Abstract
Fragile X syndrome (FXS) is the most common form of hereditary mental retardation. FXS patients have a deficit for the fragile X mental retardation protein (FMRP) that results in abnormal neuronal dendritic spine morphology and behavioral phenotypes, including sleep abnormalities. In a Drosophila model of FXS, flies lacking the dfmr1 protein (dFMRP) have abnormal circadian rhythms apparently as a result of altered clock output. In this study, we present biochemical and genetic evidence that dFMRP interacts with a known clock output component, the LARK RNA-binding protein. Our studies demonstrate physical interactions between dFMRP and LARK, that the two proteins are present in a complex in vivo, and that LARK promotes the stability of dFMRP. Furthermore, we show genetic interactions between the corresponding genes indicating that dFMRP and LARK function together to regulate eye development and circadian behavior.
Keywords: Drosophila, circadian rhythm, RNA, protein, eye, genetics
Introduction
Fragile X syndrome (FXS) (O'Donnell and Warren, 2002) is associated with sleep disorders, autism, hyperactivity, and cognitive deficits (Gould et al., 2000; Bakker and Oostra, 2003; Miano et al., 2008). FXS typically results from a CGG trinucleotide repeat expansion in the 5′ untranslated region of FMR1, resulting in loss of the FMR1 product fragile X mental retardation protein (FMRP) (O'Donnell and Warren, 2002). FMRP and its autosomal paralogs, FXR1P and FXR2P, make up a small family of RNA-binding proteins containing two KH domains and an RGG region (O'Donnell and Warren, 2002). FMRP associates with translating polyribosomes in an RNA-dependent manner (Khandjian et al., 1996; Feng et al., 1997a) and appears to function in translational control (Feng et al., 1997b; Morales et al., 2002; Jin and Warren, 2003). Putative targets of FMRP have been identified in mammals and Drosophila using biochemical and genetic approaches (Brown et al., 2001; Darnell et al., 2001; Reeve et al., 2005; Zarnescu et al., 2005).
Insights into FMRP function have been gained from Drosophila and mouse models of FXS. FMRP participates in signaling through metabotropic glutamate receptors (mGluRs) and limits protein-dependent long-term depression (LTD) in the hippocampus and cerebellum, a process responsible for activity-guided synapse removal (Huber et al., 2002; Bear et al., 2004). Fmr1 knock-out mice have exaggerated LTD, which could contribute to cognitive deficits (Huber et al., 2002; Bear et al., 2004; Koekkoek et al., 2005). These knock-out mice also display phenotypes reminiscent of those observed in human patients such as dendritic spine abnormalities and learning deficits (Greenough et al., 2001; Bakker and Oostra, 2003; Koekkoek et al., 2005).
Loss of function of the single homolog of FMR1 in Drosophila dfmr1 results in viable flies (Zhang et al., 2001). The Drosophila protein dFMRP behaves as a translational repressor (Laggerbauer et al., 2001; Li et al., 2001; Zhang et al., 2001; Reeve et al., 2005), and RNA targets have been identified using biochemical and genetic approaches (Reeve et al., 2005; Zarnescu et al., 2005). Importantly, flies lacking FMRP exhibit neuroanatomical and behavioral phenotypes that include defective neurite extension, courtship, learning, and circadian clock output (Dockendorff et al., 2002; Inoue et al., 2002; Morales et al., 2002; Pan et al., 2004). Administration of lithium or mGluR antagonists can rescue the neuroanatomical, courtship, and learning, but not the circadian, phenotypes (McBride et al., 2005). These studies indicate that, in Drosophila, as in the mouse, mGluR signaling may contribute to the altered behavior.
Another clock output component, LARK, is also an RNA-binding protein and a member of the RNA recognition motif class of proteins (Newby and Jackson, 1996). Previous studies have shown that LARK displays circadian oscillations in abundance in flies and mammals (McNeil et al., 1998; Kojima et al., 2007); in flies, overexpression of LARK in Timeless (TIM)- or pigment dispersing factor (PDF)-containing clock cells causes arrhythmic locomotor activity without affecting the molecular oscillator (Schroeder et al., 2003). These results are indicative of a role for LARK in clock output. Whereas LARK has a pan-neuronal and nuclear pattern of localization (Zhang et al., 2000), the mammalian homolog (mLARK or RBM4) has the capacity to shuttle into the cytoplasm (Lai et al., 2003), suggesting a possible role in translational control.
We report physical and functional interactions between LARK and dFMRP in vivo and suggest that the two RNA-binding proteins may cooperate to regulate the translation of certain common target RNAs.
Materials and Methods
Immunoprecipitation and immunostaining techniques.
Adult fly heads were collected and homogenized in 1 ml of ice-cold lysis buffer (10 mm Tris, pH 7.4, 150 mm NaCl, 30 mm EDTA, and 0.5% Triton X-100) with 2× complete protease inhibitors. All additional manipulations of the head lysates were performed at 4°C or on ice. Debris was pelleted, and the supernatant was collected and precleared for 1 h with 100 μl of recombinant protein G agarose (Roche Diagnostics). Anti-dFMRP or anti-LARK antibody (see source below) was incubated with recombinant protein G agarose at 4°C for 2 h. The precleared lysates were immunoprecipitated with antibody-coated recombinant protein G agarose at 4°C overnight. The precipitated complexes were used for Western blotting. Anti-dFMRP antibody was used for Western blotting at a dilution of 1:500 and anti-LARK at a dilution of 1:20,000.
Whole mounts of the adult fly brain were immunostained according to published methods (Benito et al., 2007). Antisera against PDF (anti-PDF-associated peptides from Paul Taghert, Washington University School of Medicine, St. Louis, MO), dFMRP (monoclonal 5B6 from University of Iowa Hybridoma Center, Ames, IA) and LARK (McNeil et al., 2004) were used at dilutions of 1:1000, 1:100, and 1:100, respectively.
