Abstract
Although GAPDH’s predilection for AU-rich elements has long been known, the expected connection between GAPDH and control of mRNA stability has never been made. Recently, we described GAPDH binding the AU-rich terminal 144nt of the CSF-1 3′UTR, which we showed is an mRNA decay element in ovarian cancer cells. CSF-1 is strongly correlated with poor prognosis of ovarian cancer patients. We investigated the functional significance of GAPDH’s association with CSF-1 mRNA, and found that GAPDH siRNA reduces both CSF-1 mRNA and protein levels, through destabilizing CSF-1 mRNA. CSF-1 mRNA half-lives were decreased by 50% in the presence of GAPDH siRNA. RNA footprinting analysis of the 144nt CSF-1 sequence revealed that GAPDH associates with a large AU-rich containing region. The effects of binding of GAPDH protein or ovarian extracts to mutations of the AU-rich regions within the footprint were consistent with this finding. In a tissue array containing 256 ovarian and fallopian tube cancer specimens, we found that GAPDH was regulated in these cancers, with almost 50% of specimens having no GAPDH staining. Further, we found that low GAPDH staining was associated with a low CSF-1 score (p=.008). In summary, GAPDH, a multifunctional protein, now adds regulation of mRNA stability to its repertoire. We are the first to evaluate the clinical role of GAPDH protein in cancer. In ovarian cancers, we show that GAPDH expression is regulated, and we now recognize one of the many functions of GAPDH is to promote mRNA stability of CSF-1, an important cytokine in tumor progression.
Keywords: ovarian cancer, CSF-1, GAPDH, RNA stability, tissue microarray
Introduction
While glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was originally thought of as an enzyme important only to glycolysis, and as a housekeeping gene appropriate for use as a control for the measurement of equal loading in experiments, over the last decade, every succeeding year has brought to light a new function for this protein. The many and diverse roles for this multifunctional protein (1,2) include among others: DNA replication and repair, nuclear membrane fusion, apoptosis, microtubule bundling, vesicular secretory transport, and maintenance of telomere structure, with its roles in part related to subcellular localization. GAPDH has been found in nuclear, cytoplasmic, and membrane localizations, and can shuttle between these compartments.
Importantly, GAPDH has also been implicated in having a role in transcriptional control (1), in export of nuclear tRNA (2), and in translational control (3), levels of control which regulate gene expression. GAPDH has not been previously shown to have a role in the regulation of mRNA stability, despite the fact that is has been known to have AU-rich RNA binding capabilities for over a decade. GAPDH has a predilection for regions of RNA critical for its regulation (2), such as the 5′ sequences involved in hepatitis A viral replication, and 3′ untranslated region (UTR) sequences including those containing AU-rich elements. Some RNA sequences which are recognized by GAPDH have been described to have a poly U tract (4), UC-rich region (5), or AU-rich elements (6). RNA footprinting of the GAPDH binding site has shown in hepatitis delta virus RNA, a UC-rich domain (5), and in hepatitis A virus RNA, three discontinuous AU-rich regions, one located in the 3′ translated and two in the 3′UTR (7). Computer modeling suggested that these regions were located on stem-loop structures, having the potential for tertiary interactions.
Recently, we described GAPDH to be an RNA binding protein for the AU-rich terminal 144nt of the 3′UTR of the macrophage colony stimulating factor (CSF-1) (8). CSF-1 and/or its receptor (encoded by the c-fms proto-oncogene) are expressed by the large majority of epithelial ovarian cancers (9–11), with 75% of primary tumors and 69% of the metastases expressing CSF-1, and 92% of primary tumors and 83% of metastases expressing its receptor. We further found that strong co-expression of CSF-1 and receptor by ovarian cancer metastases was found to be an independent (p=.007) poor prognostic factor, with an increased relative risk of recurrence of 2.3 fold (9). The mean time to recurrence for stage III ovarian cancer patients was shortened by 11 months, from 24.1 +/−3.9 months to 13.5 +/−4.0 months. No such co-expression of CSF-1 and its receptor was observed in any of the tumors of low malignant potential, which by definition are non-invasive. Moreover, serum CSF-1 levels proved to be a sensitive tumor marker (12), with elevated levels heralding disease recurrence or progression. Elevated levels of both serum and ascitic CSF-1 levels at diagnosis correlated with a poor outcome (13,14). This correlation with prognosis and aggressive tumor behavior suggests an etiological role for CSF-1 in ovarian cancer progression.
In our search for novel regulators of CSF-1 expression, we had noted that the terminal 144nt of the 3′ UTR in exon 10, contained in the most abundant CSF-1 transcript, was particularly AU-rich. We showed that this 144nt region served as an mRNA decay element, and identified GAPDH as an AU-rich RNA binding protein which binds with high affinity to this 144nt stretch (8). While we provided evidence by northwestern analysis that a mutant riboprobe containing sequences that differed from the wild-type 144nt sequence in most, but not all, AU-rich stretches, did not bind the 37kDa protein which we subsequently identified to be GAPDH, the region or regions within the 144nt CSF-1 RNA sequence which were recognized by GAPDH remain unknown, and the functional consequence of such binding not delineated.
Elevated levels of GAPDH RNA have been demonstrated in several human cancers, including lung, pancreas, cervical, breast cancer (as summarized in 15,16), and of GAPDH protein in prostate cancer (17). Its role in prognosis was previously investigated only in breast cancer, where the level of GAPDH RNA from breast cancer specimens correlated with decreased survival on univariate analysis. To our knowledge, the clinical role of GAPDH protein has not been explored in any cancer. We did so by immunohistochemical analysis of GAPDH expression in 256 invasive ovarian and fallopian tube cancer specimens contained in a tissue array.
In this report, we demonstrated by RNA interference that GAPDH regulates CSF-1 RNA and protein expression, which we found to be on the basis of stabilization of CSF-1 mRNA. Further, CSF-1 RNA footprinting of its 144nt 3′UTR suggested that binding by GAPDH involves a large AU-rich region, which contains several stem-loop structures predicted by computer modeling. We showed that mutating or deleting targeted areas within this region mitigates GAPDH binding. Lastly, we found that GAPDH is regulated in ovarian and fallopian tube cancers, and that GAPDH is co-expressed with CSF-1 in ovarian cancers.
