Abstract
Triptolide, an active component of the medicinal herb, lei gong teng, is a potent anti-cancer and anti-inflammatory therapeutic. It potently inhibits NFκB transcriptional activation subsequent to DNA binding, although a precise mechanism is as yet unknown. Here, we report that triptolide also induces distinct nuclear sub-structural changes in HeLa cells. These changes in the nucleolus and nuclear speckles are reversible and dependent on both time and concentration. Furthermore, nuclear changes occurred within hours of triptolide treatment and were calcium and caspase independent. Rounding of nuclear speckles, an indication of transcriptional arrest was evident and was associated with a decrease in RNA Polymerase II CTD Ser2 phosphorylation. Additionally, the nucleolus disassembled and RNA Pol I activity declined subsequent to RNA Pol II inhibition. We therefore conclude that triptolide causes global transcriptional arrest as evidenced by inactivity of RNA polymerases I and II and the subsequent alteration in nuclear sub-structure.
Keywords: triptolide, transcription, NFκB, RNA Polymerase
Introduction
The small molecule triptolide, a natural product isolated from the ‘Thunder God Vine’, has been of great interest as a therapeutic for diseases such as cancer, arthritis and autoimmune disorders. Research into its mechanism of action has revealed that it potently inhibits transcription of numerous pro-inflammatory mediators (1, 2), can activate caspases and other pro-apoptotic cascades (3–5), has separable mechanisms of action as defined by calcium concentration (6), and can activate calcium release through the polycystin-2 cation channel (7). One biological effect of triptolide that has received the most interest has been its ability to suppress cytokine mediated transcriptional activity. While there are reports in the literature citing triptolide’s transcriptional effects as both specific (2, 8) and global (9), it is clear that in many different cell lines triptolide is efficient at inhibiting expression of NFκB mediated gene targets, hence its anti-inflammatory effects. However, since triptolide has also been reported most recently to reduce total RNA levels (9), it is of interest to further characterize how triptolide might be causing global transcriptional arrest.
The nucleus is highly compartmentalized to ensure efficient cellular functions such as ribosome biogenesis and RNA Polymerase II (RNA Pol II) driven transcription. One specialized domain formed around actively transcribed rRNA genes is the nucleolus. This nuclear sub-structure is crucial not only to rRNA transcription and ribosome assembly but has also been shown to be involved in cell cycle regulation, proliferation, and the cellular stress response (10). Perturbations in cellular transcription are mirrored by changes in nucleolar structure; RNA Pol II inhibitors 5,6-dichloro-β-D-ribofuranosylbenzimidazole (DRB) and α-amanitin have previously been shown to alter nucleolar integrity, resulting in the dispersal of this sub-nuclear structure (11). Additionally, actinomycin D can be used to inhibit RNA Pol I at low concentrations (0.04–0.05 µg/ml) leading to condensation of rDNA, while high concentrations (2µg/ml) additionally inhibit RNA polymerase II activity and induce nucleolar disassembly (12, 13).
The nucleolar proteome is currently estimated to have approximately 700 proteins, many of which are of unknown function (14, 15). However, several proteins are well described such as nucleolin (C23), nucleophosmin (NPM, B23), and Upstream Binding Factor (UBF). Nucleolin is a multifunctional protein described as having roles in ribosome biogenesis, transcription, cell-cycle regulation, and nuclear-cytoplasmic shuttling capabilities (16). NPM is a phospho-protein involved in numerous cellular activities such as transcriptional regulation, centrosome duplication (17), nuclear chaperoning (18), ribosome biogenesis, and nucleic acid binding and processing (19). Additionally, NPM is frequently upregulated or is subject to genetic mutation or translocation in leukemias and lymphomas (20). UBF is a crucial co-transcriptional activator of the RNA Pol I complex; loss of UBF from the nucleolus is highly indicative of Pol I inactivity (21, 22).
