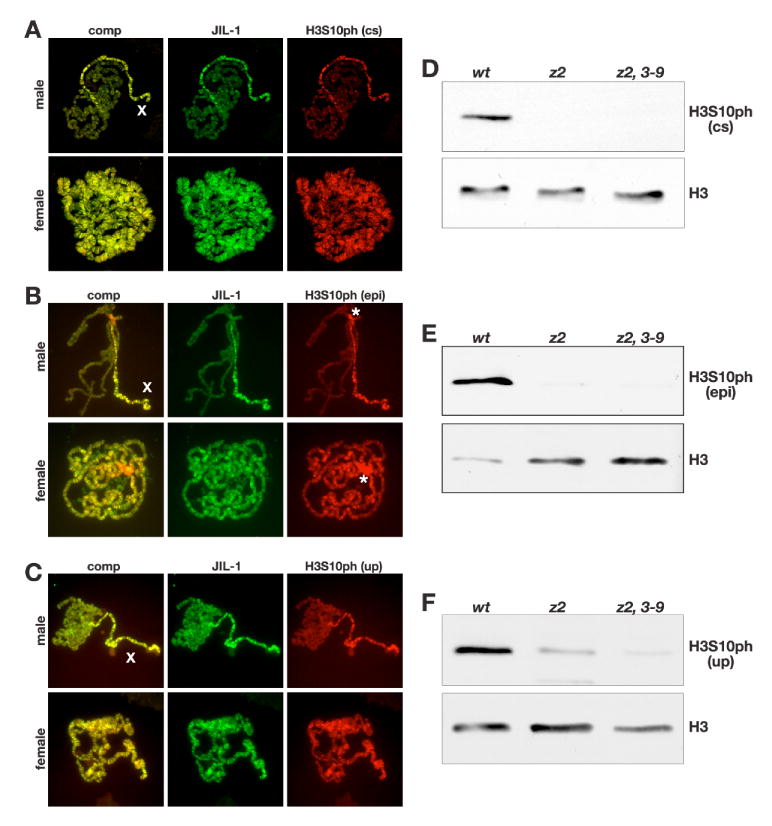
(A-C) Acid-free polytene chromosome squash preparations from male and female third instar larvae double labeled with antibodies to JIL-1 (in green) and H3S10ph (in red). H3S10ph labeling with antibody from Cell Signaling (cs) is shown in (A), from Epitomics (epi) in (B), and from Upstate (up) in (C). Composite images (comp) of the labelings are shown to the left. The labeling of all three H3S10ph antibodies shows co-localization with JIL-1 and upregulation on the male X chromosome (X). The Epitomics H3S10ph antibody in contrast to the two other antibodies showed strong labeling of the chromocenter (B, asterisks). The images in (A) are projection images from confocal sections. (D-F) Immunoblots of protein extracts from salivary glands from wild-type (wt), JIL-1z2/JIL-1z2 (z2), and JIL-1z2/JIL-1z2 Su(var)3-906 (z2, 3-9) larvae labeled with H3S10ph antibody from Cell Signaling (D), Epitomics (E), and Upstate (F). H3S10ph antibody labeling by all three antibodies is greatly reduced in JIL-1 null mutant backgrounds. Labeling with histone H3 (H3) antibody was used as a loading control.
JIL-1 and histone H3S10 phosphorylation are not upregulated at transcriptionally activated loci during heat shock
To determine the distribution of JIL-1 before and after heat shock we double labeled polytene chromosome squash preparations with the JIL-1 mAb 5C9 and with antibody to the elongating form of RNA polymerase II (Pol IIoser2), which is phosphorylated at serine 2 in the COOH-terminal domain and which serves as a marker for active transcription (Weeks et al., 1993; Boehm et al., 2003; Ivaldi et al., 2007). In non-heat shock preparations both JIL-1 and Pol IIoser2 were localized to a large number of euchromatic interband regions as illustrated in Fig. 3A (left panel). The composite image in Fig. 3A (left panel) further shows that while there may be some co-localization between JIL-1 and Pol IIoser2 as previously reported (Ivaldi et al., 2007), relatively low levels of JIL-1 were observed at many sites where there were especially high levels of Pol IIoser2 staining such as at developmental puffs. After 25 min of heat shock treatment there was a striking change in the distribution of Pol IIoser2 labeling whereas in contrast there was no appreciable redistribution of JIL-1. The Pol IIoser2 labeling was reduced at most sites while being upregulated at heat shock puffs where transcription of heat shock activated genes was occurring. This was especially prominent at the heat shock loci 87A/C and 93D as illustrated in Fig. 3B. Notably there were no indications of a concomitant upregulation of JIL-1 at these sites (Fig. 3B). This result was confirmed using two other JIl-1 antibodies, a chicken pAb as well as a rabbit pAb (data not shown).
Fig. 3. JIL-1 is not upregulated at actively transcribed regions during the heat shock response.
