Abstract
CD4 T lymphocytes regulate the adaptive immune response to most viruses, both by providing help to CD8 T cells and B cells as well as through direct antiviral activity. Currently, no mouse cytomegalovirus (MCMV)-specific CD4 T cell responses are known. In this study, we identify and characterize 15 I-Ab-restricted CD4 T cell responses specific for MCMV epitopes. CD4 T cells accumulate to high levels in the spleen and lungs during acute infection and produce multiple cytokines (IFN-γ, TNF, IL-2, IL-10, and IL-17). Interestingly, IL-17 and IFN-γ production within epitope-specific cells was found to be mutually exclusive. CD4 T cells recognizing a peptide derived from m09 were only detectable at later times of infection and displayed a unique cytokine production profile. In total, this study reveals that the MCMV-specific CD4 T cell response is complex and functionally diverse, highlighting its important role in controlling this persistent pathogen.
Cytomegalovirus (β-herpesvirus) establishes a lifelong, asymptomatic infection in immunocompetent hosts, and both innate and adaptive immunity are critical for viral control. In humans, CMV-specific CD4 T cells correlate with protection from disease (1), and CD4 T cells are required for control of mouse CMV (MCMV)3 replication in the salivary gland and lung (2-4). The MCMV-specific Ab response is also dependent upon CD4 T cells and provides protection against recurrent, but not acute, infection (2, 5). Adoptive transfer of virus-specific CD8 T cells protects against CMV-mediated disease in immunocompromised humans, and their maintenance requires human CMV-specific CD4 T cells (6). Similarly, the transfer of virus-specific CD8 T cells protects immunocompromised mice from disease (7), but acute MCMV infection is effectively controlled when CD8 T cells are absent in immunocompetent mice (3, 8), highlighting a role for CD4 T cells in healthy mice.
Approximately 4-5% of the entire CD4 and CD8 T cell compartment is specific for HCMV in seropositive humans (9) and can increase dramatically in the elderly (10, 11). Specific subpopulations of MCMV-specific CD8 T cells also increase over time (e.g., “memory inflation”) (12-15). The phenotype of inflating CD8 T cells is largely “effector-memory”-like (IL-2-CD62LlowCD127-), whereas noninflationary CD8 T cells resemble “central-memory” cells (IL-2-expressing CD62Lhigh CD127high cells) (16).
To gain more insight into the CD4 T cell response to MCMV infection, we set out to identify virus peptide-specific CD4 T cell responses restricted by MHC class II (I-Ab). Using a newly developed algorithm, we identified 15 MCMV peptide-specific CD4 T cell responses. Most of these responses displayed “classical” kinetics for the expansion/contraction phase and produced a varied pattern of cytokine expression upon reactivation. In contrast, a CD4 T cell response specific for the m09133-147 peptide was found to be inflationary and displayed a more central-memory phenotype, producing large amounts of IL-2. This study indicates that MCMV-specific CD4 T cell responses are diverse and polyfunctional and provides insight into how these lymphocytes may regulate the pathogenesis of CMV infection.
Materials and Methods
Mice and infection
C57BL/6 mice were purchased from The Jackson Laboratory. Mice aged 8-12 wk were infected i.p. with 5 × 104 PFU of MCMV (Smith strain; American Type Culture Collection). Virus was prepared from salivary glands of infected BALB/c mice and titered on 3T3 cells as described (17). All studies were conducted at the La Jolla Institute for Allergy and Immunology (La Jolla, CA) in facilities approved by the American Association for the Accreditation of Laboratory Animal Care and according to Institutional Animal Care and Use Committee-approved animal protocols.