Another sub-nuclear domain is the nuclear speckle, a region specialized in mRNA transcript splicing. These structures are highly dynamic as splicing factors are continually shuttling to sites of active transcription (23). They are characterized by their irregular shape and size and are frequently identified by the localization of the splicing factor, SC35. Nuclear speckles are thought to form on the periphery of active RNA Pol II transcriptional complexes, and in addition to splicing components may also contain Cdk/Cyclin complexes necessary for Pol II activity (24). These Cdk/Cyclin complexes phosphorylate the C-terminal Domain (CTD) of RNA Pol II, comprised of 52 heptapeptide repeats in humans, and are therefore responsible for regulation of transcriptional activity. The TFIIH complex, containing Cdk7, phosphorylates serine 5 (Ser5) in the CTD and promotes initiation of transcription and promoter clearance (25, 26). Next, serine 2 (Ser2) is phosphorylated by the positive transcription elongation factor b complex (P-TEFb) comprised of Cdk9 and cyclin T1 and promotes elongation of the mRNA transcript (27, 28). Upon RNA Pol II inhibition, nuclear speckles enlarge, become rounded, and decrease in number due to the accumulation of the splicing machinery (29, 30). As with nucleolar disruption, this effect has been observed with actinomycin D, DRB, and α-amanitin, signifying a link between active transcription and the maintenance of a structurally ordered nucleus.
In this study, we focused on triptolide’s transcriptional effect as assessed in the cervical carcinoma cell line, HeLa. Previous work with this in vitro model demonstrated that while transcriptional inhibition of NFκB is concentration dependent, calcium was not required for this effect (6). Here, we have taken a cell biological approach to correlate differences in nuclear structure to triptolide’s known inhibitory effect on NFκB mediated transcription. Therefore, we examined changes in the nucleolus and nuclear speckles, two nuclear sub-structures that are indicative of changes in the global transcriptional activity of the cell. Our results support a concentration dependent model of triptolide-induced RNA Pol II inactivity leading to transcriptional arrest and cell death.
Materials and Methods
Cell Culture and Reagents
HeLa cells were cultured as previously described (6). Triptolide was obtained from Sinobest Inc. (China) and was dissolved in DMSO. Actinomycin D, calcimycin and ionomycin were obtained from EMD Biosciences. Caspase-3 activity assay was purchased from Biomol and zVAD-fmk was purchased from Genotech. Antibodies purchased were as follows: nucleolin (sc-13057), NPM (sc-32256), UBF (sc-9131), Cdk9 (sc-484), and Cyclin T1 (sc-8127) from Santa Cruz Biotechnology; SC35 (ab11826) from Abcam; phosho-Thr199 NPM (#3541) from Cell Signaling Technologies; and both phospho-Ser2 (BL2894) and Ser5 (BL2896) RNA Pol II antibodies were from Bethyl Labs. NFκB luciferase assay was performed as previously described (6).
Generation of Clone-2
HeLa cells were incubated first with 100 nM triptolide for 96 hours, and then 1 µM triptolide for an additional 96 hours. Remaining cells were grown with 100 nM triptolide until clonal populations were evident. Triptolide resistant clones were found to grow in triptolide concentrations up to 1 µM, where 5 µM induced cell death. Routine administration of 100 nM triptolide is added to cells to confirm maintenance of triptolide resistance.
Western blotting
Total cell lysates (0.5% Triton X-100, 50mM Tris pH 7.4, 150mM NaCl, 500mM EDTA, and Complete Protease Inhibitors (Roche)) were prepared and protein levels normalized. For nuclear fractions, cytosolic extract was removed following hypotonic lysis (10mM HEPES pH 7.4, 10mM KCl, 0.1mM EDTA, 1mM DTT, 0.5mM PMSF) and 0.15% NP-40 was added immediately before centrifugation. The pellet was resuspended in nuclear extract buffer (20mM HEPES pH 7.4, 1mM EDTA, 0.4M NaCl, 1mM DTT), centrifuged and the supernatant retained. For RNAse A experiments, the cytosolic and nuclear fractions were cleared with 0.5% Triton X-100 lysis buffer, and the resulting pellet was resolubilized in nuclear lysis buffer plus 0.1mg/ml RNAse A for 30 minutes on ice. Antibodies for nucleolin, NPM, UBF, Cdk9 and Cyclin T1 (1:1000) and RNA Pol II Ser2 and Ser5 antibodies (1:5000) were incubated in 5% milk/TBST before Enhanced ECL detection (GE Healthcare).