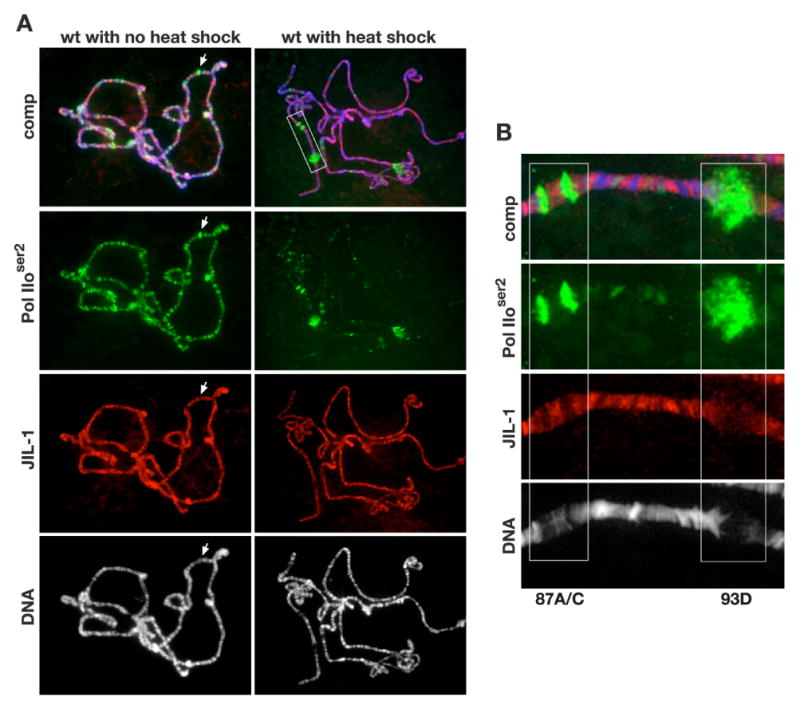
(A) Polytene chromosome squash preparations from wild type larvae (wt) triple labeled with Pol IIoser2 antibody (in green), JIL-1 mAb 5C9 (in red), and Hoechst (DNA, grey/blue) with (right panel) and without (left panel) heat shock treatment. At many sites that showed especially high levels of Pol IIoser2 staining such as at developmental puffs, there were relatively low levels of JIL-1 (arrow). After heat shock treatment (A, right panel) Pol IIoser2 labeling was reduced at most sites while being dramatically upregulated at heat shock induced puffs (A, right panel boxed area) while there was no discernable redistribution of JIL-1. (B) Higher magnification of the labeling of heat shock puffs 87A/C and 93D in the area indicated by a white box in (A, upper right panel) showing that there was no upregulation of JIL-1 at these heat shock puffs.
We next used the acid-free polytene squash technique to determine the distribution of H3S10ph before and after heat shock using three different H3S10ph antibodies (i.e., Epitomics, Cell Signaling, and Upstate). To mark heat shock puffs and other regions of enhanced transcription the preparations were double labeled with antibody to Pol IIoser2. As illustrated in Fig. 4A-D there was no obvious change in H3S10ph distribution before and after heat shock as detected by the Epitomics and Cell Signaling H3S10ph antibodies. Importantly, as also observed for JIL-1 (Fig. 3B) there was no upregulation at the 87A/C heat shock puffs although they were robustly labeled by the Pol IIoser2 antibody (Fig. 4B and D). In contrast, we found that the Upstate H3S10ph antibody strongly labeled the 87A/C puffs after heat shock (Fig. 4G, F). However, contrary to the Cell Signaling and Epitomics H3S10ph antibodies the Upstate pAb also labeled heat shock puffs in polytene chromosome squashes from JIL-1 null mutant larvae that are devoid of histone H3S10ph phosphorylation (Fig. 5). Since similar results were obtained with two different lots of the Upstate H3S10ph pAb (Table 1) we conclude that the labeling of heat shock puffs by this antibody is due to non-specific cross-reactivity possibly with proteins involved in the heat shock response. Furthermore, immunoblot analysis with all three H3S10ph antibodies of protein extracts from salivary glands before and after heat shock confirmed that there was no change in the overall level of histone H3S10 phosphorylation (Fig. 4G) as indicated by the polytene squash labelings (Fig. 4A, C, and E). In contrast, there was a clear down regulation in the levels of Pol IIoser2 in response to heat shock (Fig. 4H).
Fig. 4. Polytene chromosome distribution of H3S10ph in response to heat shock treatment.