Results
Down-regulation of GAPDH by RNA interference reduces steady-state CSF-1 mRNA and protein levels
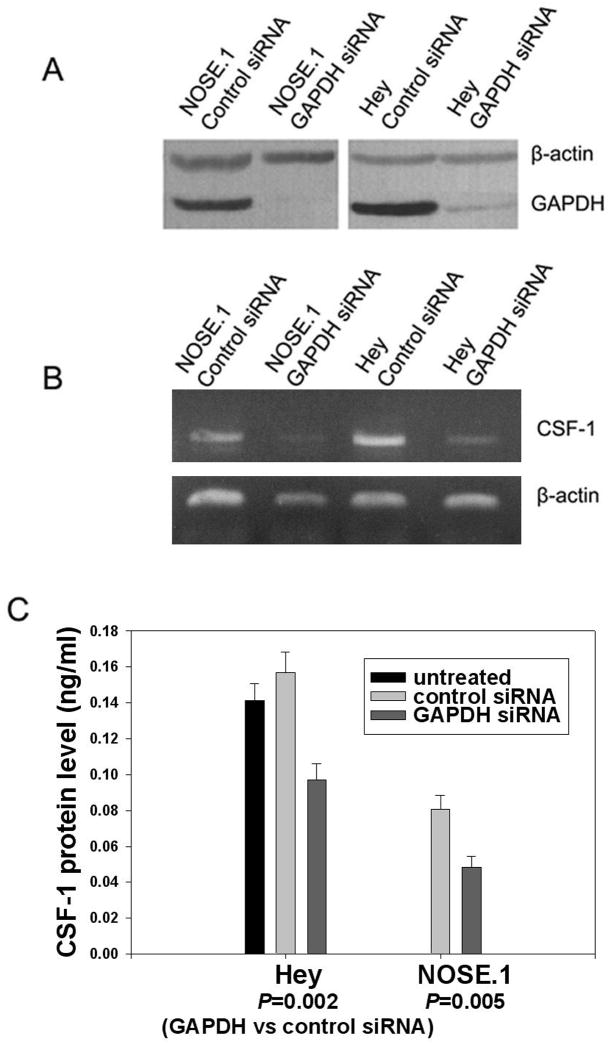
To investigate the functional significance of GAPDH’s association with CSF-1 3′UTR, siRNA targeted to GAPDH mRNA was delivered into NOSE.1 normal ovarian surface epithelial cells and Hey epithelial ovarian cancer cells. Results demonstrated the ability of GAPDH siRNA to specifically down-regulate GAPDH protein in both cell types, when compared to the effect of control siRNA which does not alter the expression of either GAPDH or β-actin (Fig. 1A). We then studied the effect of GAPDH siRNA on CSF-1 mRNA and protein levels. In both cell types GAPDH down-regulation by siRNA resulted in a significant reduction in both CSF-1 mRNA (Fig. 1B), and secreted CSF-1 levels (Fig. 1C). CSF-1 mRNA levels fell to the same degree when compared to control conditions in both cell types; in fact in NOSE.1 cells, CSF-1 RNA levels fell to almost an undetectable level. Secreted CSF-1 protein levels fell by 40% in both cell types. In NOSE.1 cells, CSF-1 protein levels fell from .0806 +/−.0078 to .0482 +/− .0064 ng/ml by GAPDH siRNA (p=.005). In Hey cells, CSF-1 protein levels fell from .1568 +/− .0116 to .0972 +/− .0091 ng/ml by GAPDH siRNA (p=.002). Thus, GAPDH positively regulated both CSF-1 mRNA and secreted protein levels. If regulation of steady-state CSF-1 mRNA by GAPDH was mediated by 3′UTR AU-rich CSF-1 mRNA binding, then this should be at a post-transcriptional level.
Figure 1. Silencing GAPDH and its effect on CSF-1 mRNA and protein expression.
(A) Immunoblot analysis of total protein extracts from control or GAPDH siRNA treated NOSE.1 or Hey cells. The extracts were harvested 48 hours post-siRNA transfection. The membrane was probed with antibodies to GAPDH and β-actin. GAPDH siRNA treatment significantly reduced or abolished GAPDH protein. (B) RT-PCR analysis of the CSF-1 and β-actin RNA expression in GAPDH or control siRNA treated NOSE.1 or Hey cells. The RNAs were harvested 24 hours post-siRNA transfection. GAPDH siRNA significantly down-regulated CSF-1 RNA compared to controls. (C) Levels of CSF-1 secreted in the conditioned media of GAPDH or control siRNA treated NOSE.1 or Hey cells (ng/ml). The media was harvested 57 hours post-siRNA transfection. There was no significant effect of control siRNA on secreted CSF-1 levels compared to untreated Hey cells (p=0.15). GAPDH siRNA was able to significantly decrease secreted CSF-1 protein levels when compared to controls.
CSF-1 mRNA stability is enhanced by GAPDH protein
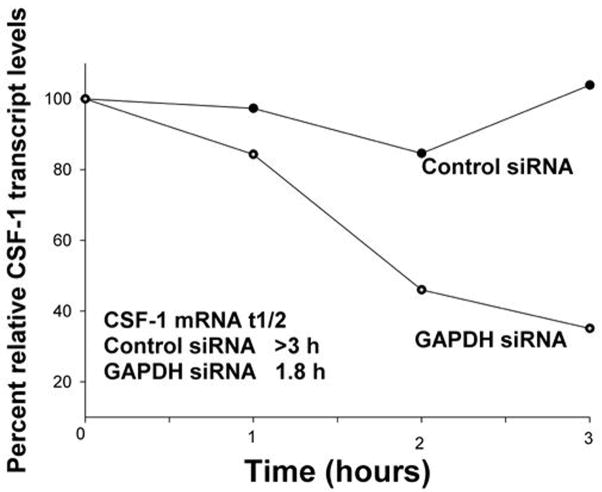
Since the majority of post-transcriptional events mediated by AU-rich regions are on the basis of altered mRNA stability, we studied the effect of GAPDH on CSF-1 mRNA half-life. Actinomycin-D chase experiments were performed in Hey ovarian cancer cells treated with either GAPDH or control siRNA. In Fig. 2, we show that in the presence of GAPDH siRNA, the CSF-1 mRNA half-life was only 1.8 hours, while in the cells treated with control siRNA, the predicted CSF-1 mRNA half-life was greater than 3 hours. A total of 3 independent experiments of CSF-1 mRNA half-life were performed. Each experiment confirmed the decrease in CSF-1 mRNA stability with GAPDH siRNA treatment. However, the measured mRNA half-life under these complex conditions differed from experiment to experiment. Collectively, the mean (+/− SEM) CSF-1 mRNA half-life with GAPDH siRNA was reduced 52%, from 3.87 +/− 0.72 hrs (control siRNA) to 1.87 +/− 0.52 hrs (GAPDH siRNA). The finding of an approximate CSF-1 mRNA half-life of 3.9 hrs in Hey ovarian cancer cells after control siRNA treatment is in line with our previous report of CSF-1 mRNA half-lives of 4.5 hrs in 2 other untreated ovarian cancer cell lines (18). We concluded that the reduction in steady-state CSF-1 RNA expression is on the basis of a decrease in CSF-1 mRNA stability, since the magnitude of the difference in CSF-1 mRNA half life resulting from treatment with GAPDH or control siRNA (Fig. 2), appeared to correlate with the difference in secreted CSF-1 protein levels (Fig. 1C).
Figure 2. Effect of GAPDH siRNA treatment on CSF-1 mRNA half-life in Hey ovarian cancer cells.
Hey cells were treated with 5 μg/ml actinomycin 48 hours after control or GAPDH siRNA transfection, and total RNA was isolated over the time period of the actinomycin-D chase for Northern Blot analysis of CSF-1 RNA and 18S RNA expression. After densitometric analysis, relative transcript levels were plotted versus time. A representative experiment is depicted. The mean difference in CSF-1 mRNA half-life is 2 fold between GAPDH and control siRNA treated Hey cells in 3 independent experiments.
A large AU-rich region within the 3′UTR of CSF-1 RNA is identified for GAPDH binding
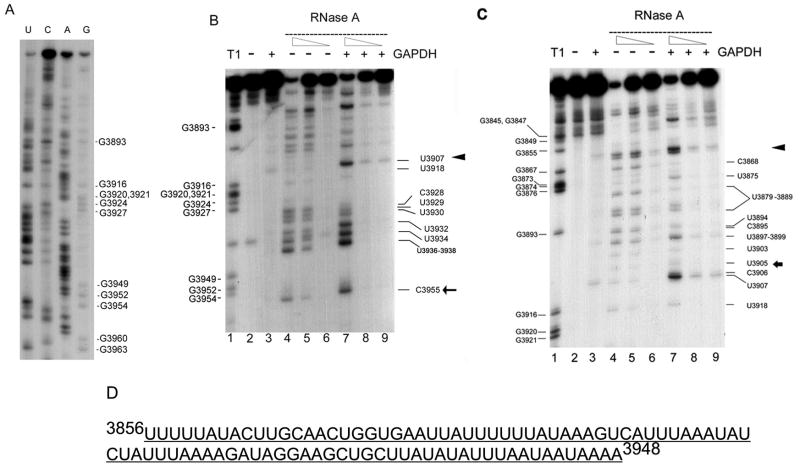
To further understand the nature of the interaction between GAPDH and the binding domains of CSF-1 mRNA within the 144nt terminal 3′UTR, enzymatic RNA footprinting analyses were performed. We had previously demonstrated that this 144nt 3′UTR CSF-1 RNA region served as an mRNA decay element (8). First, we chemically sequenced the 3′-end labeled 144nt CSF-1 RNA (Fig. 3A) in order to help interpret the footprinting experiments which followed. For the footprinting experiments, 3′-end labeled 144nt CSF-1 RNA was subjected to a standard binding reaction in the presence or absence of GAPDH, followed by treatment with RNase A or T1. Two regions for GAPDH binding lying in close proximity were identified (Fig. 3B (3′ region) and 3C (5′ region)). Since the 2 regions lie in such close proximity to each other, being separated by 2nt (C3906/U3907) only, it appears that these regions represent one large footprint with those 2 positions being susceptible to RNAseA in this in vitro reaction.