Immunofluorescence
For nucleolin, NPM, and UBF (1:200, 1.5% NGS/PBS), cells were fixed in MeOH and blocked in 5% NGS/PBS; p-Thr199 NPM (1:100, 1%BSA/TBS), MeOH fixation, blocked in 10% NGS/1%BSA/TBS; SC35 (1:1000, PBS), Ser2 and Ser5 RNA Pol II (1:400, 5% BSA/PBS) detection required 4% paraformaldehyde/0.2% Triton X-100/PBS followed by MeOH fixation. Cdk9 detection (1:600) required 4% paraformaldehyde/0.5% Triton X-100/PBS fixation and blocking in 5% Milk/TBST. Secondary antibodies were anti -rabbit or -mouse Alexa Fluor -488 (green) or -594 (red) (Molecular Probes), and all samples mounted in Vectashield (Vector Labs).
All parvalbumin-GFP constructs (31) were a gift of Anton Bennett (Yale University). Cells were transiently transfected using Lipofectamine 2000 (Invitrogen) plus 1 µg of one of the following plasmids for 24 hours: CMV-GFP, CMV-NES-parvalbumin-GFP, or CMV-NLS-parvalbumin-GFP. Following confirmation of GFP expression by microscopy, 100 nM triptolide was added for six hours. Cells were fixed and stained for nucleolin as described above.
Images were acquired using an Olympus CK40 inverted microscope and an Olympus digital camera. Confocal images were acquired under 63X oil immersion objective using a Zeiss Inverted Axiovert microscope and a BioRad MRC 1024 laser source. Images were captured in single color channels as .pic files using LaserSharp software and then converted with Image J (NIH). For co-localization studies, images were artificially merged using Adobe Photoshop CS where indicated.
RT-PCR
Total RNA was extracted using RNeasy (Qiagen) and each sample normalized. RT-PCR was performed using Superscript One-Step RT-PCR with Platinum Taq kit (Invitrogen) and S14 primers were previously described (32). 350 ng of total RNA was used for S14, RT proceeded 30 min/50°C, and the PCR amplification protocol was: 94°C/2 min; 35 cycles of 94°C/15 sec, 58°C/30 sec, 72°C/1 min; 72°C/5 min. 2 µg of total RNA was used for 5’ETS 45S amplification (33), RT proceeded at 50°C/30 min, then 5µl of the 50µl RT reaction was used with the following protocol: 95°C/3 min; 25 cycles of 95°C/1 min, 65°C/1 min, 72°C/1 min; 72°C/5 min.
Results
Triptolide Mediated Disruption of Nucleolar Structure is Time and Concentration Dependent
We began our experiments by examining the nucleolus as its integrity has been used an as indicator of both RNA Pol I and II transcriptional activity. Previous work in our laboratory has shown that in HeLa cells, 50 nM triptolide for six hours is the minimum concentration required to cause NFκB transcriptional inhibition (6). We therefore incubated HeLa cells for six hours with triptolide concentrations up to 1µM and examined nucleolar sub-structure. At concentrations of 50 nM and above, we observed that the integrity of the nucleolus was compromised as nucleolin staining became irregular in shape and diffuse through the nucleoplasm (Fig. 1A). This change in localization was prominent at 100 nM in the majority of cells and at 1 µM the nucleolus had completely dispersed (Fig. 1A). Since we have previously described that 100 nM triptolide is effective at causing robust transcriptional inhibition and cell death, and we could now correlate that with clear nucleolar disassembly, we chose to focus on this concentration in the majority of the remaining experiments. Over a 6-hour time course with 100 nM triptolide, we observed that nucleolin localization changes began after 3 hours of continual culture (Fig. 1B). Furthermore, this effect was predominantly reversible if cells were incubated for no more than four hours before triptolide removal (Fig. 1C). However, if triptolide was left in culture for 6 hours before removal, the nucleolus did not recover and cells were committed to cell death (Fig. 1C). This first set of experiments led us to believe that general transcription may be affected by triptolide, so we next examined another sub-nuclear marker that changes upon transcriptional arrest, the nuclear speckle.