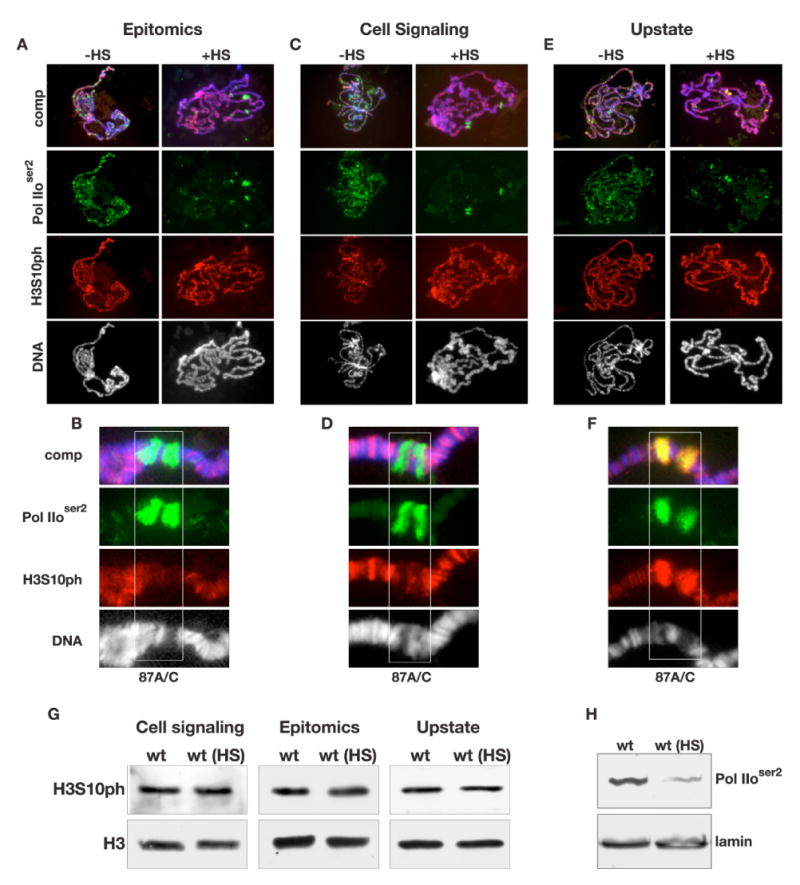
(A-F) Acid-free polytene chromosome squash preparations from female third instar larvae triple labeled with antibodies to Pol IIoser2 (in green), H3S10ph (in red), and with Hoechst (DNA, blue/grey). H3S10ph labeling with antibodies from Epitomics is shown in (A), from Cell signaling in (C), and from Upstate in (E) with (+HS) and without (-HS) heat shock treatment. (B, D, and F) Higher magnification images of the heat shock induced puffs 87A/C (boxed regions) labeled by the H3S10ph antibodies from Epitomics (B), Cell Signaling (D), and Upstate (F) respectively. (G) Immunoblots of protein extracts from salivary glands from wild-type larvae without heat shock treatment (wt) and with heat shock treatment (wt (HS)) labeled with H3S10ph antibody from Cell Signaling (D), Epitomics (E), and Upstate (F). Labeling with histone H3 (H3) antibody was used as a loading control. (H) Immunoblots of protein extracts from salivary glands from wild-type larvae without heat shock treatment (wt) and with heat shock treatment (wt (HS)) labeled with Pol IIoser2 antibody. Labeling with lamin antibody was used as a loading control.
Fig. 5. Polytene chromosome labeling by three different H3S10ph antibodies in response to heat shock treatment in JIL-1 null mutants.
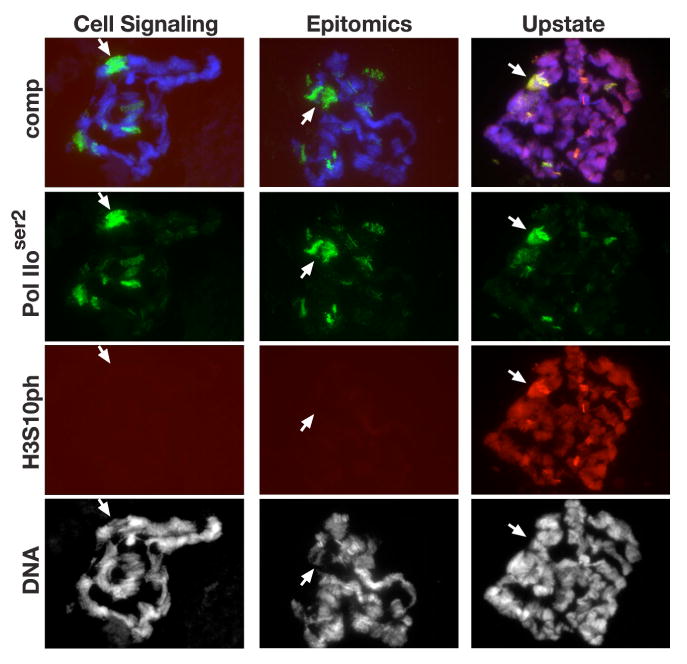
Acid-free polytene chromosome squash preparations from female JIL-1z2/JIL-1z2 third instar larvae triple labeled with antibodies to Pol IIoser2 (in green), H3S10ph (in red), and with Hoechst (DNA, blue/grey) after heat shock treatment. H3S10ph labeling by antibodies from Cell Signaling, Epitomics, and Upstate are shown. Arrows indicate the likely position of the 87A/C heat shock puff regions.
Previously, evidence has been presented that protein phosphatase 2A (PP2A) activity may regulate histone H3S10 phosphorylation at interphase (Nowak et al., 2003). Using a P-element insertion mutation into the regulating subunit of PP2A, twsP, that causes reduced catalytic activity (Mayer-Jaekel et al., 1993), Nowak et al. (2003) showed that on immunoblots of extracts from twsP mutant larvae there is a higher level of H3S10 phosphorylation than in wild-type larvae. This difference was attributed to reduced PP2A phosphatase activity indicating that PP2A may function as a H3S10ph phosphatase at interphase (Nowak et al., 2003) in addition to its role as a mitotic H3S10ph phosphatase (Mayer-Jaekel et al., 1993). However, since whole larval extracts were used it remained a possibility that the increased upregulation of H3S10ph levels was due solely to decreased dephosphorylation of mitotic H3S10ph. Using a likely null PP2A regulatory subunit P-element mutation, tws02414, we confirmed a higher level of H3S10 phosphorylation in extracts from homozygous tws02414 mutant larvae as compared to wild-type larvae (Fig. 6A). However, when extracts were compared from salivary glands which do not contain mitotic cells there was no difference (Fig. 6B). Furthermore, in extracts from salivary glands of homozygous twsP mutant larvae with or without heat shock there was no difference in H3S10 phosphorylation levels as detected by the Epitomics H3S10ph mAb (Fig. 6C). Taken together these results indicate that the PP2A phosphatase may play a role in H3S10 dephosphorylation only at mitosis and not at interphase.