Figure 3. RNA footprinting analysis of the interaction of GAPDH protein with the 3′ untranslated region of CSF-1 RNA.
(A) 3′ end-labeled 3′ UTR CSF-1 RNA (3829-3972nt) sequenced chemically. The pattern derived was used to help interpret the footprinting experiments. (B) 3′ region of GAPDH binding to the 3′UTR CSF-1 RNA. The lane numbers are at the bottom of each panel. Lane 1, labeled CSF-1 RNAs (3828-3972nt) are partially hydrolyzed by RNase T1 (cleavage site 3′ of single-stranded G’s) to provide a reference ladder. CSF-1 RNA (3828-3972 nt) was subjected to a standard binding reaction in the presence (lane 3, 7, 8, 9) (+) or absence (lane 2, 4, 5, 6) (−) of 1.4 μg human GAPDH, followed by cleavage with RNase A (lanes 4–9). Partial cleavage with the decreasing amounts of RNase A (0.1ng/ml (lanes 4, 7), 0.02ng/ml (lanes 5, 8), 0.004ng/ml (lanes 6, 9)) or control reactions without RNase (lanes 2, 3) was performed. Triangle and arrow indicate the approximate boundaries of the 3′ region of the footprint between (3908-3916nt) and 5′ to 3955nt (the 3′ end of the footprint is better defined as 3939-3948nt in the text). (C) 5′ region of GAPDH binding to the 3′UTR CSF-1 RNA. The conditions for each lane are as described in (B). Triangle and arrow indicate the approximate boundaries of the 5′ region of the footprint between (3856-3868nt) and 3905nt. The 5′ region of the footprint can also be visualized in (B), but with less detail. (D) AU-rich footprint for GAPDH binding of 3′UTR CSF-1 RNA.
5′ region
The 5′ region of GAPDH binding was described by the following findings. First, in the presence of GAPDH, RNAse A (specifically cleaves 3′of U and C residues) decreased sensitivity in U/C residues starting 3′ to G3855 (marked by ◀ in Fig. 3C). There is a run of 5U starting at U3856, so the exact start site of this protected region could not be discerned. Decreased sensitivity to RNAse A is again clearly demonstrated at position C3868 and all the U and C positions depicted in Figure 3B, ending at U3905 (marked by ← in Fig. 3C). The footprint disappears at C3906/U3907. This 5′ region is also captured in Fig. 3B, as the footprint 5′ to U3907. Thus, the 5′ region protected by GAPDH binding appears to span from positions [3856–3868nt] to 3905nt.
3′ region
The 3′ region of GAPDH binding was described by the following findings. First, in the presence of GAPDH, the CSF-1 RNA was susceptible to RNAse A at positions U3907 and C3955 (Fig. 3B), and to RNAse T1 at G3949 (not shown). Starting 3′ to U3907 (marked by ◀ in Fig. 3B), the footprint affects all the U and C positions depicted in Fig. 3B, including U3918 and U3936-U3938. RNAse T1 (specifically hydrolyzes single stranded RNA after G) showed decreased sensitivity at positions G3916 and G3927, including the G residues in between (not shown). The footprint ends before G3949. Thus, the 3′ region protected by GAPDH appears to span from positions [3908-3916nt] to [3939-3948nt].
The results from treatment with RNase A and T1 indicates a large region with reduced sensitivity to RNases, which is very AU-rich (3856-3948nt; Fig. 3D). Therefore, footprint analyses demonstrated that the interactions between GAPDH and 3′UTR of CSF-1 lie within this AU-rich region. Our findings may reflect an interaction which results from direct hindrance by GAPDH protein with or without an indirect effect secondary to altered RNA folding by GAPDH binding.
The wild type 3′ UTR CSF-1 riboprobe sequence was subjected to computer-assisted modeling with the MFold program. One of the most thermodynamically stable structures with a calculated free energy of −46.2 kcal/mole is shown in Fig. 4A. As expected, this AU-rich 3′UTR region is predicted to form several stem-loop structures. Notably, the midportion of the footprint predicts the most complex of the secondary structures, with 2 stem loops forming each arm of a ‘v’ structure.
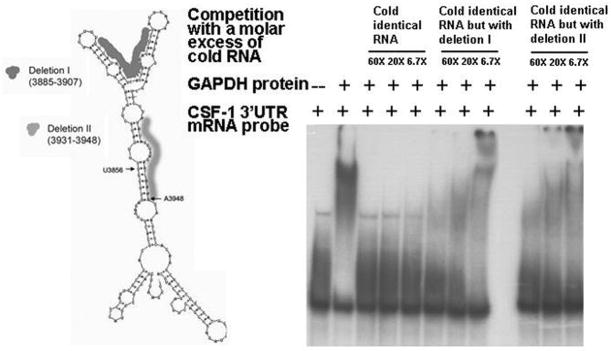
Figure 4. Predicted secondary structure of 3′UTR CSF-1 riboprobe and gel shift assays with cold competition by wildtype or mutant CSF-1 RNAs.
(A) The Mulfold RNA folding program prediction of the secondary structure of the riboprobe containing the terminal 144nt 3′ UTR CSF-1 RNA (3829-3972nt). The dark shading indicates the omitted nucleotides in deletion I (23nt: U3885-U3907, UUUAUAAAGUCAUUUAAAUAUCU) created specifically to delete the ‘v’ structure) and the light shading the omitted nucleotides in deletion II (18nt: A3931-A3948, AUAUAUUUAAUAAUAAAA) at the 3′ end of the footprint). The U3856nt and A3948nt indicated by arrows represent the approximate start and end of the footprint. (B) RNA gel shift assay of GAPDH protein binding to CSF-1 RNA sequences using the wildtype 3′UTR CSF-1 (3829-3972nt) riboprobe. The results of competition with a molar excess (6.7-60 fold) of cold RNA sequences demonstrate successful competition for binding by excess identical cold CSF-1 RNA. However, excess identical cold CSF-1 RNA but containing either deletion I or II only partially competed for binding by the labeled wildtype CSF-1 riboprobe. Therefore both regions in CSF-1 RNA appear to contribute to binding of CSF-1 RNA to GAPDH protein.
In order to explore whether this large region is important for GAPDH binding, deletions as described in Fig. 4A were introduced to assess their effects on GAPDH binding in vitro. Those AU rich deletion regions were chosen based on the footprinting data (Fig. 3), combined with information from the predicted secondary structure (Fig. 4A).
Results of RNA gel shift assays (Fig. 4B) demonstrated that the binding between CSF-1 RNA and GAPDH protein is successfully competed with a molar excess (6.7–60 fold) of unlabelled wild type RNA sequences (Fig. 4B, lanes 3–5). The binding was partially competed in reactions with a molar excess (6.7–60 fold) of either one of the unlabelled mutant RNA sequences (Fig. 4B, lanes 6–11), indicating that each mutant had weaker binding than the wildtype sequence. This data also suggested that both regions are required for efficient 3′UTR CSF-1 RNA binding to GAPDH protein.