Drosophila genetics.
Transgenic flies carrying the upstream activating sequence (UAS)–ds-lark (lark RNAiA) construct were generated using standard methods. The dfmr13 loss-of-function allele used in the experiments was a kind gift from Dr. Tom Jongens (University of Pennsylvania, Philadelphia, PA). All other lines used in this study were obtained from the Bloomington Drosophila Stock Center (Bloomington, IN). Fly strains and genetic crosses were reared on standard medium with added yeast paste at 25°C.
Bioinformatics.
To identify potential LARK/FMRP common targets, we searched the published mRNA target lists for both proteins (Zarnescu et al., 2005; Huang et al., 2007). The expression data reported by Zarnesu et al. were filtered to contain only those probe sets that have the Affymetrix “Present” call. The average value of expression was calculated for each sample condition present in the expression data. The fold change for Fmrp immunoprecipitation (IP) versus input mRNA was calculated for both the wild-type (WT) and the mutant. Probe sets showing a WT fold change of 1.4 and enriched by at least twofold compared with the mutant fold change were selected. The Flybase gene (FBgn) identifications of these probe sets were matched to the 216 known LARK target FBgn identifications identify common targets.
Behavioral analysis.
Locomotor activity was monitored in single flies using monitors from Trikinetics as described previously (Levine et al., 1994). Activity records were visualized and statistically analyzed using a Matlab-based package known as Fly Toolbox (Levine et al., 2002). Individuals were considered rhythmic if there was statistically significant evidence of periodicity using the Fly Toolbox package. The robustness of rhythmicity was assessed using the rhythmicity index (RI) of Levine et al. (2002). For behavioral analysis, flies were maintained in 12 h light/dark cycle at 25°C for ∼6 d and then transferred to constant darkness, for ∼10 d.
Scanning electron microscopy.
Scanning electron microscopy was performed using whole flies that were dehydrated in ethanol before microscopy.
Reverse transcription-PCR.
RNA samples were prepared from third-instar larvae (y w and lark null) using Trizol (Invitrogen). RNA was reverse transcribed with oligo-dT12–18 and SuperScript II (Invitrogen). PCR reactions were performed using dFMRP-specific primers.
Results
dFMRP and LARK are present in a complex in vivo
We performed a yeast two-hybrid screen to identify proteins in a Drosophila library that interact with dFMRP. Such interacting proteins might provide additional insights about the cellular and biological functions of dFMRP. We used four different dfmr1 constructs as bait against a fly library. These baits expressed the KH domains of the protein (60-1), the first 500 bp of N-terminal coding sequence (60-2), the N-terminal half of the protein (60-3), or the C-terminal half of the protein (60-4). We found that bait 60-3, which contains the FMR1/FXR interaction domain, bound to ∼30 different proteins, consistent with the known role of the dFMRP N terminus in mediating protein–protein interactions (Reeve et al., 2008). One of these proteins, detected twice in our experiments, was Drosophila LARK (supplemental Fig. 1, available at www.jneurosci.org as supplemental material).
To verify that LARK and dFMRP are found in a complex in vivo, we attempted to coimmunoprecipitate the two proteins from protein extracts of adult fly heads using anti-LARK and anti-dFMRP antibodies. In two independent experiments, anti-dFMRP precipitated dFMRP and a small amount of LARK, whereas anti-LARK brought down LARK and a small amount of dFMRP (Fig. 1 A). Neither protein was precipitated in negative control experiments (beads alone and no antibody; data not shown). The nonstoichiometric ratio of the proteins in these co-IP experiments may reflect the steady-state localization patterns of the two proteins (LARK in the nucleus, dFMRP in the cytoplasm) (Wan et al., 2000; Zhang et al., 2000; Morales et al., 2002). We postulate that the two proteins may interact only transiently when one of the two proteins shuttles to the other intracellular compartment. Studies of mLARK and FMRP shuttling behavior are consistent with this idea (Feng et al., 1997b; Lai et al., 2003).