Fig. 6. Immunoblot analysis of histone H3S10 phosphorylation in mutants of the 55 kd regulatory subunit of protein phosphatase 2A.
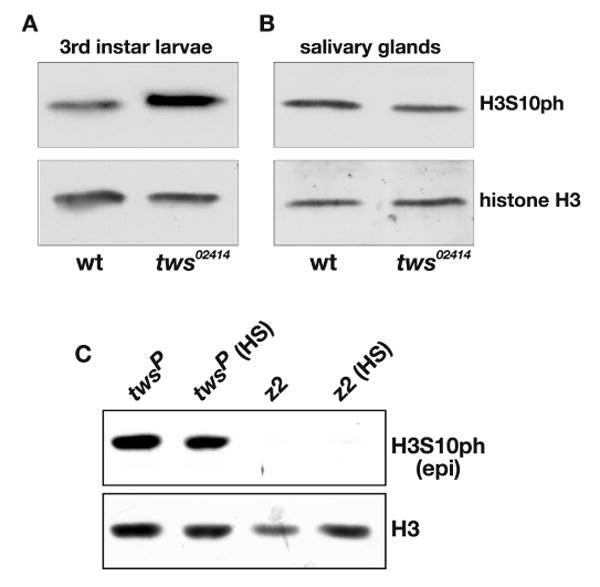
(A) H3S10ph antibody labeling of protein extracts from wild type (wt) and tws02414/tws02414 (tws02414) whole third instar larvae. (B) H3S10ph antibody labeling of protein extracts from wild type (wt) and tws02414/tws02414 (tws02414) salivary glands. (C) H3S10ph antibody labeling of protein extracts from twsP/twsP (twsP) and JIL-1z2/JIL-1z2 (z2) salivary glands with (HS) and without heat shock treatment. The H3S10ph antibody was from Epitomics (epi) and labeling with histone H3 (H3) antibody was used as a loading control.
JIL-1 and H3S10 phosphorylation are not required for transcription at active loci during heat shock
Deng et al. (2007) have recently provided evidence that the lethality as well as some of the chromosome defects associated with the JIL-1 null phenotype can be substantially rescued by reducing the dose of the Su(var)3-9 gene. This suggests that the Pol II transcriptional machinery has the capacity to function more or less normally in the complete absence of JIL-1 mediated interphase histone H3S10 phosphorylation. We therefore investigated the distribution of Pol IIoser2 labeling and heat shock induced transcription in both JIL-1z2/JIL-1z2 null as well as in JIL-1z2/JIL-1z2 Su(var)3-906 double mutant backgrounds. In JIL-1z2/JIL-1z2 Su(var)3-906 larvae the adult eclosion rate increases to 60% that of wild type larvae as compared to 0% for JIL-1z2/JIL-1z2 null larvae (Deng et al., 2007). Fig. 7A shows robust antibody labeling of Pol IIoser2 in polytene chromosome squashes from both genotypes even though the chromatin structure was greatly perturbed. This included the characteristic “puffed” male X chromosome in JIL-1 null larvae (Fig. 7A, upper panel). Furthermore, on immunoblots of extracts from wild-type, JIL-1z2/JIL-1z2, and JIL-1z2/JIL-1z2 Su(var)3-906 salivary glands there was no detectable difference in Pol IIoser2 levels (Fig. 7B). This indicates that transcript elongation by the Pol II machinery is likely to be functional in JIL-1 null mutant backgrounds. To further investigate this possibility we double labeled JIL-1 mutant polytene chromosome squashes after they were heat shock treated with Pol IIoser2 antibody and with antibody to the heat shock transcription factor HSF (Fig. 8A). When inactive, HSF is diffusely distributed at very low levels; however, following heat shock HSF redistributes very prominently to heat shock induced puffs (Westwood et al., 1991; Ivaldi et al., 2007). As shown in Fig. 8A although the chromosome morphology is greatly disrupted, puffed regions can be clearly identified in JIL-1 null mutant backgrounds as defined by the presence of decondensed chromatin and strong HSF antibody labeling. Importantly, these heat shock induced puffs are also strongly labeled by Pol IIoser2 antibody in an overlapping pattern with that of the HSF antibody (Fig. 8A, arrows). Furthermore, as also confirmed by immunoblot analysis (Fig. 8B), Pol IIoser2 levels are greatly reduced at non-heat shock sites (Fig. 8A). This suggests that Pol IIoser2 is actively involved in heat shock induced transcription in JIL-1 null mutants. To test this directly we used quantitative RT-PCR to measure the transcription of the six nearly identical genes that encode Hsp70, the major heat shock protein in Drosophila (Gong and Golic, 2004) under heat shock and non-heat shock conditions. Primers were designed that would amplify transcripts from all six hsp70 genes whereas primers specific to the gene encoding the ribosomal non-heat shock sensitive protein Rp49 was used for normalization. We performed two independent experiments where total RNA was isolated from wild-type, JIL-1z2/JIL-1z2, and JIL-1z2/JIL-1z2 Su(var)3-906 third instar larvae and where qRT-PCR determination of transcript levels was performed in duplicate. As illustrated in Fig. 8C, very low levels of hsp70 mRNA transcripts in both wild type and JIL-1 mutant backgrounds were detected under non-heat shock conditions. However, a robust increase in hsp70 mRNA transcript levels was detected in response to heat shock treatment in all three genotypes relative to rp49 transcript levels (Fig. 8C). The increase in JIL-1z2/JIL-1z2 null mutant larvae was at least two orders of magnitude larger than under non-heat shock conditions and was about 1/3 that observed in wild type larvae. Interestingly, the heat shock-induced increase in hsp70 mRNA levels improved considerably to almost 2/3 of wild type levels in larvae where Su(var)3-9 levels were reduced by half (e.g. JIL-1z2/JIL-1z2 Su(var)3-906 larvae).