Peptide predictions and synthesis
Candidate 15-mer class II epitopes were predicted as described (18), by converting the predictions into a ranking of candidate peptides. From all of the predicted MCMV open reading frames (ORFs) (19), the top 100 predicted I-Ab restricted peptides from the entire MCMV genome were synthesized. In addition, the top 100 peptides from five MCMV proteins that elicit immunodominant CD8 T cell responses (i.e., M45, M57, m139, M38, and IE3) (16) were also synthesized (∼80% pure) (Pepscan Systems). Of these 200 peptides, 12 were predicted by both methods and the resulting 188 unique “crude” peptides were tested individually. The 15 MCMV-specific peptides that showed specific CD4 T cell reactivity after this initial analysis were then synthesized as highly purified peptides by A&A Systems, confirmed by mass spectrometry, and used in future experiments.
Intracellular cytokine staining and flow cytometry
Single-cell suspensions of spleens, lymph nodes, and salivary glands from MCMV-infected mice were stimulated in 96-well plates (1.5 × 106/well) with 3 μg/ml peptide for 8 h (the last 6 h in the presence of 1 μg/ml brefeldin A (Golgiplug; BD Pharmingen) at 37°C. Subsequently, cells were stained in PBS containing 1% FCS and 0.05% sodium azide (FACS buffer) with mAbs specific for CD4 (clone RM4-5) and CD8 (clone 53-6.7), fixed and permeabilized according to Cytofix/Cytoperm kit instructions (BD Pharmingen), and intracellular cytokine staining was performed for IFN-γ (clone XMG1.2), IL-2 (clone JES6-5H4), IL-10 (clone JES5-16E3), IL-17 (clone TC11-18H10.1), and/or TNF (clone MP6-XT22). All Abs were purchased from either BD Pharmingen or eBioscience. A LSRII flow cytometer with Diva software was used (BD Pharmingen) and data were analyzed with FlowJo software (Tree Star). Two independent experiments were performed for each peptide in multiple mice.
Results
Identification of MCMV-specific I-Ab-restricted CD4 T cell epitopes
To date, no MCMV epitope-specific CD4 T cell responses have been described. To identify class II (I-Ab)-restricted MCMV peptide epitopes, all of the MCMV ORFs were used for algorithm-based predictions, and the 100 most highly ranked 15-mer peptides were synthesized. In addition, the top 100 ranked 15-mer peptides predicted from five MCMV proteins that elicit immunodominant CD8 T cell responses were also synthesized (M45, M57, m139, M38, and IE3) (16). The ability of these peptides to induce MCMV-specific CD4 T cell activation was determined by the restimulation of splenocytes and intracellular cytokine staining for IFN-γ 8 days after MCMV infection of C57BL/6 mice (Fig. 1A). A total of 14 peptides derived from 10 distinct MCMV proteins activated CD4 T cells above the threshold value of 0.015% (3× over background) and did not activate CD8 T cells (Table I and Fig. 1). The responses specific for peptides m18872-886, M25409-423, m139560-574, m141181-195, and m14224-38 comprised >0.1% of total splenic CD4 T cells. Additional responses ranging from 0.02 to 0.1% were detected for M45102-116, M45192-206, M45239-253, M45796-810, M83229-307, M104261-275, M122/IE3390-404, m139497-511, and m1461-15. The sum of these individual responses represented ∼1.5% of the total splenic CD4 T cells following MCMV infection. The entire MCMV-specific CD4 T cell population in the spleen was estimated to be ∼6% of the total population 8 days after MCMV infection based on restimulation with PMA/ionomycin (∼9.5% IFN-γ+ CD4 T cells in MCMV-infected mice and ∼3.5% IFN-γ+ in naive mice), indicating that ∼25% of the MCMV-specific CD4 T cell response is likely detected using these 14 epitopes.
FIGURE 1.