Figure 1.
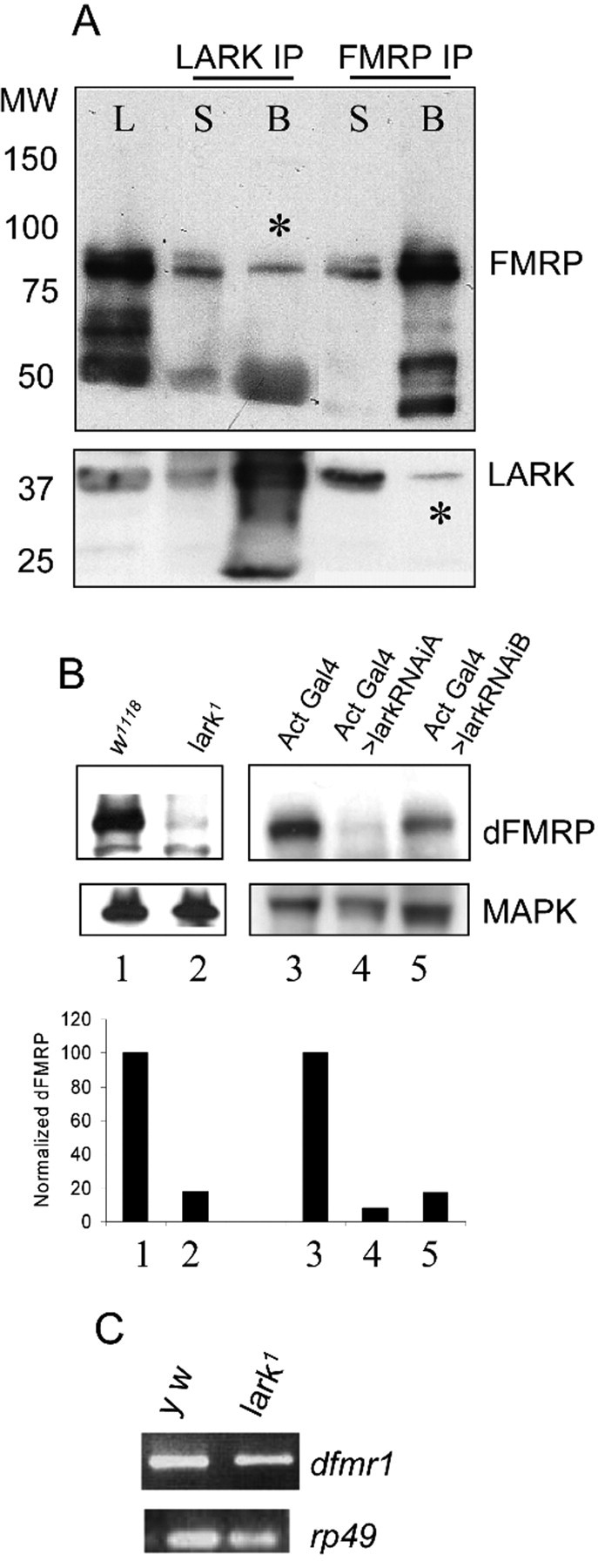
Biochemical and genetic interactions between dFMRP and LARK. A, Co-IP was performed on fly head lysates using either anti-dFMRP or anti-LARK antibody. The immunoprecipitates were then analyzed by immunoblotting using the same antibodies. Asterisks indicate dFMRP (middle lane) or LARK (far right lane) that was precipitated by antibody against the other protein. L, Lysate; B, beads plus antibody; S, supernatant after IP. B, dFMRP levels are reduced in larvae missing LARK and in pupae with reduced LARK amounts (lanes 2, 4, 5), MAPK was used as a control for protein loading. C, dfmr1 RNA levels are comparable in wild-type flies and those with reduced LARK amount. rp49 was used as control for RNA loading.
dFMRP levels are reduced in larval extracts concomitant with decreased LARK
In the course of pursuing coimmunoprecipitation experiments, we examined LARK abundance in dfmr1 mutants and dFMRP levels in larvae lacking LARK protein (null lark1 mutants survive until early pupal stages) or expressing a lark RNA interference (lark RNAiA) transgene that knocks down but does not eliminate LARK protein (V. Sundram and F. R. Jackson, unpublished results). Whereas dfmr1 mutants had normal levels of LARK (data not shown), we were surprised to discover that larvae lacking LARK and adults with reduced abundance for the protein had significantly decreased dFMRP levels (Fig. 1B, lanes 2, 4, 5) relative to controls. Interestingly, dfmr1 RNA abundance appears to be normal in lark null larvae (Fig. 1C), indicating that LARK posttranscriptionally regulates dFMRP, perhaps by controlling the translation of dfmr1 RNA or by stabilizing dFMRP protein.
Genetic interactions between dfmr1 and lark
We used two different phenotypic assays, eye morphology and circadian activity, to determine whether dfmr1 and lark genetically interact in vivo. Because both mutations alter circadian locomotor activity rhythms, we examined this behavior in different dfmr1/lark genotypes.
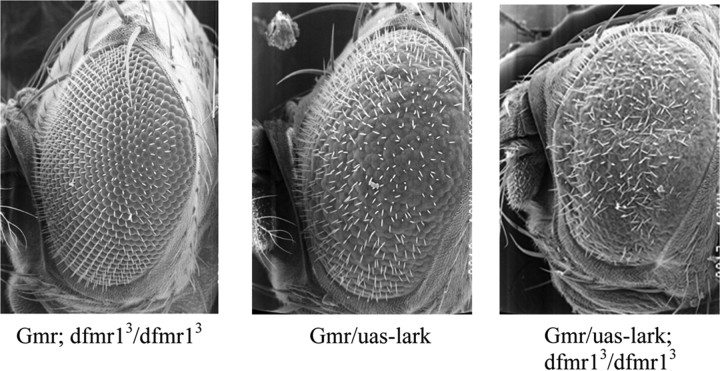
Overexpression of LARK, using an eye-specific driver [glass multimer receptor (GMR)–Gal4], leads to a rough eye phenotype in which ommatidia are fused and there is disorganization of inter-ommatidial bristles (Fig. 2 B). Whereas dfmr1 null flies have wild-type eye morphology (Fig. 2 A), flies simultaneously lacking dFMRP and overexpressing LARK have a more severe eye phenotype than control flies overexpressing LARK alone (Fig. 2, compare B, C). Figure 2 shows this interaction using the dfmr13 allele, but similar results were obtained using dfmr1113 (data not shown). These results indicate that eliminating dFMRP enhances the LARK-induced eye phenotype and suggest that dfmr1 dosage is important for LARK function. To validate the genetic interaction between dfmr1 and lark in a more biologically relevant assay, we investigated the effects of altering the levels of dFMRP and LARK on circadian rhythms.
Figure 2.
The eye phenotype resulting from LARK overexpression is enhanced by the elimination of dFMRP. A, dfmr1 null fly, displaying a wild-type eye phenotype. B, gmr–Gal4; UAS–lark fly. High level expression of LARK in the eye, using gmr–Gal4, causes a disorganized eye phenotype. Note the fused ommatidia and general disruption of inter-ommatidial bristles. C, gmr–Gal4/UAS–lark; dfmr1/dfmr1 fly with severe eye phenotype.