Fig. 7. Immunocytochemical and immunoblot labeling of Pol IIoser2 in JIL-1 null mutant backgrounds.
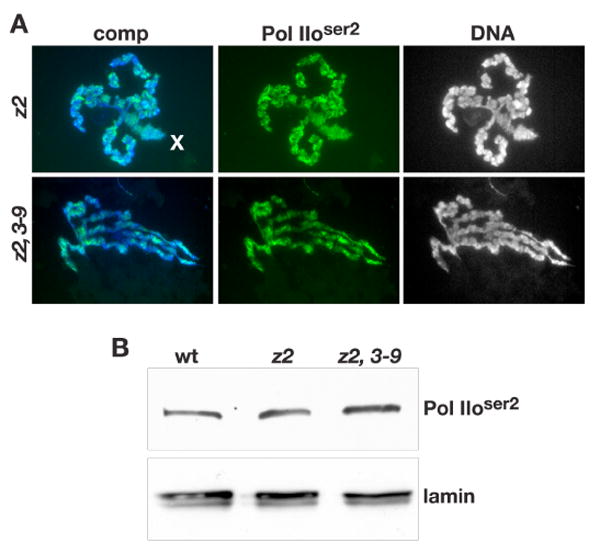
(A) Robust Pol IIoser2 antibody labeling (in green) of chromosomes including the male X (X) chromosome in polytene chromosome squash preparations from JIL-1z2/JIL-1z2 (z2), and JIL-1z2/JIL-1z2 Su(var)3-906 (z2, 3-9) third instar larvae. The DNA (blue/grey) was labeled by Hoechst. (B) Immunoblot of protein extracts from wild-type (wt), JIL-1z2/JIL-1z2 (z2), and JIL-1z2/JIL-1z2 Su(var)3-906 (z2, 3-9) larvae labeled with Pol IIoser2 antibody. Labeling with lamin antibody was used as a loading control.
Fig. 8. Analysis of Pol IIoser2 distribution and transcription at active loci during the heat shock response in JIL-1 mutant backgrounds.
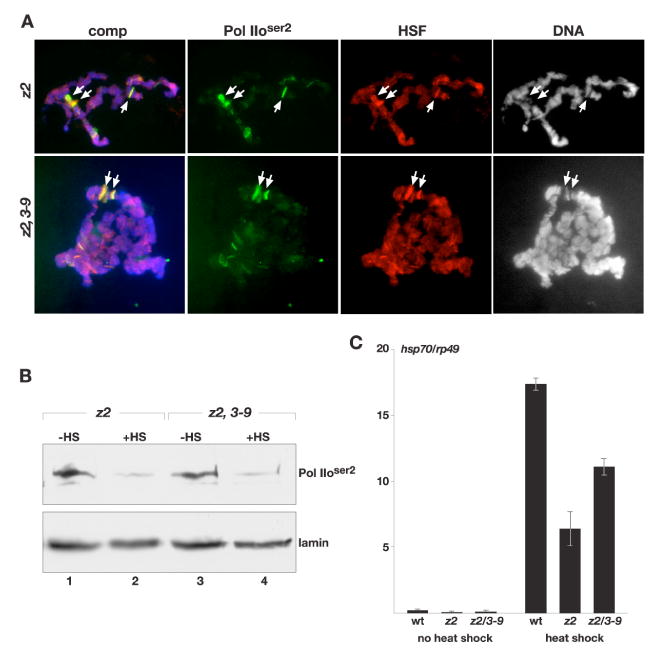
(A) Polytene chromosome squash preparations from JIL-1z2/JIL-1z2 (z2), and JIL-1z2/JIL-1z2 Su(var)3-906 (z2, 3-9) larvae triple labeled with Pol IIoser2 antibody (in green), HSF antibody (in red), and Hoechst (DNA, grey/blue) after heat shock treatment. Arrows point to heat shock puff regions. (B) Immunoblot of protein extracts from salivary glands from JIL-1z2/JIL-1z2 (z2) and JIL-1z2/JIL-1z2 Su(var)3-906 (z2, 3-9) larvae with (+HS) and without (-HS) heat shock treatment labeled with Pol IIoser2 antibody. Labeling with lamin antibody was used as a loading control. (C) Transcript levels of hsp70 mRNA in JIL-1 mutant null backgrounds in response to heat shock treatment. hsp70 transcript levels were determined by qRT-PCR and normalized to the mRNA levels of the control non-heat shock protein Rp49 (ribosomal protein 49) both without and after heat shock treatment. The data shown are the average from two independent experiments where total RNA was isolated from wild-type (wt), JIL-1z2/JIL-1z2 (z2), and JIL-1z2/JIL-1z2 Su(var)3-906 (z2, 3-9) larvae and where each determination of transcript levels was performed in duplicate. The error bars indicate the SDM.