Before we performed footprinting analyses, we empirically created a mutant of the 144nt 3′UTR CSF-1 RNA (8), where within 4 AU-rich regions (3850-3861nt, 3881-3888nt, 3908-3912nt, and 3935-3939nt), the A was mutated to a C, and T to a G. Northwestern analysis showed that this mutant abolished binding visualized by wildtype CSF-1 RNA to a 37kD protein in Hey ovarian cancer cells subsequently identified by us to be GAPDH (8). Comparison of the mutant sequence with that of the region identified by enzymatic footprinting shows that 3 of the 4 regions mutated and part of the fourth lie within this region. One (3881-3888nt) partially overlaps with deletion I, and the other (3935-3939nt) was completely included in Deletion II. We performed RNA gel shift assays with this mutant in Hey and NOSE.1 cells (Fig. 5), and show that these mutations largely abrogate 3′UTR CSF-1 RNA binding, when compared with the wildtype 144nt 3′ UTR CSF-1 sequence in NOSE.1 cells, and partly interfere with such binding in Hey cells. Our results in Fig. 4 suggest that a large AU-rich region is important to GAPDH binding. Our results in Fig. 5 along with the previously published northwestern analysis (8) are in line with this. They also suggest in Hey ovarian cancer cells, that other proteins such as the so far unidentified 75kD protein visualized on northwestern analysis (8) may have different binding affinities or sequence requirements within the 144nt CSF-1 RNA. Alternatively, mutations of most but not all of the AU-rich stretches may not be adequate to abrogate GAPDH binding in Hey ovarian cancer cells.
Figure 5. RNA gel shift assays comparing binding of wild type and mutated form of CSF-1 144nt 3′UTR to protein extract from Hey or NOSE.1 cells.

Mutations were created in 4 AU-rich regions within the terminal 144nt CSF-1 3′UTR as described in the text. Panels A&B represent results of binding of labeled wild type CSF-1 riboprobe to Hey (A) or NOSE.1 (B) protein extracts; Panels C&D represent results of binding of labeled mutant CSF-1 riboprobe to Hey (C) or NOSE.1 (D) protein extracts. Lanes 1, free riboprobe control; lanes 2, 1X probe mixed with 8 ug of protein extract; lanes 3, 3X probe mixed with 8ug of protein extract; lanes 4, 5 and 6, 1X probe mixed with 8 ug of protein extract in presence of 1500X, 770X or 390X excess cold probe, respectively.
GAPDH immunohistochemical staining of a tissue microarray cohort of epithelial ovarian and fallopian tube cancer specimens
The levels of expression of nuclear and cytoplasmic GAPDH in ovarian cancer cells was examined in an epithelial ovarian and fallopian tube cancer tissue microarray (TMA) consisting of 322 specimens. This array also contained benign epithelial ovarian neoplasms (N=4), and ovarian tumors of low malignant potential (N=5), and duplicate tissue cores (N=16). All cases were reviewed for histologic diagnosis by one gynecologic pathologist as described previously (19). The primary peritoneal cancers were included as part of the invasive epithelial ovarian cancers.
The analysis in this report was restricted to the invasive ovarian (N=243) and fallopian tube cancer (N=13) patients who underwent primary surgery, and excluded those who were treated with neoadjuvant chemotherapy or who had concurrent cancers. Thus 256 invasive cancer specimens were available for analysis of the clinical role of GAPDH. Of these 256 cores, 248 were interpretable for nuclear and cytoplasmic immunoreactivity. Tissue cores which were deemed non-interpretable usually had insufficient tumor cells, or the core was not representative of the cancer. Follow-up information was missing in only one patient. The median follow-up time was 38 months (range <1–222 months). Of the 256 invasive cancers, the primary adjuvant treatment was known in 82% of cases. Of these cases, 14% received no adjuvant treatment, and 62% were treated with platinum-based adjuvant chemotherapy, of which 46% also received a taxane. The remaining 24% received either non-platinum based chemotherapy or radiation therapy. Within this array were 16 duplicate cores. There was perfect concordance in the scoring for GAPDH in 14 of 16 of these duplicates; in only one case would the discrepancy have resulted in a change in the grouping of the score.
Analysis of clinical prognostic factors
Analysis of standard prognostic factors in epithelial ovarian and fallopian tube cancer was performed to confirm that studies using this tissue array will have clinical validity. As expected, we found that stage (I&II vs III&IV), grade (1&2 vs 3), age (≤60 vs >60 years), and histologic subtype (serous vs non-serous) were all highly significant factors for overall and progression-free survival (P<.009 for all analyses).
GAPDH staining of non-invasive cases
Regarding GAPDH immunostaining, we found that all 4 benign ovarian tumors had a nuclear and cytoplasmic score of 0. Among the 5 ovarian tumors of LMP, 3 had nuclear and cytoplasmic scores of 0, while one had a nuclear score of 20 and a cytoplasmic score of 100, and the other had a nuclear score of 180 and a cytoplasmic score of 90.
GAPDH staining of invasive cases
Overall, 52% of specimens expressed GAPDH in cytoplasmic and/or nuclear locations, with 24% of the whole group expressing GAPDH in both compartments. Expression of cytoplasmic GAPDH was highly associated with that of nuclear GAPDH (p<.0001), in line with the known ability of GAPDH to shuttle between compartments. Only 23% of specimens had moderate to strong immunoreactivity (score >100) for cytoplasmic GAPDH, and 9% for nuclear GAPDH. Notably, 55% of ovarian cancer cases had no staining (score=0) for GAPDH in the cytoplasm, and 70% with those findings in the nucleus. Forty-eight percent had no staining for GAPDH in either compartment.
We studied GAPDH staining in another cohort of 53 primary epithelial ovarian cancer cases using conventional slides. In this cohort, 92% had zero nuclear staining. We found 26 of 53 cases (50%) to have a GAPDH score of 0–50 out of a total score of up to 300, i.e., zero to extremely weak staining. Out of those 26 cases, 15 (29% of the entire cohort) had zero cytoplasmic and nuclear staining for GAPDH.
Thus the findings in the second cohort validate the findings of the TMA, in that the large majority of cases (70–90%) have no nuclear staining for GAPDH, and that approximately half of the cases have zero to very weak GAPDH staining in the cytoplasm. This implies that if GAPDH was expressed by these tumors, it is at a very low level undetectable or barely detectable by immunohistochemistry. Representative examples of GAPDH staining in the TMA are shown in Fig. 6.
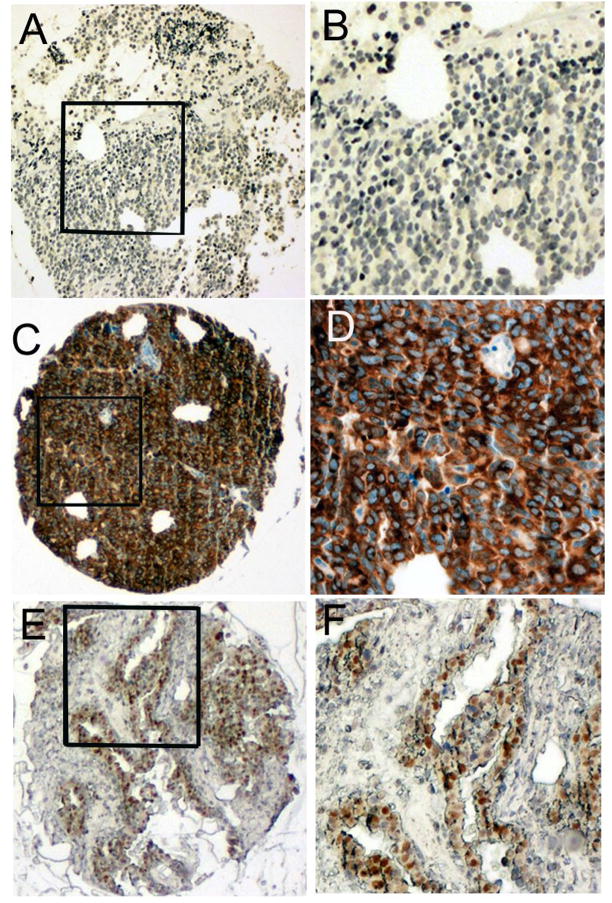
Figure 6. Representative pictures of GAPDH from an ovarian and fallopian tube cancer tissue microarray.