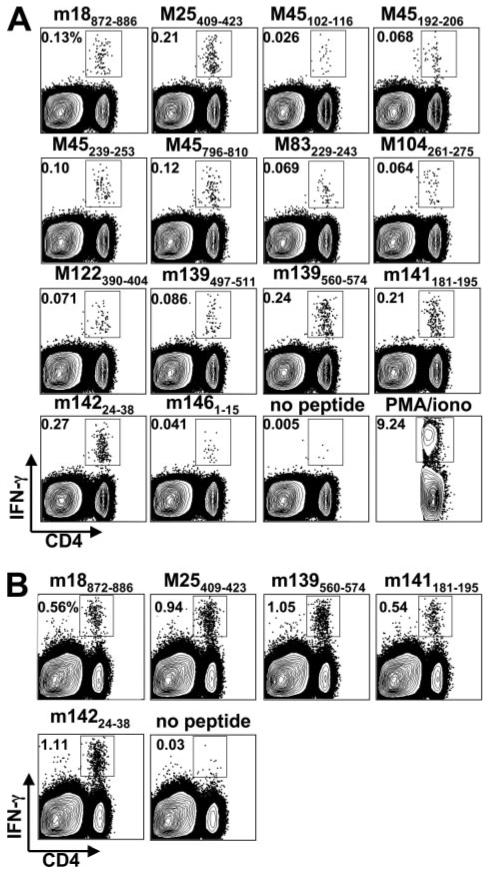
Identification of MCMV-specific CD4 T cells. A, Intracellular cytokine staining for IFN-γ after stimulation with the indicated MHCII binding peptides was performed on splenocytes from MCMV-infected mice. B, Lymphocytes were isolated from the lung and IFN-γ production by CD4 T cells was assessed after stimulation with the indicated MCMV peptides. Analysis was done 8 days postinfection. Flow plots show the IFN-γ-positive cells within the total cell population except in the case of PMA/ionomycin, where only the gated CD4 T cell population is shown. Histogram numbers indicate the number of IFN-γ-producing cells within the CD4 T cell population. All data are representative of three or four mice per group from at least two separate experiments for this and the other two figures.
Table I.
MCMV peptide-specific CD4 T cell responses
| Protein | Epitope | Sequence | ICCSa | Protein Family/Function | Human CMV Homologue |
|---|---|---|---|---|---|
| m09 | 133-147 | GYLYIYPSAGNSFDL | ND | Glycoprotein family, nonessential | |
| m18 | 872-886 | NERAKSPAAMTAEDE | 0.177 ± 0.050 | Early phase gene | |
| M25 | 409-423 | NHLYETPISATAMVI | 0.264 ± 0.072 | UL25 family homologue, Tegument protein | UL25(GF1) |
| M45 | 102-116 | AVSAANAAVNAAAAA | 0.026 ± 0.006 | Ribonucleotide reductase homologue | UL45 |
| M45 | 192-206 | TPAATTPAATAVENR | 0.046 ± 0.017 | UL45 | |
| M45 | 239-253 | QATPSTPIPIPAPRC | 0.086 ± 0.022 | UL45 | |
| M45 | 796-810 | RPAVCGPGVSVVSGG | 0.073 ± 0.037 | UL45 | |
| M83 | 229-243 | TLRYAKANGTPPDSL | 0.048 ± 0.016 | Virion matrix phosphoprotein | UL83 (pp65) |
| M104 | 261-275 | LKRFIYAEPTILEEE | 0.038 ± 0.018 | Structural protein | UL104 |
| M122/IE3 | 390-404 | DRTAGGYVAPNAHKK | 0.041 ± 0.016 | Essential immediate early gene | UL122(IE2) |
| m139 | 497-511 | GSPWKTSAVTVSRKA | 0.059 ± 0.021 | US22 family homologue | US22 |
| m139 | 560-574 | TRPYRYPRVCDASLS | 0.221 ± 0.051 | US22 family homologue | US22 |
| m141 | 181-195 | LVVFSDPNADAATSV | 0.173 ± 0.047 | US22 family homologue | US24 |
| m142 | 24-38 | RSRYLTAAAVTAVLQ | 0.261 ± 0.038 | US22 family homologue, with m143 block of PKR activation | US26 |
| m146 | 1-15 | MTTPSPIRVRAIAVW | 0.029 ± 0.009 | Member of MGP family m145 |
Intracellular cytokine staining (ICCS) at day 8 postinfection; showing percentage of IFN-γ+ within CD4 T ± cells SD. ND, Not detectable at day 8.