It has been shown previously that GAL4-driven overexpression of LARK within pacemaker cells results in arrhythmic locomotor activity for many flies (Schroeder et al., 2003). Expression in all clock cells, for example, using the tim–Gal4 driver, causes most flies to be arrhythmic. In contrast, pdf–Gal4-driven expression results in ∼50% arrhythmicity for the population (Schroeder et al., 2003). Consistent with these previous results, we found that pdf–Gal4; UAS–lark fly populations are only 42–67% rhythmic in five independent experiments, whereas pdf–Gal4 control flies (which carry only the driver transgene) are 100% rhythmic (Table 1 and data not shown). Note also that the RI, a measure of the robustness of rhythmicity, was significantly lower in pdf–Gal4; UAS–lark flies than in pdf–Gal4 controls and that circadian period was ∼1 h shorter than normal (Table 1; Fig. 3, compare A, B). In contrast, pdf–Gal4; UAS–lark fly populations that only carry one functional copy of dfmr1 (pdf–Gal4; UAS–lark/dfmr1) were 83–92% rhythmic in three different experiments. As shown in Figure 3 and Table 1, these flies are more similar to control pdf–Gal4 and pdf–Gal4; dfmr1 populations, with regard to the RI, than to pdf–Gal4; UAS–lark populations. However, the average circadian period was slightly short, similar to the rhythmic pdf–Gal4; UAS–lark flies (Table 1). These results indicate that reducing dFMRP function (i.e., heterozygosity for a dfmr1 loss-of-function allele) suppresses at least one aspect of the LARK overexpression phenotype (arrhythmicity). One interpretation of these data are that dFMRP normally promotes LARK activity in vivo, and therefore reducing dFMRP results in decreased LARK function and more robust rhythmicity.
Table 1.
Genetic interactions between dfmr and lark for circadian behavior
| Genotype | n | % rhythmic | RI ± SEM | Period ± SEM (h) |
|---|---|---|---|---|
| pdf–Gal4 | 17 | 100 | 0.43 ± 0.03 | 24.0 ± 0.07 (n = 17) |
| pdf–Gal4; UAS–larka | 26 | 50 | 0.24 ± 0.02 | 23.2 ± 0.17 (n = 13) |
| pdf–Gal4;UAS–lark/dfmr1a* | 23 | 83 | 0.40 ± 0.03c | 23.2 ± 0.10 (n = 19) |
| pdf–Gal4; dfmr1/+a | 23 | 96 | 0.51 ± 0.03 | 24.2 ± 0.04 (n = 22) |
| pdf–Gal4; UAS–lark-RNAib | 28 | 93 | 0.39 ± 0.03 | 24.6 ± 0.12 (n = 26) |
| pdf–Gal4; UAS–dfmrb | 23 | 70 | 0.19 ± 0.03 | 26.2 ± 0.42 (n = 16) |
| pdf–Gal4; UAS–dfmr/UAS–lark–RNAib | 22 | 37d | 0.13 ± 0.02 | 23.7 ± 0.29 (n = 8) |
Populations assessed behaviorally in the same experiment.
Populations assessed behaviorally in the same experiment.
Different from pdf–Gal4; UAS–lark, p < 0.001 and from pdf–Gal4; dfmr1/+, p < 0.05, ANOVA with Bonferroni's multiple-comparison test.
Different from pdf–Gal4; UAS–lark–RNAi (p < 0.01) and pdf–Gal4; UAS–dfmr (p < 0.02).
Figure 3.
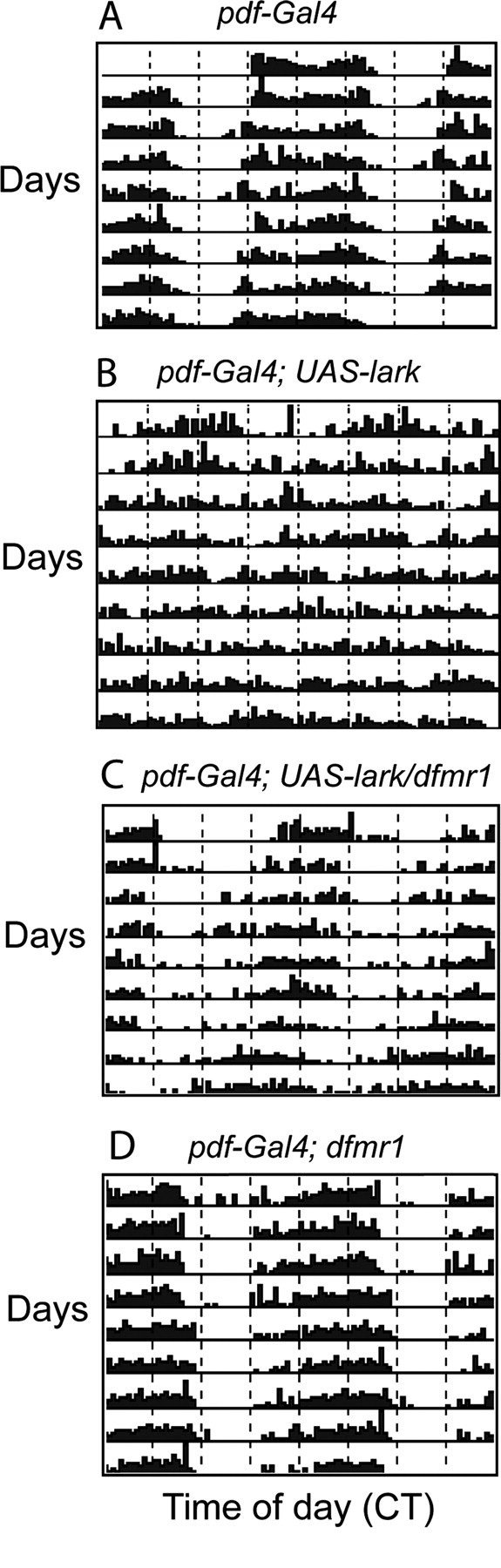
Reducing dfmr1 expression suppresses the rhythm phenotype of flies overexpressing LARK. A, Control pdf–Gal4 fly with a wild-type locomotor activity rhythm. B, Overexpression of LARK causes arrhythmicity. C, Heterozygosity for a dfmr1 null mutation ameliorates the rhythm phenotype of flies overexpressing LARK. D, pdf–Gal4; dfmr1 fly (heterozygous for dfmr1 null) with a wild-type activity rhythm. CT, Circadian time.