Figure 1.
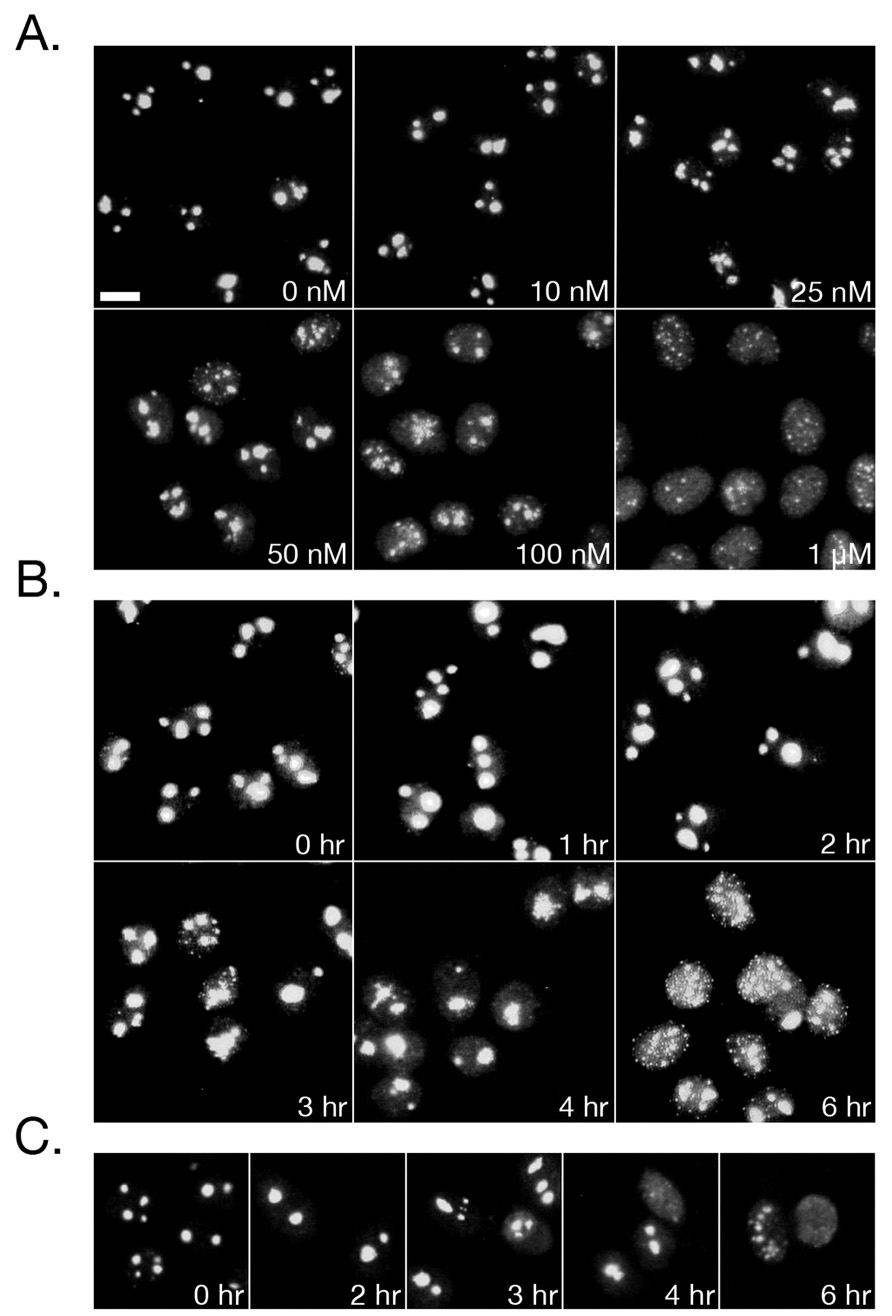
Triptolide Induces Nucleolar Disassembly. HeLa cells were treated with triptolide before fixation and staining for nucleolin by indirect immunofluorescence under the following conditions: A) Increasing concentrations of triptolide for 6 hours B) 100 nM triptolide over a 6-hour time course C) Cells were pre-treated with 100 nM triptolide for the indicated times, washed, and allowed to recover for 16 hours. All images were acquired under 25X magnification and scale bar is 10 µm.