Organ distribution of MCMV-specific CD4 T cells
Cellular immune control of MCMV replication during infection is organ specific, with CD4 T cells playing a very important role in both the lungs and salivary gland (2-4). Consequently, the numbers of MCMV-specific CD4 T cells were examined in the lungs, salivary glands, and their draining lymph nodes 8 days postinfection. A high frequency of these cells were found to be present in the lung at day 8 postinfection (Fig. 1B) and made up a significantly higher percentage of the total CD4 T cells when compared with the spleen. In contrast, only very few MCMV-specific CD4 T cells could be detected in the salivary glands and the draining lymph nodes at this time point (10- to 20-fold lower percentages than in the spleen; data not shown). Thus, MCMV-specific CD4 T cells reside in both lymphoid and nonlymphoid organs and are present in high numbers at times in the lung, where they are critical for viral control.
Cytokine expression profiles of MCMV-specific CD4 T cells
Diverse subsets of CD4 T cells can be identified based on the spectrum of cytokines they produce when encountering Ag (i.e., Th1, Th2, Th17, and regulatory T cells). To identify which subsets develop during acute MCMV infection, we examined the ability of MCMV-specific CD4 T cells to produce TNF, IL-2, IL-10, and IL-17 after peptide stimulation (Fig. 2). During the acute phase of infection, we found that 30-40% of the MCMV-specific, IFN-γ expressing CD4 T cells also produced TNF. Upon restimulation, the percentage of peptide-specific CD4 T cells that produced IL-2 varied between ∼0.1 and 0.2% (tested for m18872-886, M25409-423, m139560-574, m141181-195, and m14224-38); IL-2 was expressed preferentially in cells that did not express IFN-γ, but a significant percentage of cells did coexpress these two cytokines (∼20-30%). IL-10, a cytokine known to dampen Th1 responses and promote persistent MCMV replication in the salivary gland of B6 mice (20), showed a very similar expression pattern to that of IL-2. In contrast to IL-2 and IL-10, CD4 T cells that expressed IL-17 were never found to coexpress IFN-γ and were present at 40-60% as compared with IFN-γ-expressing cells. Collectively, these data show that polyfunctional subsets of Ag-specific CD4 T cells develop during MCMV infection.
FIGURE 2.
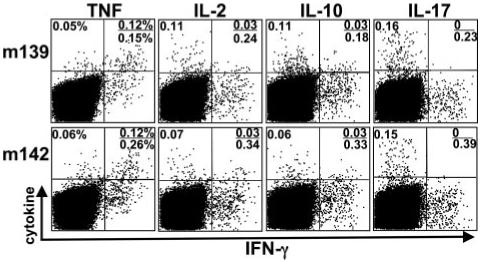
Cytokine expression by MCMV-specific CD4 T cells. The production of various cytokines by splenic CD4 T cells was assessed after stimulation with m139560-574 and m14224-38 peptides 8 days following MCMV infection. Flow plots are gated on CD4 T cells and the numbers indicate the frequency of IFN-γ-, TNF-, IL-2-, IL-10-, and IL-17-producing cells within this population.
Profile of the CD4 T cell memory pool during the course of MCMV infection
The MCMV-specific, polyclonal memory CD8 T cell pool displays several phenotypes. These range from “classical” responses that expand maximally by ∼8 days postinfection, contract, and develop into stable long-term memory cells to so-called “inflationary” CD8 T cell memory responses that are virtually undetectable at day 8 but increase dramatically in frequency during chronic/latent infection (15, 21). Consequently, we measured the time course kinetics of the MCMV-specific CD4 T cell response periodically through day 100 postinfection. We found that all 14 responses originally identified at day 8 postinfection peaked at this time and then contracted and established a stable memory cell pool (Fig. 3A).
FIGURE 3.