Discussion
A number of studies have suggested that regulation of early stages of transcriptional elongation may be a relatively common phenomenon in higher eukaryotes (reviewed in Hartzog and Tamkun, 2007) and that histone H3S10 phosphorylation may play an important role in specific transcriptional responses to signaling stimuli (Mahadewan et al., 1991; Lo et al., 2001; Ivaldi et al., 2007). In this study we characterized three commercially available histone H3S10ph antibodies and used an acid-free squash protocol to revisit the role of histone H3S10 phosphorylation in transcription in Drosophila under both heat shock and non-heat shock conditions. We show that there is no change in the levels of the elongating form of RNA polymerase II in larvae from JIL-1 null mutant backgrounds compared to wild-type. Furthermore, we provide evidence that heat shock induced puffs in JIL-1 null mutant backgrounds are strongly labeled by Pol IIoser2 antibody in an overlapping pattern with that of HSF antibody indicating that Pol IIoser2 is actively involved in heat shock induced transcription in the absence of H3S10 phosphorylation. That mRNA from the six genes that encode the major heat shock protein Hsp70 in Drosophila is transcribed at robust levels in JIL-1 null mutants was directly demonstrated by qRT-PCR. Thus these data strongly suggest that histone H3S10 phosphorylation by JIL-1 is not involved in transcriptional elongation in Drosophila. The finding that there is no redistribution of JIL-1 or H3S10 phosphorylation to transcriptionally active puffs in wild-type polytene squash preparations during the heat shock response further supports this conclusion. These results are contrary to those reported by Nowak and Corces (2000), Nowak et al. (2003), and Ivaldi et al. (2007). However, the discrepancies may largely be due to the reliance in these studies on the Upstate H3S10ph pAb which our study indicates is unsuitable for analysis of heat shock induced transcription due to non-specific cross-reactivity at heat shock induced puffs. Nonetheless, it should be noted that the present study confirms the findings of Ivaldi et al. (2007) that the chromatin remodeling associated with heat shock puff formation still occurs in JIL-1 null mutants despite the disruption of chromatin structure and the absence of H3S10 phosphorylation.
Previous studies indicated that the lethality of JIL-1 null mutants may be due to ectopic Su(var)3-9 activity and the disruption of chromatin structure (Zhang et al., 2006; Deng et al., 2007). At interphase JIL-1 phosphorylates the histone H3S10 residue in euchromatic regions of polytene chromosomes (Jin et al., 1999; Wang et al., 2001) suggesting as a plausible model that this phosphorylation during interphase prevents Su(var)3-9 mediated heterochromatization and gene repression at these sites (Zhang et al., 2006; Deng et al., 2007). However, the present results clearly indicate that such repression is not global and that the Pol II machinery is functional as indicated by the robust transcription of heat shock induced genes in JIL-1 null mutants. Furthermore, while dosage compensation of the white locus in males is impaired in hypomorphic JIL-1 mutant backgrounds, white expression in females is relatively unaffected (Lerach et al., 2005). Thus, the lethality may be caused instead by a severe repression of a few essential genes and/or a more graded decrease in expression of a larger number of genes due to the altered chromatin structure. The latter scenario is supported by the finding that the expression of heat shock induced genes in JIL-1 null mutant backgrounds increases when chromatin structure is improved by reducing the levels of the heterochromatic factor Su(var)3-9 (Deng et al., 2007; this study). That JIL-1 levels can directly affect gene expression was recently demonstrated by experiments that showed that loss-of-function JIL-1 alleles act as enhancers of position-effect-variegation (PEV) whereas the gain-of-function JIL-1Su(var)3-1 allele acts as a suppressor of PEV at pericentric sites (Bao et al., 2007). The JIL-1Su(var)3-1 allele is one of the strongest suppressors of PEV so far described (Ebert et al., 2004) and it generates truncated proteins with COOH-terminal deletions that mislocalize to ectopic chromosome sites (Ebert et al., 2004; Zhang et al., 2006). Thus, the dominant gain-of-function effect of the JIL-1Su(var)3-1 alleles may be attributable to JIL-1 kinase activity at ectopic locations leading to misregulated localization of the phosphorylated histone H3S10 mark counteracting the spreading and gene repression of Su(var)3-9. This is supported by the finding of Deng et al. (2008) that ectopic H3S10 phosphorylation at interphase can function as a causative regulator of higher-order chromatin structure in vivo. Furthermore, studies of PEV of the wm4 allele have indicated that loss of JIL-1 function also can cause a change in the levels of heterochromatic factors at the chromocenter that indirectly can affect gene expression at nearby loci (Lerach et al., 2006; reviewed in Girton and Johansen, 2008). In future experiments it will be of interest to further explore a possible mechanistic link between H3S10 phosphorylation and the regulation of chromatin structure and gene expression in Drosophila.