In a second set of experiments, we examined flies overexpressing dfmr1 (pdf–Gal4; UAS–dfmr1). We found that most were rhythmic (in four experiments, 68–87% rhythmic, one experiment shown in Table 1) and that circadian period was lengthened as reported previously (Dockendorff et al., 2002); however, the percentage rhythmicity and robustness of rhythms were slightly reduced relative to flies carrying only the pdf–Gal4 driver (Table 1). We also found that pdf–Gal4; UAS–lark–RNAi flies, which express a lark RNAi transgene that knocks down but does not eliminate LARK within the PDF cells (Sundram and Jackson, unpublished results), had normal rhythmicity (93 and 100% rhythmic in two independent experiments; one dataset shown in Table 1). In contrast, pdf–Gal4; UAS–dfmr1/UAS–lark–RNAi flies, which express both transgenes, were poorly rhythmic (37–47% rhythmic in two independent experiments; one shown in Table 1). Thus, partial loss of lark function enhances the overexpression phenotype of pdf–Gal4; UAS–dfmr1 flies, which is consistent with the idea that LARK normally antagonizes dFMRP function.
The genetic interactions between lark and dfmr1, based on circadian behavior, strongly suggest that LARK and FMRP cooperate, in vivo, to regulate behavior. However, our previous Western blotting experiments had suggested that LARK stabilized dFMRP (Fig. 1B), whereas the behavioral genetic interactions (enhancement of dFMRP overexpression by decreased LARK and suppression of LARK overexpression by decreased dFMRP) are not in accord with such a simple model. Because the behavioral results used the pdf–Gal4 driver, we wanted to determine how LARK and FMRP biochemically interact within PDF neurons. To examine potential protein interactions in the PDF cells, we conducted immunostaining experiments with LARK, dFMRP, and PDF antibodies using flies with increased (pdf > UAS–lark) or decreased (pdf > larki) LARK levels (supplemental Fig. 2, available at www.jneurosci.org as supplemental material). We found that knockdown of LARK in either the large or small PDF neurons (small LNv or large LNv of supplemental Fig. 2, available at www.jneurosci.org as supplemental material) resulted in an increase in dFMRP immunosignal (green signal) within PDF cells (blue signal) rather than a decrease, although the increased signal within the small LNvs was not statistically significant given the sample sizes in our experiments (supplemental Fig. 2B, available at www.jneurosci.org as supplemental material). Furthermore, overexpression of LARK in the PDF cells was associated with decreased FMRP immunosignal rather than an increase (supplemental Fig. 2A,B, available at www.jneurosci.org as supplemental material). It is clear from these experiments that LARK does not simply stabilize dFMRP in the PDF neurons and that decreases in dFMRP observed in whole animal extracts, with LARK knockdown or knock-out, must reflect stabilization of the protein by LARK in non-PDF cells (and perhaps in non-neural tissues). We address the interpretation of these results in Discussion.
Discussion
Our studies demonstrate both physical and genetic interactions between dFMRP and LARK. On the basis of circadian behavioral phenotypes, we infer that dFMRP promotes LARK function in vivo, whereas LARK may antagonize dFMRP function. Larvae lacking LARK or with severe deficits for the protein in all tissues have significantly decreased dFMRP levels. However, a knockdown of LARK specifically within the PDF clock neurons leads to increased dFMRP immunosignal in that cell type. We interpret these results to mean that LARK stabilizes dFMRP in certain tissues but apparently not in the PDF cells. It is expected that a knockdown of LARK and the concomitant increased dFMRP within PDF cells will genetically enhance the effects of dFMRP overexpression (the observed result for behavior). If LARK normally antagonizes dFMRP function, then decreased amounts of LARK are predicted to lead to increased dFMRP activity in addition to increasing dFMRP amount. It is difficult to rationalize the suppression of LARK overexpression that is observed in pdf–Gal4; UAS–lark/dfmr1 flies (i.e., better rhythmicity) (Fig. 3) based simply on decreased dFMRP amounts. In principle, such a decrease ought to result in worse rhythmicity unless dFMRP regulates LARK activity. Thus, we favor a model wherein LARK activity is promoted by dFMRP binding; the reduced dFMRP in pdf–Gal4;UAS–lark/dfmr1 flies is then expected to suppress the effects of LARK overexpression. Alternative models specifying differential effects of the two RNA-binding proteins on target RNA molecules may also explain the interactions, although it is premature to specify explicit biochemical models.
With respect to eye development, our results suggest that loss of dFMRP enhances a LARK-induced rough-eye phenotype, as if dFMRP normally represses LARK function in this tissue. The different genetic interactions observed for behavior versus eye development may reflect the regulation of distinct target RNAs in retinal tissue versus the PDF cell population. Alternatively, the presence of additional proteins in the LARK/dFMRP complex may modulate the activity of either or both of the RNA-binding proteins in a tissue-dependent manner. Given the plethora of functions to which RNA-binding proteins contribute in different cellular compartments, such as processing, export, stability, localization, and translation of mRNAs (Dreyfuss et al., 2002), which can result in promotion or repression of gene expression, it is impossible to assign meaning to the different genetic interactions in the eye versus clock cells without additional mechanistic understanding of the LARK–dFMRP interaction and knowledge about the relevant target mRNAs.