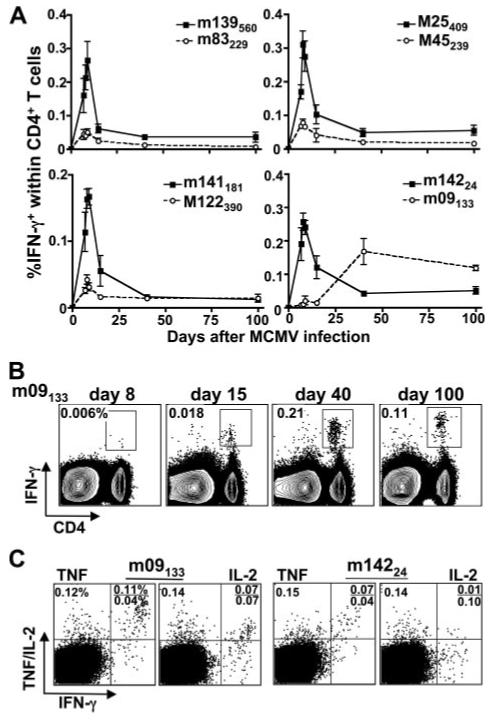
MCMV-specific CD4 T cell responses vary in their time of induction. A, The frequency of splenic IFN-γ-producing cells after stimulation with the indicated peptide epitopes was determined on days 7, 8, 9, 15, 40, and 100 after MCMV infection. Plots represent the percentage of IFN-γ-producing cells within the CD4 T cell population. B, The frequency of splenic IFN-γ-producing cells within the total CD4 T cell population specific for the inflating MCMV epitope m09133-147 was determined on days 8, 15, 40, and 100 after MCMV infection. C, Cytokine production of splenic CD4 T cells specific for m09133-147 or m14224-38 was assessed on day 100 after MCMV infection. Flow plots are gated on CD4 T cells and numbers indicate the frequency of IFN-γ-, TNF-, and IL-2-producing cells within the CD4 T cell population.
In addition to analyzing the kinetics of the 14 epitope-specific CD4 T cell responses originally identified at day 8 postinfection, the original 188 peptides were all tested again at day 100 postinfection. Strikingly, at this time CD4 T cells specific for a peptide epitope derived from the m09 protein (m09133-147) were identified. Subsequent kinetic analysis of the CD4 T cells specific for the m09133-147 epitope revealed that reactivity was undetectable at day 8, very low at day 15, but increased dramatically at day 40 and at day 100 postinfection (Fig. 3A). Interestingly, when m09-specific CD4 T cells were compared with m142-specific cells, a significantly higher percentage produced IL-2 and TNF within the IFN-γ+ population and also produced higher levels of these cytokines (Fig. 3C). Therefore, m09-specific CD4 T cells display a significantly altered time course of induction than that of most MCMV-specific CD4 responses, somewhat resembling inflationary CD8 T cells, but their cytokine expression profiles differ significantly from those of previously characterized MCMV-specific CD8 T cells that increase in number over time.
Discussion
The consensus prediction approach used in this study was highly successful in identifying MCMV-specific CD4 T cell responses. Although the peptide candidates tested covered <1% of the total predicted ORFs encoded by the MCMV genome, they uncovered ∼25% of the IFN-γ-producing CD4 T cell population specific for the virus. This result is a marked improvement over our past experiences predicting H-2 I-Ab-restricted, vaccinia virus-derived CD4 T cell epitopes (22). The performance improvements are due to more available public data sets that can be incorporated into next generation algorithms that use a consensus-based approach.
Two H-2b-restricted CD8 T cell responses specific for peptides derived from the immediate early (IE) 3 protein increase in number throughout the latent infection of C57BL/6 mice, and these cells display an effector-memory phenotype (IL-2-CD62LlowCD127-) (16). In contrast, the CD4 T cell population specific for the IE3390-404 epitope identified in this study was not found to increase over time. One potential explanation for the inflation of CD8 T cell responses specific for CMV IE proteins is that periodic, stochastic reactivation of a latent virus is a source of chronic Ag(s) for these CD8 T cells (23). For MCMV it has been shown that a CD8 T cell population specific for a H-2d-restricted IE1-derived epitope localizes to the lung and regulates “abortive transcriptional reactivation” (24). Why then do IE3390-404 specific CD4 T cells not inflate if this process is merely driven by chronic Ag? Currently, we have no definitive explanation for this, but the increase in specific CD8 T cell populations during chronic CMV infection cannot likely be exclusively explained by the above model.