Acknowledgments
We thank members of the laboratory for discussion, advice, and critical reading of the manuscript. We also wish to acknowledge Ms. V. Lephart for maintenance of fly stocks and Mr. Laurence Woodruff for technical assistance. We especially thank Dr. C. Wu for providing the HSF antibody and Dr. D.M. Glover for the twsP allele. This work was supported by National Institutes of Health grant GM62916 to K.M.J.
References
- Bao X, Deng H, Johansen J, Girton J, Johansen KM. Loss-of-function alleles of the JIL-1 histone H3S10 kinase enhance position-effect-variegation at pericentric sites in Drosophila heterochromatin. Genetics. 2007;176:1355–1358. doi: 10.1534/genetics.107.073676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm AK, Saunders A, Werner J, Lis JT. Transcription factor and polymerase recruitment, modification, and movement on dhsp70 in vivo in the minutes following heat shock. Mol Cell Biol. 2003;23:7628–7637. doi: 10.1128/MCB.23.21.7628-7637.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Zhang W, Bao X, Martin JN, Girton J, Johansen J, Johansen KM. The JIL-1 kinase regulates the structure of Drosophila polytene chromosomes. Chromosoma. 2005;114:173–182. doi: 10.1007/s00412-005-0006-8. [DOI] [PubMed] [Google Scholar]
- Deng H, Bao X, Zhang W, Girton J, Johansen J, Johansen KM. Reduced levels of Su(var)3-9 but not Su(var)2-5 (HP1) counteract the effects on chromatin structure and viability in loss-of-function mutants of the JIL-1 histone H3S10 kinase. Genetics. 2007;177:79–87. doi: 10.1534/genetics.107.075143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H, Bao X, Cai W, Blacketer MJ, Belmont AS, Girton J, Johansen J, Johansen KM. Ectopic histone H3S10 phosphorylation causes chromatin structure remodeling in Drosophila. Development. 2008;135:699–705. doi: 10.1242/dev.015362. [DOI] [PubMed] [Google Scholar]
- DiMario P, Rosby R, Cui Z. Direct visualization of GFP-fusion proteins on polytene chromosomes. Dros Inf Serv. 2006;89:115–118. [Google Scholar]
- Ebert A, Schotta G, Lein S, Kubicek S, Krauss V, Jenuwein T, Reuter G. Su(var) genes regulate the balance between euchromatin and heterochromatin in Drosophila. Genes Dev. 2004;18:2973–2983. doi: 10.1101/gad.323004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elgin SCR, Reuter G. Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila. In: Allis CD, Jenuwein T, Reinberg D, Caparros ML, editors. Epigenetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. pp. 81–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giet R, Glover DM. Drosophila Aurora B kinase is required for histone H3 phosphorylation and condensin recruitment during chromosome condensation and to organize the central spindle during cytokinesis. J Cell Biol. 2001;152:669–681. doi: 10.1083/jcb.152.4.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girton JR, Johansen KM. Chromatin structure and the regulation of gene expression: the lessons of PEV in Drosophila. Adv Genet. 2008;61:1–43. doi: 10.1016/S0065-2660(07)00001-6. [DOI] [PubMed] [Google Scholar]
- Gong WJ, Golic KG. Genomic deletions of the Drosophila melanogaster Hsp70 genes. Genetics. 2004;168:1467–1476. doi: 10.1534/genetics.104.030874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruenbaum Y, Landesman Y, Drees B, Bare JW, Saumweber H, Paddy MR, Sedat JW, Smith DE, Benton BM, Fisher PA. Drosophila nuclear lamin precursor Dm0 is translated from either of two developmentally regulated mRNA species apparently encoded by a single gene. J Cell Biol. 1988;106:585–596. doi: 10.1083/jcb.106.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartzog GA, Tamkun JW. A new role for histone tail modifications in transcription elongation. Genes Dev. 2007;23:3209–3213. doi: 10.1101/gad.1628707. [DOI] [PubMed] [Google Scholar]
- Hendzel MJ, Wei Y, Mancini MA, Van Hooser A, Ranalli T, Brinkley BR, Bazett-Jones DP, Allis CD. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma. 1997;106:348–360. doi: 10.1007/s004120050256. [DOI] [PubMed] [Google Scholar]
- Ivaldi MS, Karam CS, Corces VG. Phosphorylation of histone H3 at Ser10 facilitates RNA polymerase II release from promoter-proximal pausing in Drosophila. Genes Dev. 2007;21:2818–2831. doi: 10.1101/gad.1604007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Y, Wang Y, Walker DL, Dong H, Conley C, Johansen J, Johansen KM. JIL-1: a novel chromosomal tandem kinase implicated in transcriptional regulation in Drosophila. Mol Cell. 1999;4:129–135. doi: 10.1016/s1097-2765(00)80195-1. [DOI] [PubMed] [Google Scholar]