Target RNAs have been reported for dFMRP (Reeve et al., 2005; Zarnescu et al., 2005), although only a few of them have been validated in vivo by genetic analysis (Reeve et al., 2005). Similarly, target RNAs were recently reported for fly LARK; this RNA-binding protein is thought to be associated with as many as 200 different RNAs in vivo (Huang et al., 2007). For both dFMRP and LARK, a number of putative targets are known to have circadian functions or to display circadian changes in abundance. In addition, there are at least 12 targets that overlap from the two independent studies (Y. Huang, and F. R. Jackson, unpublished results) (for data mining procedures, see Materials and Methods); it is possible that these RNAs represent common targets that are modulated by both proteins. Finally, because dFMRP is known to be in a complex with lethal giant larvae (Lgl) (Zarnescu et al., 2005) and interacts with it to regulate eye development, it might be the case that all three proteins, LARK, dFMRP, and Lgl, execute certain functions together, in vivo, perhaps by regulating common targets. Presumably, certain targets encode proteins that function as elements of the clock output pathways regulating behavior, whereas others may mediate functions relevant for eye development.
Of related interest, it has been shown that mammalian and fly FMRP are associated with Argonaute (Ago) proteins and micro-RNAs (miRNAs) in vivo and that Ago1 is necessary for FMRP function in vivo in Drosophila (Caudy et al., 2002; Jin and Warren, 2003). More recently, it was shown that human RBM4 (hLARK) is associated with Ago/FMRP/miRNA complexes and that knockdown of RBM4 in human cells results in altered miRNA-mediated gene regulation for several different mRNA targets (Höck et al., 2007). Another recent study demonstrated that a significant number of fly LARK RNA targets contain consensus binding sites for miRNAs, suggesting that the fly protein may also be required for miRNA-mediated gene regulation (Huang et al., 2007). Thus, dFMRP and LARK may function together with miRNAs in the regulation of certain RNA targets.
How might LARK and dFMRP function together to regulate development or behavior? Although the proteins are predominantly located in distinct intracellular compartments, LARK in the nucleus and dFMRP in the cytoplasm, both vertebrate homologs are known to shuttle between the two compartments (Feng et al., 1997b; Lai et al., 2003), and they may function together in the cytoplasm to regulate the translation of target RNAs. Indeed, cell-based studies have suggested recently that mouse LARK (mLARK or Rbm4) becomes phosphorylated, in a mitogen-activated protein kinase (MAPK)-dependent manner, in response to cell stress and then shuttles into the cytoplasm to regulate the internal ribosomal entry site-dependent translation of target RNAs (Lin et al., 2007). mLARK can bind to a sequence within the 3′ untranslated region of the mouse Per1 mRNA (a known clock factor), and it has been suggested that the observed rhythm in LARK abundance within the mouse suprachiasmatic nuclei serves to modulate the translation and circadian expression of PER1 protein (Kojima et al., 2007). Of interest, mLARK overexpression in cell-based assays increases PER (Period) level and slightly lengthens circadian period, whereas increased fLARK expression in vivo seems to shorten period (Table 1). We do not understand this species difference, but the fLARK result is consistent with a promotion of PER expression, which in flies is predicted to shorten circadian period (Baylies et al., 1987). Nonetheless, our results suggest that LARK and dFMRP cooperate within certain cell types to posttranscriptionally regulate target RNAs that are relevant for circadian and other biological functions.
A role for Fmr1 in mammalian circadian behavior has been established recently (Zhang et al., 2008). Mice that lack Fmr1 exhibit a short circadian period in free-running conditions, whereas those lacking both Fmr1 and Fxr2 display arrhythmic locomotor activity as measured by wheel running during light/dark conditions. This anomaly appears to be attributable to clock output, because the suprachiasmatic nuclei of double knock-out animals appear to function normally. These data underscore the relevance of the Fmr/Fxr gene family to circadian control. The finding that LARK and dFMRP interact in the fly suggests that mLARK and FMRP/FXR2P interactions may also be relevant in the mouse in exerting circadian control.
Finally, the importance of sleep in learning and memory consolidation and also in the generation/formation of new memory (Walker and Stickgold, 2004; Stickgold, 2005; Yoo et al., 2007) suggests that the abnormal circadian clock and sleep patterns in FXS patients could contribute to their cognitive impairment.
Footnotes
D.L.N. was supported by National Institutes of Health Grant RO1 HD038038, the Baylor College of Medicine Mental Retardation and Developmental Disabilities Research Center Grant P50 HD024064, and the Baylor College of Medicine–Emory Fragile X Research Center. J.B was supported by National Institutes of Health Grant NS42179. Work on dFMRP by F.R.J. was supported by National Institutes of Health Grants R01 HL59873 and P30 NS047243 to the Tufts Center for Neuroscience Research. We thank Dr. Jim Barrish for help with scanning electron microscopy. dFMRP antibody was a kind gift from Dr. Gideon Dreyfuss. We also thank members of the Jackson, Nelson, and Botas laboratories for assistance, Jackie Lane (Tufts University Genetics Program) for help with co-IP experiments, and Lax Iyer and Chris Parkin (Tufts Center for Neuroscience Research Computational Genomics Core) for help with statistics and bioinformatics.