Four of the 15 CD4 T cell responses identified in this study are specific for the M45 protein, and M45 is the target of the immunodominant CD8 T cell response during acute MCMV infection in C57BL/6 mice (25). M45 is required for MCMV to inhibit apoptosis and replicate efficiently in endothelial cells (26), cells that express both MHC class I and II. Stromal cells are the primary targets of MCMV infection in the spleen (27) and likely in other organs, suggesting that infected endothelial cells may serve as an important source of viral Ag to promote both CD4 and CD8 T cell responses.
Strikingly, a substantial percentage of the MCMV-specific CD4 T cell population produced IL-17 upon peptide stimulation. To date, very little is known about the role of this cytokine in the context of viral infection. IL-17-producing CD4 T cells are critical for controlling Mycobacterium tuberculosis infection in the lung and do so by regulating the recruitment of IFN-γ-producing CD4 T cells (28). As we have observed a significant number of IFN-γ-producing, MCMV-specific CD4 T cells in lung, it is tempting to speculate that IL-17 might play a similar role in controlling MCMV infection. In total, this study characterizes specific CD4 T cell responses that arise during MCMV infection and will help to further our understanding of the ways CD4 T cells regulate immunity to infection with this persistent virus.
Acknowledgments
We thank Dr. M. Croft and C. F. Ware for reading the manuscript. This is publication no.989 from the La Jolla Institute for Allergy and Immunology.
Footnotes
This work was supported in part by a Veni Grant from The Netherlands Organization for Scientific Research (to R.A.), National Institutes of Health (NIH) Grants AI048073 and AI057840, NIH Contract HHSN26620040006C, Pacific Southwest Regional Center of Excellence Grant U54AI065359, and NIH Contract N01-AI-40023 (to A.S. and B.P.), Deutsche Forschungsgemeinschaft (DFG) fellowship (to A.L.), and NIH Grants CA081261 and AI076972 (to S.P.S.).
- MCMV
- mouse CMV
- IE
- immediate early
- ORF
- open reading frame
Disclosures
The authors have no financial conflict of interest.
References
- 1.Gamadia LE, Rentenaar RJ, van Lier RA, ten Berge IJ. Properties of CD4+ T cells in human cytomegalovirus infection. Hum. Immunol. 2004;65:486–492. doi: 10.1016/j.humimm.2004.02.020. [DOI] [PubMed] [Google Scholar]
- 2.Jonjic S, Mutter W, Weiland F, Reddehase MJ, Koszinowski UH. Site-restricted persistent cytomegalovirus infection after selective long-term depletion of CD4+ T lymphocytes. J. Exp. Med. 1989;169:1199–1212. doi: 10.1084/jem.169.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jonjic S, Pavic I, Lucin P, Rukavina D, Koszinowski UH. Efficacious control of cytomegalovirus infection after long-term depletion of CD8+ T lymphocytes. J. Virol. 1990;64:5457–5464. doi: 10.1128/jvi.64.11.5457-5464.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lucin P, Pavic I, Polic B, Jonjic S, Koszinowski UH. Gamma interferon-dependent clearance of cytomegalovirus infection in salivary glands. J. Virol. 1992;66:1977–1984. doi: 10.1128/jvi.66.4.1977-1984.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jonjic S, Pavic I, Polic B, Crnkovic I, Lucin P, Koszinowski UH. Antibodies are not essential for the resolution of primary cytomegalovirus infection but limit dissemination of recurrent virus. J. Exp. Med. 1994;179:1713–1717. doi: 10.1084/jem.179.5.1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Riddell SR, Greenberg PD. T cell therapy of human CMV and EBV infection in immunocompromised hosts. Rev. Med. Virol. 