Nuclear Speckles Become Enlarged and Rounded Following Triptolide Treatment
We used an antibody to the splicing factor SC35 to assess nuclear speckle morphology in response to 100 nM triptolide or DMSO over a 6-hour time course. While DMSO control cells had numerous speckles of various size and shape, cells treated with triptolide displayed a distinct rounding and enlargement of speckles beginning after 2 hours of incubation (Fig. 2A). This morphological change, a marker of transcriptional arrest, became more pronounced over the 6-hour time course that was in marked contrast to DMSO control (Fig. 2A). We next examined the concentration dependent effect of triptolide on speckle morphology after 6 and 16 hours of continual incubation. After 6 hours (the time at which NFκB inhibition has been studied), only 50 and 100 nM triptolide caused observable speckle rounding. Following 16 hours, 25–100 nM triptolide (concentrations shown to cause cell death in HeLa cells) (6) induced prominent speckle rounding and an increase in size (Fig. 2B). Therefore, our data indicate that nuclear speckle rounding begins at 50 nM with acute triptolide addition (6 hours) while 25 nM is also sufficient for this effect although over a longer time course.
Figure 2.
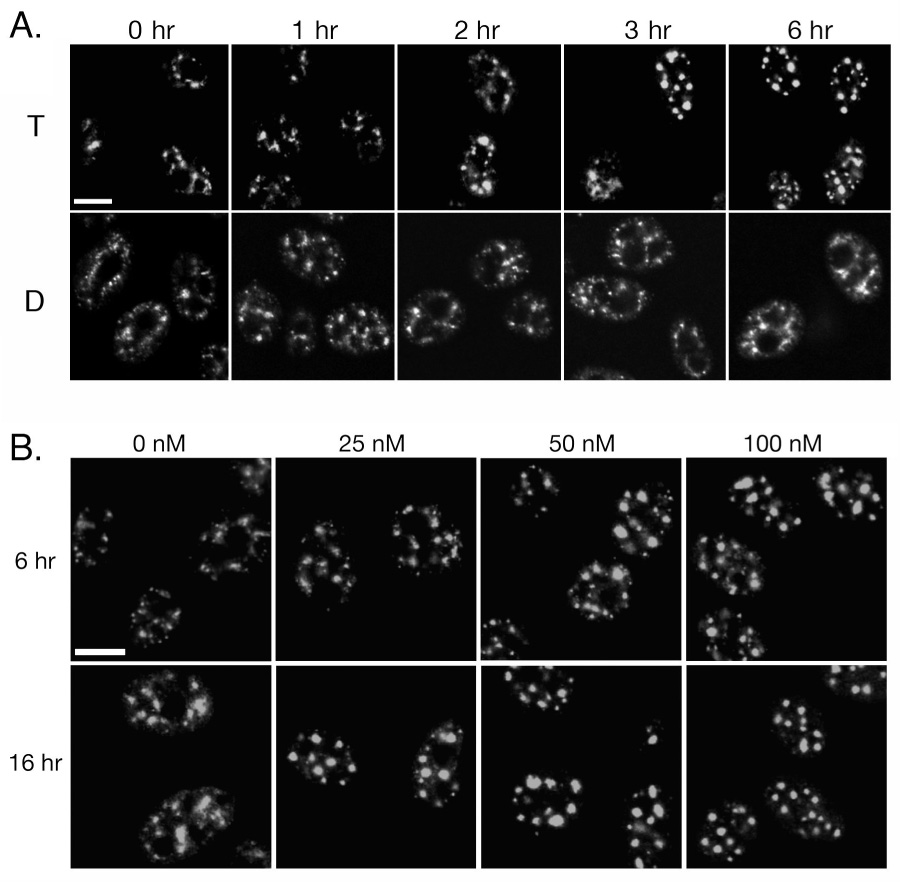
Triptolide Induces Nuclear Speckle Rounding. A) HeLa cells were incubated with 100 nM triptolide (T) or DMSO (D) over a 6-hour time course. Following fixation, cells were stained for the splicing factor SC35 by indirect immunofluorescence. B) Cells were incubated with 0, 25, 50, or 100 nM triptolide for either 6- or 16- hours of continual culture before assessing SC35 localization. All images were acquired by confocal microscopy under 63X magnification and scale bars are 10 µm.