Panels A,C&E (original magnification, 40X) and B,D&F (magnified from squared area). Panels A&B: A case of ovarian serous carcinoma with both cytoplasmic and nuclear GAPDH score of zero. Hematoxlin was used as a counterstaining agent, therefore the blue color in the nuclei indicates negative staining. Panels C&D show an ovarian serous carcinoma case which had cytoplasmic (score 300) but no nuclear GAPDH staining. Panels E&F show an ovarian serous carcinoma case which had nuclear (score 100) but no cytoplasmic GAPDH staining.
Analysis of association of GAPDH with clinicopathologic factors and with survival
In the TMA, among all stages, presence of either cytoplasmic or nuclear GAPDH staining (scores >0) was associated with non-serous histologic subtype (p=.0007 for cytoplasmic, and p=.0073 for nuclear staining), with cytoplasmic GAPDH staining also associated with early stage disease (p=.0004). Neither cytoplasmic nor nuclear GAPDH was associated with any other clinicopathologic factor, such as grade or age. Moreover, neither nuclear nor cytoplasmic GAPDH proved to be a prognostic factor for either progression-free or overall survival.
Association of GAPDH with CSF-1
Thirty-eight of the second cohort of 53 ovarian cancer cases had previously been analyzed for CSF-1 and c-fms expression (9). We analyzed this subset for association between GAPDH and CSF-1. We found a highly significant association (p=.008) between a low cytoplasmic GAPDH score and a low CSF-1 score (Table 1). In fact, we found that 93% of ovarian cancer cases with low CSF-1 staining, had a low GAPDH score. In contrast, and as expected, there was no significant association between GAPDH and c-fms expression.
Table 1.
The association between cytoplasmic GAPDH score and CSF-1 staining in ovarian cancer cases (n=38). The association is highly significant (p=0.008; Fisher’s Exact Test).
| CSF-1 score | |||
|---|---|---|---|
| < 200 | ≥ 200 | ||
| Cytoplasmic
GAPDH score |
≤ 80 | 26 | 5 |
| > 80 | 2 | 5 | |
Summary of immunohistochemical findings for GAPDH
Thus, our findings demonstrate significant regulation of GAPDH protein in ovarian and fallopian tube cancers. Further, our observation that GAPDH and CSF-1 were co-expressed in ovarian cancer cases is in line with our finding that GAPDH positively regulates CSF-1 expression.
Discussion
Although GAPDH’s predilection for AU-rich elements has long been known, the expected connection between GAPDH and control of mRNA stability has never been made. Our results now document a functional role for GAPDH in controlling mRNA decay rates. The key observation supporting this claim is that siRNA mediated knockdown of GAPDH levels leads to increased degradation of the CSF-1 mRNA. Moreover, GAPDH binds to a region in CSF-1 mRNA 3′UTR that has been shown to promote mRNA degradation. Taken together, this suggests that GAPDH can bind to the CSF-1 3′UTR and enhance the stability of the mRNA. We do not believe that this effect of GAPDH on control of mRNA stability is limited to CSF-1 as a target. Indeed, GAPDH can bind the AU-rich regions of the 3′UTR of c-myc and GM-CSF RNA (6). It would not be surprising to find that GAPDH too regulates these and other AU-rich containing messages. A role for GAPDH in regulating mRNA turnover is also important in the context of the increasing recognition of the contribution of mRNA stability to control of gene expression. It is estimated that at least 50% of steady-state gene expression is controlled by stability regulation (20), and GAPDH now adds this critical level of regulation to its repertoire.
We show by enzymatic footprinting that there is a large AU-rich region within the 144nt 3′UTR of CSF-1 RNA which is protected by GAPDH protein, and appears to be necessary for GAPDH binding. As previously suggested, it is likely that there does not exist an RNA concensus binding motif for GAPDH (21). Instead, localization of binding to secondary structures such as stem loop regions likely plays a strong contributory role (7,21,22). The predicted secondary structure of the AU-rich 3′UTR of CSF-1 RNA contains several stem-loop structures, in line with other reports showing GAPDH predilection for AU-rich regions and those with complex secondary structures (6,7,21). We show that deletions or mutations targeting AU-rich regions within these CSF-1 3′UTR structures mitigate binding to GAPDH protein and ovarian cell extracts.
Our results demonstrate that GAPDH expression levels vary in ovarian and fallopian tube cancers, with almost 50% of tumors showing no detectable staining. It makes sense that GAPDH is regulated specifically in cancers. It has been proposed that cancers may have a glycolytic phenotype (23), in that hypoxia, a common feature of malignancy, can stimulate anaerobic glucose metabolism. Our finding that GAPDH is not detectable by immunohistochemistry in both nuclear and cytoplasmic compartments in almost 50% of ovarian and fallopian tube cancers deserves some note. We are the first to make this observation in a large array of cancers, in part because there is a surprising paucity of literature on the study of GAPDH protein levels in cancer specimens. In one study of 13 prostate cancer specimens, heterogeneous nuclear and/or cytoplasmic GAPDH staining was observed, with GAPDH not detectable in the normal secretory epithelium (24).
In our TMA, the results of a highly significant association between nuclear and cytoplasmic expression of GAPDH are in line with GAPDH’s known shuttling function between the nuclear and cytoplasmic compartments. GAPDH has numerous functions, some specifically related to its nuclear localization, such as in transcriptional control, DNA repair, and apoptosis (1). In the cytoplasm, GAPDH also has many roles, such as in translational control (2,3), and we now add regulation of mRNA turnover as one of GAPDH’s cytoplasmic functions.
Our finding that GAPDH staining is associated more with early stage may be counterintuitive, in that we show one of its functions if to stabilize CSF-1 mRNA, an important factor in tumor progression. However, among advanced stage disease, GAPDH was still detectable almost 50% of the time, and its functions in the cell are innumerable. Thus, it comes as no surprise that GAPDH protein has little prognostic value. Prior to this report, the only study to examine prognosis related to GAPDH expression, measured GAPDH RNA by RT-PCR in breast cancers, and found it to be associated with reduced survival (15). Importantly, we showed that cytoplasmic GAPDH is significantly co-expressed with CSF-1 in ovarian cancer cases, which is in line with our finding that CSF-1 RNA and protein is regulated by GAPDH.
In conclusion, (1) we are the first to show that GAPDH, a multifunctional protein, has the capacity to regulate gene expression at the level of mRNA stability. We find that GAPDH promotes CSF-1 RNA and protein expression in ovarian cancer cells, by stabilizing its message; (2) we show that GAPDH protein interacts with a large region of AU-rich CSF-1 RNA, which is predicted to have a complex secondary structure; (3) we are the first to evaluate the clinical role of nuclear and cytoplasmic expression of GAPDH protein in any cancer. Our finding that GAPDH is not detectable by immunohistochemistry in both nuclear and cytoplasmic compartments in almost 50% of ovarian and fallopian tube cancers is notable, and underscore that fact that GAPDH protein is regulated in cancers. We also found that GAPDH is co-expressed with CSF-1 in ovarian cancers, which makes sense, since we now recognize one of the many functions of GAPDH is to promote mRNA stability of CSF-1, an important cytokine in tumor progression.