1997;7:181–192. doi: 10.1002/(sici)1099-1654(199709)7:3<181::aid-rmv200>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 7.Reddehase MJ, Mutter W, Munch K, Buhring HJ, Koszinowski UH. CD8-positive T lymphocytes specific for murine cytomegalovirus immediateearly antigens mediate protective immunity. J. Virol. 1987;61:3102–3108. doi: 10.1128/jvi.61.10.3102-3108.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Polic B, Jonjic S, Pavic I, Crnkovic I, Zorica I, Hengel H, Lucin P, Koszinowski UH. Lack of MHC class I complex expression has no effect on spread and control of cytomegalovirus infection in vivo. J. Gen. Virol. 1996;77:217–225. doi: 10.1099/0022-1317-77-2-217. [DOI] [PubMed] [Google Scholar]
- 9.Sylwester AW, Mitchell BL, Edgar JB, Taormina C, Pelte C, Ruchti F, Sleath PR, Grabstein KH, Hosken NA, Kern F, et al. Broadly targeted human cytomegalovirus-specific CD4+ and CD8+ T cells dominate the memory compartments of exposed subjects. J. Exp. Med. 2005;202:673–685. doi: 10.1084/jem.20050882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khan N, Hislop A, Gudgeon N, Cobbold M, Khanna R, Nayak L, Rickinson AB, Moss PA. Herpesvirus-specific CD8 T cell immunity in old age: cytomegalovirus impairs the response to a coresident EBV infection. J. Immunol. 2004;173:7481–7489. doi: 10.4049/jimmunol.173.12.7481. [DOI] [PubMed] [Google Scholar]
- 11.Vescovini R, Biasini C, Fagnoni FF, Telera AR, Zanlari L, Pedrazzoni M, Bucci L, Monti D, Medici MC, Chezzi C, et al. Massive load of functional effector CD4+ and CD8+ T cells against cytomegalovirus in very old subjects. J. Immunol. 2007;179:4283–4291. doi: 10.4049/jimmunol.179.6.4283. [DOI] [PubMed] [Google Scholar]
- 12.Holtappels R, Thomas D, Podlech J, Reddehase MJ. Two antigenic peptides from genes m123 and m164 of murine cytomegalovirus quantitatively dominate CD8 T-cell memory in the H-2d haplotype. J. Virol. 2002;76:151–164. doi: 10.1128/JVI.76.1.151-164.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karrer U, Sierro S, Wagner M, Oxenius A, Hengel H, Koszinowski UH, Phillips RE, Klenerman P. Memory inflation: continuous accumulation of antiviral CD8+ T cells over time. J. Immunol. 2003;170:2022–2029. doi: 10.4049/jimmunol.170.4.2022. [DOI] [PubMed] [Google Scholar]
- 14.Munks MW, Gold MC, Zajac AL, Doom CM, Morello CS, Spector DH, Hill AB. Genome-wide analysis reveals a highly diverse CD8 T cell response to murine cytomegalovirus. J. Immunol. 2006;176:3760–3766. doi: 10.4049/jimmunol.176.6.3760. [DOI] [PubMed] [Google Scholar]
- 15.Sierro S, Rothkopf R, Klenerman P. Evolution of diverse antiviral CD8+ T cell populations after murine cytomegalovirus infection. Eur. J. Immunol. 2005;35:1113–1123. doi: 10.1002/eji.200425534. [DOI] [PubMed] [Google Scholar]
- 16.Munks MW, Cho KS, Pinto AK, Sierro S, Klenerman P, Hill AB. Four distinct patterns of memory CD8 T cell responses to chronic murine cytomegalovirus infection. J. Immunol. 2006;177:450–458. doi: 10.4049/jimmunol.177.1.450. [DOI] [PubMed] [Google Scholar]