Sub-Nuclear Structures are Unaltered in a Triptolide Resistant Clonal Line
We have isolated a triptolide-resistant clonal cell line derived from HeLa, named Clone-2 that remain viable in the presence of up to 1 µM triptolide. Since 100 nM triptolide causes rapid death in HeLa, but is permissive for growth in Clone-2 cells, we compared nucleolar and nuclear speckle differences in these lines as an indicator of cell death and transcriptional activity. After 6 hours of incubation with 100 nM triptolide, HeLa cells exhibited the characteristic nucleolar disassembly as assessed by both Differential Interference Contrast (DIC) microscopy and localization of nucleolin (Fig. 3A). This was in contrast to Clone-2 cells that retained nucleolar integrity however nucleolar borders became slightly irregular in shape (Fig. 3A). To confirm that Clone-2 cells are capable of a cellular stress response and RNA Pol I inhibition, we incubated cells with 10 nM actinomycin D, and found that in both HeLa and Clone-2 cells, nucleolar structure was affected (Fig. 3A).
Figure 3.
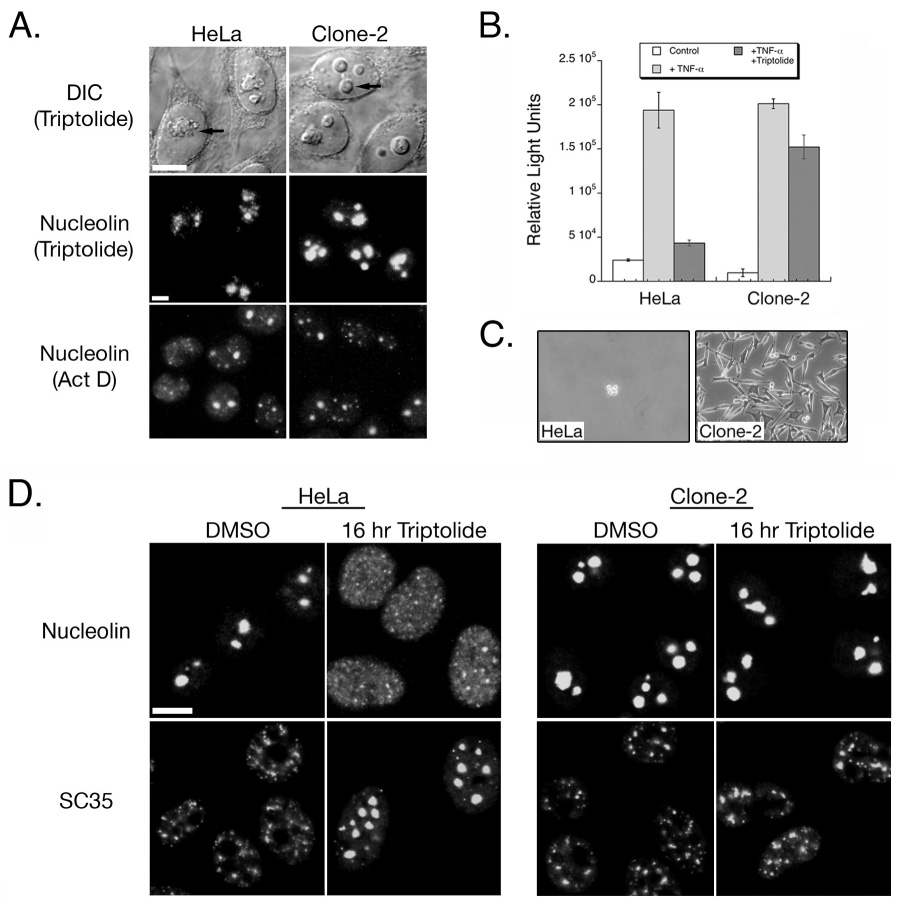
A HeLa – Derived Triptolide Resistant Clonal Population Does Not Undergo Nuclear Structure Changes. A) HeLa and Clone-2 cells were assessed for nucleolar changes by either DIC or indirect immunofluorescence of nucleolin. Cells were treated with either 100 nM triptolide or 10 nM actinomycin D (Act D) for 6 hours. Arrow indicates nucleolus, DIC images were acquired by confocal microscopy using 63X magnification, and immunofluorescence images were acquired under 25X magnification. B) Transcriptional activity was assessed using an NFκB-luciferase construct. HeLa or Clone-2 cells were incubated with vehicle only (control), TNF-α, or with TNF-α plus 100 nM triptolide for a total of 6 hours. C) HeLa and Clone-2 cells were assessed for survival following 72 hours of 100 nM triptolide incubation. D) Cells were incubated with DMSO or 100 nM triptolide for 16 hours before fixation and indirect immunofluorescent detection of nucleolin or SC35. Images were acquired by confocal microscopy under 63X magnification. All scale bars represent 10 µm.