Materials and Methods
Cell culture and protein extraction
Hey (8) human epithelial ovarian carcinoma cells were maintained in DMEM (ATCC, Manassas, VA) supplemented with 1.5 g/L sodium bicarbonate and 10% fetal calf serum (GIBCO, Grand Island, NY). Normal human ovarian surface epithelial cell line (NOSE.1) (8) was cultured in M199 and MCDB1051 medium (v/v; Sigma-Aldrich, St. Louis, MO) supplemented with 15% fetal calf serum and 1.5 g/L sodium bicarbonate. Total cellular protein extract for immunoblot analysis was prepared from cells using 50 mM TrisCl pH 7.4, 100 mM NaCl, 2 mM EDTA, 1% Igepal (Sigma-Aldrich, St. Louis, MO), and Protease Inhibitor Cocktail Set 1 at 1:100 dilution (Calbiochem, San Diego, CA). S100 and total extracts for gel shift assays were prepared as described (8). Protein concentrations were determined by BCA assay (Pierce, Rockford, IL) using BSA as the standard.
Preparation of RNA probes
The terminal 144 nt wildtype sequence of CSF-1 exon 10 (3829-3972 nt) was PCR amplified and subcloned into the KpnI and XbaI restriction sites of the PGEM-3Z (Promega, Madison, WI) transcription vector (PGEM-3Z-WT) as described previously (8). The original mutant pGEM-3Z-MT was created as described previously (8). This mutant sequence which contained mutations within 4 AU-rich areas within the 144nt 3′UTR was designed empirically prior to the footprinting experiments.
The mutated sequences containing Deletion I or Deletion II which differed from the wild-type sequence in AU-rich regions contained within the footprint (as depicted in Fig. 4A) were inserted into PGEM-3Z, to create PGEM-3Z-MT.1 and MT.II. For construction of mutant sequences, the deletions were introduced by PCR using two overlapping oligonucleotides (5′-CCCGGGGTACCCCATTGGCTCACGCACTGTGAGATTTTGTTTTTATACTTGCAACTGGTGAATTATTTATTTAAAAGATAGGA-3′ and 5′-CCTGCTCTAGAGCGTCAACGGCAGCTTGTGCACTTCTTTTATTATTAAATATATAAGCAGCTTCCTATCTTTTAAATAAATAA-3′) for Deletion I and (5′-CCCGGGGTACCCCATTGGCTCACGCACTGTGAGATTTTGTTTTTATACTTGCAACTGGTGAATTATTTTTTATAAAGTCATTTAAA-3′ and 5′-CCTGCTCTAGAGCGTCAACGGCAGCTTGTGCACTTCAAGCAGCTTCCTATCTTTTAAATAGATATTTAAATGACTTTATAAAAAAT-3′) for Deletion II. The wild type and mutant constructs were linearized with HindIII endonuclease. PCR reactions were performed using Taq polymerase(Perkin-Elmer, Norwalk, CT) and were carried out at 95°C for 1 min, 42°C for 1 min, and 72°C for 1 min for 40 cycles (72°C at end).
Unlabelled RNAs, which were used for competition experiments and 3′-end labelling, were generated with the MEGAscript In vitro transcription kit (Ambion, Austin, TX).
For sequencing of 3′UTR of CSF-1 RNA and footprinting of GAPDH, 50 pmol of the 3′UTR CSF1 RNA was labeled with 32Ppcp (Amersham Biosciences, Piscataway, NJ) at the 3′-end using T4 RNA ligase (Ambion, Austin, TX) according to the manual.
Direct chemical method for sequencing RNA
G, A, C, U reactions were carried out as previously described (25). For each reaction, 3×104 cpm of 3′-end labeled CSF-1 RNA was used. After specific chemical modification of each RNA base, aniline was used for strand scission. Samples containing those labeled fragments were dissolved in 2–3 μl of 8 M urea/20 mM TrisHCl, Ph7.4/1 mM EDTA/0.05% xylene cyanol/0.05% bromphenol blue. The samples were heated at 90°C for 2 minutes, chilled on ice immediately, and then layered on 8% polyacrylamide, 8 M urea gel.
Enzymatic footprinting of GAPDH
For enzymatic footprinting (7,26), 1 μl (6 × 103 cpm) of 3′-end labeled CSF-1 RNA was incubated with 1.4 μg purified human heart GAPDH (Advanced Immunochemical Inc., Long Beach, CA) in reaction buffer (5 mM HEPES, pH 7.6, 40 mM KCl, 2.5 mM MgCl2, 3.8% glycerol, 1.5 mM ATP, 0.1 mM DTT, 5 μg yeast tRNA) at 30°C for 10 min, then placed on ice. Next, RNase A (0.1 or 0.02 or 0.004 ng/ml) or RNase T1 (1.25×10−5 or 1.25×10−6 U/μL) (Ambion, Austin, TX) was added and incubated at 30°C for 10 min. Reactions were stopped with 300 μl 0.2 mg/ml proteinase K, 0.03 mg/ml yeast tRNA, 50 mM Tris pH 7.5, 50 mM NaCl, 5 mM EDTA, 0.5% SDS and incubated 30 min at 55°C. Following phenol extraction and ethanol precipitation, samples were fractionated in 8% polyacrylamide, 8 M urea gel.
RNA gel shift and cold competition assays
Reaction mixtures (10 μl) containing 1 μl (6 × 103 cpm, 2ng) of 3′-end labeled RNA and 0.7 μg purified human heart GAPDH in reaction buffer (5 mM HEPES, pH 7.6, 40 mM KCl, 2.5 mM MgCl2, 3.8% glycerol, 1.5 mM ATP, 0.1 mM DTT, 5 μg yeast tRNA) were incubated for 10 min at 30°C. The complexes were resolved by electrophoresis through non-denaturing 3.2% polyacrylamide gels and autoradiographed. RNA gel shift assays with Hey S100 or NOSE.1 total cell extracts were performed as described (8). For competition analysis, excess unlabeled wild type RNAs or mutant RNAs were used in addition to the labeled RNAs.
siRNA treatment
The GAPDH siRNA (5′-UGGUUUACAUGUUCCAAUAUU3′ UUACCAAAUGUACAAGGUUAU P-5′) used was obtained from Dharmacon (Lafayette, CO), and serves as their positive control for other siRNA experiments. Transfection of siRNA into Hey or NOSE.1 was performed in 6-well plates or T75 flasks. Cells were plated at a density of 1.7×104 cells/cm2. Pilot experiments of dose response were performed first to determine the lowest effective dose of GAPDH siRNA, in order to decrease off target effects. Transfection of 20nM GAPDH siRNA or negative control RISC-Free siRNA (Dharmacon, Lafayette, CO) was performed for 24, 48, or 57 hours. For all transfections, DharmaFECT-1 siRNA Transfection Reagent was used.
Immunoblot
Twenty μg of proteins were loaded per lane on 10% SDS-PAGE gels, electrophoresed, transferred to Immobilon P membranes (Millipore, Billerica, MA) and Ponceau S (Pierce, Rockford, IL) stained. Membranes were probed with GAPDH mAb (Abcam Inc., Cambridge, MA), and β-actin mAb (ACTN05, Lab Vision, Fremont, CA). Immunoblot processing and ECL protein detection were performed according to the manufacturer’s instructions using SuperSignal West Pico (Pierce, Rockford, IL) and HRP-conjugated 2° antibodies (Amersham GE, Fairfield, CT).
Preparation of total RNA and analysis by RT-PCR
Total RNA extracts from cells were prepared with a RNeasy Mini kit (Qiagen, Valencia, CA). Reverse transcription was done using 0.7 ug RNA according to the manufacturer’s protocol (Fermentas, Glen Burnie, MD) in a final volume of 20 μl. Subsequently, 3 μl of cDNA was PCR-amplified using HotMaster DNA Taq polymerase (Eppendorf, Westbury, NY). The PCR primers corresponding to the 4-kb CSF-1 were used (5′-GACAGAGCTTTCCCCCTCCAG-3′ and 5′-TGGCAGCATGGGGAGCATCCT-3′). CSF-1 PCR reactions were carried out for 40 cycles according to a temperature-cycling profile of 94°C for 1 min, 60°C for 2 min, and 72°C for 3 min. The PCR conditions and the sequences of the β-actin primers (27) have been described.