- 17.Schneider K, Loewendorf A, De Trez C, Fulton J, Rhode A, Shumway H, Ha S, Patterson G, Pfeffer K, Nedospasov SA, et al. Lymphotoxin-mediated crosstalk between B cells and splenic stroma promotes the initial type I interferon response to cytomegalovirus. Cell Host Microbe. 2008;3:67–76. doi: 10.1016/j.chom.2007.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moutaftsi M, Peters B, Pasquetto V, Tscharke DC, Sidney J, Bui HH, Grey H, Sette A. A consensus epitope prediction approach identifies the breadth of murine T(CD8+)-cell responses to vaccinia virus. Nat. Biotechnol. 2006;24:817–819. doi: 10.1038/nbt1215. [DOI] [PubMed] [Google Scholar]
- 19.Rawlinson WD, Farrell HE, Barrell BG. Analysis of the complete DNA sequence of murine cytomegalovirus. J. Virol. 1996;70:8833–8849. doi: 10.1128/jvi.70.12.8833-8849.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Humphreys IR, de Trez C, Kinkade A, Benedict CA, Croft M, Ware CF. Cytomegalovirus exploits IL-10-mediated immune regulation in the salivary glands. J. Exp. Med. 2007;204:1217–1225. doi: 10.1084/jem.20062424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Munks MW, Pinto AK, Doom CM, Hill AB. Viral interference with antigen presentation does not alter acute or chronic CD8 T cell immunodominance in murine cytomegalovirus infection. J. Immunol. 2007;178:7235–7241. doi: 10.4049/jimmunol.178.11.7235. [DOI] [PubMed] [Google Scholar]
- 22.Moutaftsi M, Bui HH, Peters B, Sidney J, Salek-Ardakani S, Oseroff C, Pasquetto V, Crotty S, Croft M, Lefkowitz EJ, et al. Vaccinia virus-specific CD4+ T cell responses target a set of antigens largely distinct from those targeted by CD8+ T cell responses. J. Immunol. 2007;178:6814–6820. doi: 10.4049/jimmunol.178.11.6814. [DOI] [PubMed] [Google Scholar]
- 23.Kurz SK, Reddehase MJ. Patchwork pattern of transcriptional reactivation in the lungs indicates sequential checkpoints in the transition from murine cytomegalovirus latency to recurrence. J. Virol. 1999;73:8612–8622. doi: 10.1128/jvi.73.10.8612-8622.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simon CO, Holtappels R, Tervo HM, Bohm V, Daubner T, Oehrlein-Karpi SA, Kuhnapfel B, Renzaho A, Strand D, Podlech J, et al. CD8 T cells control cytomegalovirus latency by epitope-specific sensing of transcriptional reactivation. J. Virol. 2006;80:10436–10456. doi: 10.1128/JVI.01248-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gold MC, Munks MW, Wagner M, Koszinowski UH, Hill AB, Fling SP. The murine cytomegalovirus immunomodulatory gene m152 prevents recognition of infected cells by M45-specific CTL but does not alter the immunodominance of the M45-specific CD8 T cell response in vivo. J. Immunol. 2002;169:359–365. doi: 10.4049/jimmunol.169.1.359. [DOI] [PubMed] [Google Scholar]
- 26.Brune W, Menard C, Heesemann J, Koszinowski UH. A ribonucleotide reductase homolog of cytomegalovirus and endothelial cell tropism. Science. 2001;291:303–305. doi: 10.1126/science.291.5502.303. [DOI] [PubMed] [Google Scholar]
- 27.Benedict CA, De Trez C, Schneider K, Ha S, Patterson G, Ware CF. Specific Remodeling of Splenic Architecture by Cytomegalovirus. PLoS Pathog. 2006;2:e16. doi: 10.1371/journal.ppat.0020016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khader SA, Bell GK, Pearl JE, Fountain JJ, Rangel-Moreno J, Cilley GE, Shen F, Eaton SM, Gaffen SL, Swain SL, et al. IL-23 and IL-17 in the establishment of protective pulmonary CD4+ T cell responses after vaccination and during Mycobacterium tuberculosis challenge. Nat. Immunol. 2007;8:369–377. doi: 10.1038/ni1449. [DOI] [PubMed] [Google Scholar]