References
- Bakker CE, Oostra BA. Understanding fragile X syndrome: insights from animal models. Cytogenet Genome Res. 2003;100:111–123. doi: 10.1159/000072845. [DOI] [PubMed] [Google Scholar]
- Baylies MK, Bargiello TA, Jackson FR, Young MW. Changes in abundance or structure of the per gene product can alter periodicity of the Drosophila clock. Nature. 1987;326:390–392. doi: 10.1038/326390a0. [DOI] [PubMed] [Google Scholar]
- Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- Benito J, Zheng H, Hardin PE. PDP1epsilon functions downstream of the circadian oscillator to mediate behavioral rhythms. J Neurosci. 2007;27:2539–2547. doi: 10.1523/JNEUROSCI.4870-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown V, Jin P, Ceman S, Darnell JC, O'Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, Darnell RB, Warren ST. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- Caudy AA, Myers M, Hannon GJ, Hammond SM. Fragile X-related protein and VIG associate with the RNA interference machinery. Genes Dev. 2002;16:2491–2496. doi: 10.1101/gad.1025202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- Dockendorff TC, Su HS, McBride SM, Yang Z, Choi CH, Siwicki KK, Sehgal A, Jongens TA. Drosophila lacking dfmr1 activity show defects in circadian output and fail to maintain courtship interest. Neuron. 2002;34:973–984. doi: 10.1016/s0896-6273(02)00724-9. [DOI] [PubMed] [Google Scholar]
- Dreyfuss G, Kim VN, Kataoka N. Messenger-RNA-binding proteins and the messages they carry. Nat Rev Mol Cell Biol. 2002;3:195–205. doi: 10.1038/nrm760. [DOI] [PubMed] [Google Scholar]
- Feng Y, Absher D, Eberhart DE, Brown V, Malter HE, Warren ST. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol Cell. 1997a;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- Feng Y, Gutekunst CA, Eberhart DE, Yi H, Warren ST, Hersch SM. Fragile X mental retardation protein: nucleocytoplasmic shuttling and association with somatodendritic ribosomes. J Neurosci. 1997b;17:1539–1547. doi: 10.1523/JNEUROSCI.17-05-01539.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould EL, Loesch DZ, Martin MJ, Hagerman RJ, Armstrong SM, Huggins RM. Melatonin profiles and sleep characteristics in boys with fragile X syndrome: a preliminary study. Am J Med Genet. 2000;95:307–315. [PubMed] [Google Scholar]
- Greenough WT, Klintsova AY, Irwin SA, Galvez R, Bates KE, Weiler IJ. Synaptic regulation of protein synthesis and the fragile X protein. Proc Natl Acad Sci USA. 2001;98:7101–7106. doi: 10.1073/pnas.141145998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höck J, Weinmann L, Ender C, Rüdel S, Kremmer E, Raabe M, Urlaub H, Meister G. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Genova G, Roberts M, Jackson FR. The LARK RNA-binding protein selectively regulates the circadian eclosion rhythm by controlling E74 protein expression. PLoS ONE. 2007;2:e1107. doi: 10.1371/journal.pone.0001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber KM, Gallagher SM, Warren ST, Bear MF. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc Natl Acad Sci USA. 2002;99:7746–7750. doi: 10.1073/pnas.122205699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue S, Shimoda M, Nishinokubi I, Siomi MC, Okamura M, Nakamura A, Kobayashi S, Ishida N, Siomi H. A role for the Drosophila fragile X-related gene in circadian output. Curr Biol. 2002;12:1331–1335. doi: 10.1016/s0960-9822(02)01036-9. [DOI] [PubMed] [Google Scholar]
- Jin P, Warren ST. New insights into fragile X syndrome: from molecules to neurobehaviors. Trends Biochem Sci. 2003;28:152–158. doi: 10.1016/S0968-0004(03)00033-1. [DOI] [PubMed] [Google Scholar]
- Khandjian EW, Corbin F, Woerly S, Rousseau F. The fragile X mental retardation protein is associated with ribosomes. Nat Genet. 1996;12:91–93. doi: 10.1038/ng0196-91. [DOI] [PubMed] [Google Scholar]
- Koekkoek SK, Yamaguchi K, Milojkovic BA, Dortland BR, Ruigrok TJ, Maex R, De Graaf W, Smit AE, VanderWerf F, Bakker CE, Willemsen R, Ikeda T, Kakizawa S, Onodera K, Nelson DL, Mientjes E, Joosten M, De Schutter E, Oostra BA, Ito M, De Zeeuw CI. Deletion of FMR1 in Purkinje cells enhances parallel fiber LTD, enlarges spines, and attenuates cerebellar eyelid conditioning in Fragile X syndrome. Neuron. 2005;47:339–352. doi: 10.1016/j.neuron.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Kojima S, Matsumoto K, Hirose M, Shimada M, Nagano M, Shigeyoshi Y, Hoshino S, Ui-Tei K, Saigo K, Green CB, Sakaki Y, Tei H. LARK activates posttranscriptional expression of an essential mammalian clock protein, PERIOD1. Proc Natl Acad Sci USA. 2007;104:1859–1864. doi: 10.1073/pnas.0607567104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laggerbauer B, Ostareck D, Keidel EM, Ostareck-Lederer A, Fischer U. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum Mol Genet. 2001;10:329–338. doi: 10.1093/hmg/10.4.329. [DOI] [PubMed] [Google Scholar]
- Lai MC, Kuo HW, Chang WC, Tarn WY. A novel splicing regulator shares a nuclear import pathway with SR proteins. EMBO J. 2003;22:1359–1369. doi: 10.1093/emboj/cdg126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine JD, Casey CI, Kalderon DD, Jackson FR. Altered circadian pacemaker functions and cyclic AMP rhythms in the Drosophila learning mutant dunce. Neuron. 1994;13:967–974. doi: 10.1016/0896-6273(94)90262-3. [DOI] [PubMed] [Google Scholar]