Actinomycin D chase experiment
Hey cells were transfected with GAPDH siRNA or control RISC-Free siRNA (0.02 μM) for 48 hours. Total cellular RNA was isolated using RNeasy Mini Kit (Qiagen, Valencia, CA). The RNAs (10 μg per well) were dissolved in glyoxal/dimethylsulfoxide buffer (Ambion, Austin, TX), electrophoresed in a 1% agarose gel, and transferred onto BrightStar-Plus membranes (Ambion, Austin, TX). The Northern blots were then hybridized with a (α-32P) dCTP-labeled 500-bp fragment of the human CSF-1 coding region, a kind gift from Eleanor C. Weir (Yale University) and 18s rRNA (Ambion, Austin TX) . For actinomycin-D chase experiments (8), 5 μg/ml actinomycin D (Sigma) was added to inhibit new transcription at time=0. Cells were harvested at various intervals after actinomycin treatment, total RNA extracted, and CSF-1 RNA and 18S RNA level analyzed by Northern blot. Graphs of relative CSF-1 mRNA were derived by densitometry (Image Quant, Molecular Dynamics, Inc, Fairfield, CT) and normalization of CSF-1 RNA to its 18S RNA control. Half-lives were determined with the maximum intensity normalized for each half-life to 100%. A representative experiment of 3 independent experiments was shown.
Detection of CSF-1 in the culture supernatants of post-siRNA transfected cells by ELISA
Hey or NOSE.1 cells were plated at a density of 1.7×104 cells/cm2. Transfection with GAPDH siRNA or control RISC-Free siRNA was performed as above. The concentration of secreted CSF-1 in the culture supernatants was measured by ELISA (R&D Systems Inc, Minneapolis, MN) 57 hours post siRNA transfection.
Construction of Tissue Microarray
To construct the ovarian and fallopian tube cancer tissue microarray used in this study, formalin-fixed, paraffin-embedded archival tissue blocks and their matching hematoxylin and eosin-stained slides were retrieved, reviewed and screened for representative tumor regions by a gynecologic pathologist as described previously (19). The tissue cores were 0.6mm in diameter and spaced 0.8mm apart using a precision instrument (Beecher Instruments, Sun Prairie, WI, USA); the TMA was constructed by the Yale University Cancer Center TMA Shared Resource, with approval from the Yale University Institutional Review Board. Microarrays were stored in a nitrogen chamber at room temperature. Samples used in this study were from patients mainly with primary epithelial ovarian cancers who had undergone initial surgery at Department of Obstetrics and Gynecology of Yale University between 1980 and 2001. The TMA consisting of 322 specimens including 297 invasive epithelial ovarian cancers, 16 fallopian tube cancers, 5 ovarian tumors of low malignant potential, 4 benign epithelial ovarian neoplasms and 16 duplicate tissue cores.
Demographic and survival data for each specimen were entered into a comprehensive database created with Microsoft Excel (version 2002). Histopathologic diagnoses and grading were based on World Health Organization criteria. Disease stage was assigned according to the International Federation of Gynecology and Obstetrics (FIGO) system.
Validation of the Tissue Microarray
This TMA was validated with p53 immunohistochemical staining (19). A total of 30 ovarian cancer tissue cores with a corresponding set of 30 conventional paraffin sections were stained with p53 antibody (anti-human monoclonal antibody, D07, (Dako, Carpinteria, CA) in separate batches. Each tissue core and tissue section were scored blindly and recorded individually. Ten of the 30 tissue cores had negative p53 staining, with 20 cores positive for p53. Correlation coefficient analysis was performed after individual staining score was obtained. The correlation coefficient index was 0.95, supporting that the staining results obtained from the TMA are representative as compared with the results obtained from the conventional paraffin sections.
Immunohistochemistry of GAPDH
Immunohistochemical analysis was performed using the streptavidin-biotin-peroxidase technique with a mouse monoclonal anti-human GAPDH antibody (Biogenesis Inc., Kingston, NH). The procedure is briefly described as follows. Tissue microarray slides were incubated at 65°C overnight and rinsed with xylene and ethanol to remove paraffin. GAPDH antigen was unmasked with the heat-mediated antigen retrieval method described previously. The primary anti-GAPDH antibody was coated on the slides overnight at 4°C in a moist chamber. The final concentration of the antibody was 1 μg/ml. Visualization was carried out using a secondary antibody conjugated to HRP and diaminobenzidine tetrahydrochloride as the chromogen. Sections from conventional invasive ovarian carcinoma served as positive controls for GAPDH staining. Negative controls were prepared by replacing primary antibodies with class-matched mouse IgG immunoglobulins on parallel sections. To confirm the specificity of the GAPDH staining, 10 additional specimens were restained with and without the antigenic peptide for GAPDH for 2 h at room temperature before the staining procedure.
To validate our findings with this TMA, we studied another cohort of 53 conventional slides of primary epithelial ovarian cancer for GAPDH staining using the same antibody as used for the TMA. The technique was the same with the exception that the final concentration used for the conventional slides was 3 μg/ml. Negative controls were prepared by replacing primary antibodies with class-matched mouse IgG immunoglobulins. Thirty-eight of the 53 cases had been previously analyzed for CSF-1 and c-fms expression (9).
Evaluation of immunohistochemical staining
The nuclear and cytoplasmic staining was determined separately for each stained tissue core. Specimens that were non-interpretable or did not contain carcinoma were not included in the analysis. Scoring was performed by two individuals who were blinded to the clinical data. A few discrepancies were found between the two scores. A consensus score was generated after the slides were reviewed again. The staining intensity was graded on the following scale: 0 = no staining; 1 = weak staining; 2 = moderate staining; and 3 = intense staining. The scores used for analysis were generated by multiplying the intensity by the percentage of positive cells in a defined tissue core, yielding scores ranging from 0 to 300.
Statistical analysis
All analyses were performed using SAS Proprietary Software version 9.0 (SAS Institute Inc., Cary, NC). The difference between levels of secreted CSF-1 was determined by the student’s t test. Survival curves were calculated using the Kaplan-Meier method, with the significance evaluated using the Mantel-Cox Log-rank test. The association between GAPDH staining scores and clinicopathological parameters was calculated using the Chi-square test, or Fisher’s Exact Test as appropriate. Spearman’s correlation coefficient method was used for validation of the TMA with conventional slides.
Acknowledgments
We thank Elisabetta Ullu, Lara Weinstein and Ghadiyaram Chakshusmathi for helpful advice, and Roy Parker both for his advice and comments on the manuscript. We thank Wenxin Zheng for scoring and for optimizing the immunohistochemical staining. Immunohistochemical staining was generated in part by the TACMASS Core (Tissue Acquisition and Cellular/Molecular Analysis Shared Service) at the Arizona Cancer Center, supported by NIH CA2304. This work was supported by NIH CA60665 (to S.K.C).
Footnotes
S.K.C., Y.Z, and M.G.H. designed research; Y.Z., X.Y., N.B., J.M., and J.B.S. performed research; Y.Z. and S.K.C. analyzed data; and S.K.C. and Y.Z. wrote the paper.