- Jin Y, Wang Y, Johansen J, Johansen KM. JIL-1, a chromosomal kinase implicated in regulation of chromatin structure, associates with the MSL dosage compensation complex. J Cell Biol. 2000;149:1005–1010. doi: 10.1083/jcb.149.5.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley RL, Meller VH, Gordadze PR, Roman G, Davis RL, Kuroda MI. Epigenetic spreading of the Drosophila dosage compensation complex from roX RNA genes into flanking chromatin. Cell. 1999;98:513–522. doi: 10.1016/s0092-8674(00)81979-0. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lerach S, Zhang W, Deng H, Bao X, Girton J, Johansen J, Johansen KM. JIL-1 kinase, a member of the male-specific lethal (MSL) complex, is necessary for proper dosage compensation of eye pigmentation in Drosophila. Genesis. 2005;43:213–215. doi: 10.1002/gene.20172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerach S, Zhang W, Bao X, Deng H, Girton J, Johansen J, Johansen KM. Loss-of-function alleles of the JIL-1 kinase are strong suppressors of position effect variegation of the wm4 allele in Drosophila. Genetics. 2006;173:2403–2406. doi: 10.1534/genetics.106.059253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsley DL, Zimm GG. The genome of Drosophila melanogaster. Academic Press; New York, NY: 1992. [Google Scholar]
- Lo WS, Duggan L, Emre NC, Belotserkovskya R, Lane WS, Shiekhattar R, Berger SL. Snf1 - A histone kinase that works in concert with the histone acetyltransferase Gcn5 to regulate transcription. Science. 2001;293:1142–1146. doi: 10.1126/science.1062322. [DOI] [PubMed] [Google Scholar]
- Mahadevan LC, Willis AC, Barratt MJ. Rapid histone H3 phosphorylation in response to growth factors, phorbol esters, okadaic acid, and protein synthesis inhibitors. Cell. 1991;65:775–783. doi: 10.1016/0092-8674(91)90385-c. [DOI] [PubMed] [Google Scholar]
- Mayer-Jaekel RE, Ohkura H, Gomes R, Sunkel CE, Baumgartner S, Hemmings BA, Glover DM. The 55 kd regulatory subunit of Drosophila protein phosphatase 2A is required for anaphase. Cell. 1993;72:621–633. doi: 10.1016/0092-8674(93)90080-a. [DOI] [PubMed] [Google Scholar]
- Nowak SJ, Corces VG. Phosphorylation of histone H3 correlates with transcriptionally active loci. Genes Dev. 2000;14:3003–3013. doi: 10.1101/gad.848800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak SJ, Pai CY, Corces VG. Protein phosphatase 2A activity affects histone H3 phosphorylation and transcription in Drosophila melanogaster. Mol Cell Biol. 2003;23:6129–6138. doi: 10.1128/MCB.23.17.6129-6138.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts DB. Drosophila: A Practical Approach. IRL Press; Oxford, UK: 1998. [Google Scholar]
- Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, Rea S, Jenuwein T, Dorn R, Reuter G. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhang W, Jin Y, Johansen J, Johansen KM. The JIL-1 tandem kinase mediates histone H3 phosphorylation and is required for maintenance of chromatin structure in Drosophila. Cell. 2001;105:433–443. doi: 10.1016/s0092-8674(01)00325-7. [DOI] [PubMed] [Google Scholar]
- Weeks JR, Hardin SE, Shen J, Lee JM, Greenleaf AL. Locus-specific variation in phoshorylation state of RNA polymerase II in vivo: correlations with gene activity and transcript processing. Genes Dev. 1993;7:2329–2344. doi: 10.1101/gad.7.12a.2329. [DOI] [PubMed] [Google Scholar]
- Wei Y, Mizzen CA, Cook RG, Gorovsky MA, Allis CD. Phosphorylation of histone H3 at serine 10 is correlated with chromosome condensation during mitosis and meiosis in Tetrahymena. Proc Natl Acad Sci USA. 1998;95:7480–7484. doi: 10.1073/pnas.95.13.7480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler KS, Wakimoto BT. Heterochromatin and gene expression in Drosophila. Annu Rev Genet. 1995;29:577–605. doi: 10.1146/annurev.ge.29.120195.003045. [DOI] [PubMed] [Google Scholar]
- Westwood JT, Clos J, Wu C. Stress-induced oligomerization and chromosomal relocalization of heat-shock factor. Nature. 1991;353:822–827. doi: 10.1038/353822a0. [DOI] [PubMed] [Google Scholar]
- Zhang W, Jin Y, Ji Y, Girton J, Johansen J, Johansen KM. Genetic and phenotypic analysis of alleles of the Drosophila chromosomal JIL-1 kinase reveals a functional requirement at multiple developmental stages. Genetics. 2003;165:1341–1354. doi: 10.1093/genetics/165.3.1341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Deng H, Bao X, Lerach S, Girton J, Johansen J, Johansen KM. The JIL-1 histone H3S10 kinase regulates dimethyl H3K9 modifications and heterochromatic spreading in Drosophila. Development. 2006;133:229–235. doi: 10.1242/dev.02199. [DOI] [PubMed] [Google Scholar]
- Zink B, Paro R. In vivo binding pattern of a trans-regulator of homoeotic genes in Drosophila melanogaster. Nature. 1989;337:468–471. doi: 10.1038/337468a0. [DOI] [PubMed] [Google Scholar]