- Levine JD, Funes P, Dowse HB, Hall JC. Signal analysis of behavioral and molecular cycles. BMC Neurosci. 2002;3:1. doi: 10.1186/1471-2202-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhang Y, Ku L, Wilkinson KD, Warren ST, Feng Y. The fragile X mental retardation protein inhibits translation via interacting with mRNA. Nucleic Acids Res. 2001;29:2276–2283. doi: 10.1093/nar/29.11.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JC, Hsu M, Tarn WY. Cell stress modulates the function of splicing regulatory protein RBM4 in translation control. Proc Natl Acad Sci USA. 2007;104:2235–2240. doi: 10.1073/pnas.0611015104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride SM, Choi CH, Wang Y, Liebelt D, Braunstein E, Ferreiro D, Sehgal A, Siwicki KK, Dockendorff TC, Nguyen HT, McDonald TV, Jongens TA. Pharmacological rescue of synaptic plasticity, courtship behavior, and mushroom body defects in a Drosophila model of fragile X syndrome. Neuron. 2005;45:753–764. doi: 10.1016/j.neuron.2005.01.038. [DOI] [PubMed] [Google Scholar]
- McNeil GP, Zhang X, Genova G, Jackson FR. A molecular rhythm mediating circadian clock output in Drosophila. Neuron. 1998;20:297–303. doi: 10.1016/s0896-6273(00)80457-2. [DOI] [PubMed] [Google Scholar]
- McNeil GP, Smith F, Galioto R. The Drosophila RNA-binding protein Lark is required for the organization of the actin cytoskeleton and Hu-li tai shao localization during oogenesis. Genesis. 2004;40:90–100. doi: 10.1002/gene.20069. [DOI] [PubMed] [Google Scholar]
- Miano S, Bruni O, Elia M, Scifo L, Smerieri A, Trovato A, Verrillo E, Terzano MG, Ferri R. Sleep phenotypes of intellectual disability: a polysomnographic evaluation in subjects with Down syndrome and Fragile-X syndrome. Clin Neurophysiol. 2008;119:1242–1247. doi: 10.1016/j.clinph.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Morales J, Hiesinger PR, Schroeder AJ, Kume K, Verstreken P, Jackson FR, Nelson DL, Hassan BA. Drosophila fragile X protein, DFXR, regulates neuronal morphology and function in the brain. Neuron. 2002;34:961–972. doi: 10.1016/s0896-6273(02)00731-6. [DOI] [PubMed] [Google Scholar]
- Newby LM, Jackson FR. Regulation of a specific circadian clock output pathway by lark, a putative RNA-binding protein with repressor activity. J Neurobiol. 1996;31:117–128. doi: 10.1002/(SICI)1097-4695(199609)31:1<117::AID-NEU10>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- O'Donnell WT, Warren ST. A decade of molecular studies of fragile X syndrome. Annu Rev Neurosci. 2002;25:315–338. doi: 10.1146/annurev.neuro.25.112701.142909. [DOI] [PubMed] [Google Scholar]
- Pan L, Zhang YQ, Woodruff E, Broadie K. The Drosophila fragile X gene negatively regulates neuronal elaboration and synaptic differentiation. Curr Biol. 2004;14:1863–1870. doi: 10.1016/j.cub.2004.09.085. [DOI] [PubMed] [Google Scholar]
- Reeve SP, Bassetto L, Genova GK, Kleyner Y, Leyssen M, Jackson FR, Hassan BA. The Drosophila fragile X mental retardation protein controls actin dynamics by directly regulating profilin in the brain. Curr Biol. 2005;15:1156–1163. doi: 10.1016/j.cub.2005.05.050. [DOI] [PubMed] [Google Scholar]
- Reeve SP, Lin X, Sahin BH, Jiang F, Yao A, Liu Z, Zhi H, Broadie K, Li W, Giangrande A, Hassan BA, Zhang YQ. Mutational analysis establishes a critical role for the N terminus of fragile X mental retardation protein FMRP. J Neurosci. 2008;28:3221–3226. doi: 10.1523/JNEUROSCI.5528-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder AJ, Genova GK, Roberts MA, Kleyner Y, Suh J, Jackson FR. Cell-specific expression of the lark RNA-binding protein in Drosophila results in morphological and circadian behavioral phenotypes. J Neurogenet. 2003;17:139–169. [PubMed] [Google Scholar]
- Stickgold R. Sleep-dependent memory consolidation. Nature. 2005;437:1272–1278. doi: 10.1038/nature04286. [DOI] [PubMed] [Google Scholar]
- Walker MP, Stickgold R. Sleep-dependent learning and memory consolidation. Neuron. 2004;44:121–133. doi: 10.1016/j.neuron.2004.08.031. [DOI] [PubMed] [Google Scholar]
- Wan L, Dockendorff TC, Jongens TA, Dreyfuss G. Characterization of dFMR1, a Drosophila melanogaster homolog of the fragile X mental retardation protein. Mol Cell Biol. 2000;20:8536–8547. doi: 10.1128/mcb.20.22.8536-8547.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo SS, Hu PT, Gujar N, Jolesz FA, Walker MP. A deficit in the ability to form new human memories without sleep. Nat Neurosci. 2007;10:385–392. doi: 10.1038/nn1851. [DOI] [PubMed] [Google Scholar]
- Zarnescu DC, Jin P, Betschinger J, Nakamoto M, Wang Y, Dockendorff TC, Feng Y, Jongens TA, Sisson JC, Knoblich JA, Warren ST, Moses K. Fragile X protein functions with lgl and the par complex in flies and mice. Dev Cell. 2005;8:43–52. doi: 10.1016/j.devcel.2004.10.020. [DOI] [PubMed] [Google Scholar]
- Zhang J, Fang Z, Jud C, Vansteensel MJ, Kaasik K, Lee CC, Albrecht U, Tamanini F, Meijer JH, Oostra BA, Nelson DL. Fragile X related proteins regulate mammalian circadian behavioral rhythms. Am J Hum Genet. 2008;83:43–52. doi: 10.1016/j.ajhg.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, McNeil GP, Hilderbrand-Chae MJ, Franklin TM, Schroeder AJ, Jackson FR. Circadian regulation of the lark RNA-binding protein within identifiable neurosecretory cells. J Neurobiol. 2000;45:14–29. doi: 10.1002/1097-4695(200010)45:1<14::aid-neu2>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- Zhang YQ, Bailey AM, Matthies HJ, Renden RB, Smith MA, Speese SD, Rubin GM, Broadie K. Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell. 2001;107:591–603. doi: 10.1016/s0092-8674(01)00589-x. [DOI] [PubMed] [Google Scholar]