References
- 1.Sirover MA. New nuclear functions of the glycolytic protein, glyceraldehyde-3-phosphate dehydrogenase, in mammalian cells. J Cell Biochem. 2005;95:45–52. doi: 10.1002/jcb.20399. [DOI] [PubMed] [Google Scholar]
- 2.Sirover MA. New insights into an old protein: the functional diversity of mammalian glyceraldehyde-3-phosphate dehydrogenase. Biochimica Biophysica Acta. 1999;1432:159–84. doi: 10.1016/s0167-4838(99)00119-3. [DOI] [PubMed] [Google Scholar]
- 3.Yi M, Schultz DE, Lemon SM. Functional significance of the interaction of hepatitis A virus RNA with glyceraldehyde 3-phosphate dehydrogenaase (GAPDH): opposing effects of GAPDH and polypyrimidine tract binding protein on internal ribosome entry site function. J Virol. 2000;74:6459–68. doi: 10.1128/jvi.74.14.6459-6468.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Petrik J, Parker H, Alexander GJM. Human hepatic glyceraldehyde-3-phosphate dehydrogenase binds to the poly(U) tract of the 3′ non-coding region of hepatitis C virus genomic RNA. J Gen Virol. 1999;80:3109–13. doi: 10.1099/0022-1317-80-12-3109. [DOI] [PubMed] [Google Scholar]
- 5.Lin S-S, Chang SC, Wang Y-H, Sun C-Y, Chang M-F. Specific interaction between the hepatitis delta virus RNA and glyceraldehyde 3-phosphate dehydrogenase: an enhancement on ribozyme catalysis. Virology. 2000;271:46–57. doi: 10.1006/viro.2000.0302. [DOI] [PubMed] [Google Scholar]
- 6.Nagy E, Rigby WFC. Glyceraldehyde-3-phosphate dehydrogenase selectively binds AU-rich RNA in the NAD+-binding region (Rossmann Fold) J Biol Chem. 1995;270:2755–63. doi: 10.1074/jbc.270.6.2755. [DOI] [PubMed] [Google Scholar]
- 7.Dollenmaier G, Weitz M. Interaction of glyceraldehyde-3-phosphate dehydrogenase with secondary and tertiary RNA structural elements of the hepatitis A virus 3′ translated and non-translated regions. J Gen Virol. 2003 Feb;84(Pt 2):403–14. doi: 10.1099/vir.0.18501-0. [DOI] [PubMed] [Google Scholar]
- 8.Bonafe N, Gilmore-Hebert M, Folk NL, Azodi M, Zhou Y, Chambers SK. Glyceraldehyde-3-phosphate dehydrogenase binds to the AU-Rich 3′ untranslated region of colony-stimulating factor-1 (CSF-1) messenger RNA in human ovarian cancer cells: possible role in CSF-1 posttranscriptional regulation and tumor phenotype. Cancer Res. 2005 May 1;65(9):3762–71. doi: 10.1158/0008-5472.CAN-04-3954. [DOI] [PubMed] [Google Scholar]
- 9.Chambers SK, Kacinski BM, Ivins CM, Carcangiu ML. Overexpression of epithelial macrophage colony-stimulating factor (CSF-1) and CSF-1 receptor: a poor prognostic factor in epithelial ovarian cancer, contrasted with a protective effect of stromal CSF-1. Clin Cancer Res. 1997;3(6):999–1007. [PubMed] [Google Scholar]
- 10.Kacinski BM, Carter D, Mittal K, et al. Ovarian adenocarcinomas express fms-complementary transcripts and fms antigen, often with coexpression of CSF-1. Am J Pathol. 1990;137:135–47. [PMC free article] [PubMed] [Google Scholar]
- 11.Toy EP, Chambers JT, Kacinski BM, Flick MB, Chambers SK. The activated macrophage colony-stimulating factor (CSF-1) receptor as a predictor of poor outcome in advanced epithelial ovarian cancer. Gynecol Oncol. 2001;80:1–7. doi: 10.1006/gyno.2000.6070. [DOI] [PubMed] [Google Scholar]
- 12.Kacinski BM, Stanley ER, Carter D, et al. Circulating levels of CSF-1 (M-CSF) a lymphohematopoietic cytokine may be a useful marker of disease status in patients with malignant ovarian neoplasms. Int J Radiat Oncol Biol Phys. 1989;17:159–64. doi: 10.1016/0360-3016(89)90383-0. [DOI] [PubMed] [Google Scholar]
- 13.Price FV, Chambers SK, Chambers JT, et al. CSF-1 concentration in primary ascites of ovarian cancer is a significant predictor of survival. Am J Obstet Gynecol. 1993;168:520–7. doi: 10.1016/0002-9378(93)90485-2. [DOI] [PubMed] [Google Scholar]
- 14.Scholl S, Bascou CH, Mosseri V, et al. Circulating levels of colony-stimulating factor 1 as a prognostic indicator in 82 patients with epithelial ovarian cancer. Br J Cancer. 1994;62:342–6. doi: 10.1038/bjc.1994.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Revillion F, Pawlowski V, Hornez L, Peyrat J-P. Glyceraldehyde-3-phosphate dehydrogenase gene expression in human breast cancer. Eur J Cancer. 2000;36:1038–42. doi: 10.1016/s0959-8049(00)00051-4. [DOI] [PubMed] [Google Scholar]
- 16.Lu H, Zhang Y, Roberts DD, Osborne CK, Templeton NS. Enhanced gene expression in breast cancer cells in vitro and tumors in vivo. Mol Therapy. 2002;6:783–92. doi: 10.1006/mthe.2002.0813. [DOI] [PubMed] [Google Scholar]
- 17.Rondinelli RH, Epner DE, Tricoli JV. Increased glyceraldehyde-3-phosphate dehydrogenase gene expression in late pathological stage human prostate cancer. Prostate Cancer & Prostatic Diseases. 1997;1:66–72. doi: 10.1038/sj.pcan.4500208. [DOI] [PubMed] [Google Scholar]
- 18.Chambers SK, Kacinski BM. Messenger RNA decay of macrophage colony-stimulating factor in human ovarian carcinomas in vitro. J Soc Gynecol Invest. 1994;1:310–6. doi: 10.1177/107155769400100411. [DOI] [PubMed] [Google Scholar]
- 19.McAlpine JN, Baron AT, Kim M-O, et al. Soluble EGFR isoform expression levels improve the prognostic significance of EGFR in ovarian cancer. Gynecol Oncol. 2008 (submitted) [Google Scholar]
- 20.Cheadle C, Fan J, Cho-Chung YS, et al. Stability regulation of mRNA and the control of gene expression. Ann NY Acad Sci. 2005;1058:196–204. doi: 10.1196/annals.1359.026. [DOI] [PubMed] [Google Scholar]
- 21.De BP, Gupta S, Zhao H, Drazba JA, Banerjee AK. Specific interaction in vitro and in vivo of glyceraldehyde-3-phosphate dehydrogenase and LA protein with cis-acting RNAs of human parainfluenza virus type 3. J Biol Chem. 1996;271:24728–35. doi: 10.1074/jbc.271.40.24728. [DOI] [PubMed] [Google Scholar]
- 22.Kiri A, Goldspink G. RNA-protein interactions of the 3′ untranslated regions of myosin heavy chain transcripts. Journal of Muscle Research & Cell Motility. 2002;23:119–29. doi: 10.1023/a:1020211729728. [DOI] [PubMed] [Google Scholar]
- 23.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nature Rev Cancer. 2004;4:891–9. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 24.Epner DE, Coffey DS. There are multiple forms of glyceraldehyde-3-dehydrogenase in prostate cancer cells and normal prostate tissue. Prostate. 1996;28:372–8. doi: 10.1002/(SICI)1097-0045(199606)28:6<372::AID-PROS6>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 25.Peattie DA. Direct chemical method for sequencing RNA. Proc Natl Acad Sci USA. 1979 Apr;76(4):1760–4. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chakshusmathi G, Kim SD, Rubinson DA, Wolin SL. A La protein requirement for efficient pre-tRNA folding. EMBO J. 2003 Dec 15;22(24):6562–72. doi: 10.1093/emboj/cdg625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jindal SK, Ishii E, Letarte M, Vera S, Teerds KJ, Dorrington JH. Regulation of transforming growth factor alpha gene expression in an ovarian surface epithelial cell line derived from a human carcinoma. Biol Reprod. 1995 May;52(5):1027–37. doi: 10.1095/biolreprod52.5.1027. [DOI] [PubMed] [Google Scholar]