Since our triptolide-resistant cell line showed only a minor change in the shape of the nucleolar compartment, we next determined if NFκB mediated transactivation was affected. Following TNF-α incubation alone, both HeLa and Clone-2 cells responded with similar levels of NFκB-luciferase induction (Fig. 3B). Co-incubation of 100 nM triptolide plus TNF-α resulted in a four-fold reduction of activity in HeLa, while Clone-2 cells retained the majority of its transcriptional activation (Fig. 3B). The resistance of Clone-2 to both nucleolar disassembly and transcriptional inhibition ultimately correlated with cell survival. While 72 hours of incubation with 100 nM triptolide resulted in cell detachment and death for HeLa cells, Clone-2 cells survived and continued to proliferate with a doubling time of approximately 28 hours (Fig. 2C and data not shown). Since only minor effects on nucleolar integrity or transcription were observed for Clone-2 cells with an acute triptolide time course, we next assessed nucleolar changes after 16 hours of continual culture with triptolide. After 16 hours of 100 nM triptolide, many HeLa cells detached indicating cell death and those remaining adherent were characterized by a dispersal of nucleolin throughout the nucleoplasm, and several large rounded nuclear speckles (Fig. 3D). Clone-2 cells remained adherent, the nucleolus was intact, nuclear speckles looked similar to DMSO treated control and cells continued to grow (Fig. 3D). Taken together, our results so far indicate that changes in the structure of both the nucleolus and nuclear speckles correlates to transcriptional inactivation and cell death due to high triptolide concentrations.
Triptolide Induced Changes in Nucleolar and Nuclear Speckle Morphology are Calcium, Stress and Caspase Independent
To begin examining a putative mechanism for the observed sub-nuclear changes due to triptolide we next focused on the role of calcium. We have previously shown that NFκB transcriptional inhibition is calcium independent, and therefore wanted to determine if changes to both the nucleolus and nuclear speckles (and their role in transcriptional regulation) were also calcium independent. Since triptolide can elicit calcium release (7), we first wanted to establish if calcium ionophores could mimic triptolide induced nucleolar disassembly. While incubation with 100 nM triptolide for 6 hours led to a dramatic loss of nucleolar integrity, neither calcimycin nor ionomycin could mimic the effect (Fig. 4A). We next incubated cells in the absence of extracellular calcium and still observed triptolide induced nucleolin dispersal and speckle rounding (Fig. 4B). As a third method to test the requirement of calcium mediated signaling on triptolide mediated nuclear changes, we used GFP-parvalbumin (PV) constructs (31) that were localized to either the nucleus (NLS-PV-GFP) or cytosol (NES-PV-GFP). Expression of these constructs in their specific cellular compartments act to buffer the response of a calcium signal. Expression of the GFP-constructs alone did not affect nucleolar morphology as assessed by nucleolin immunofluorescent localization (Fig. 4C). In the presence of triptolide, nucleolar structure dispersed regardless of either cytosolic or nuclear calcium buffering (Fig. 4C). We therefore conclude that the sub-nuclear changes we observe with high triptolide concentrations are calcium independent.
Figure 4.
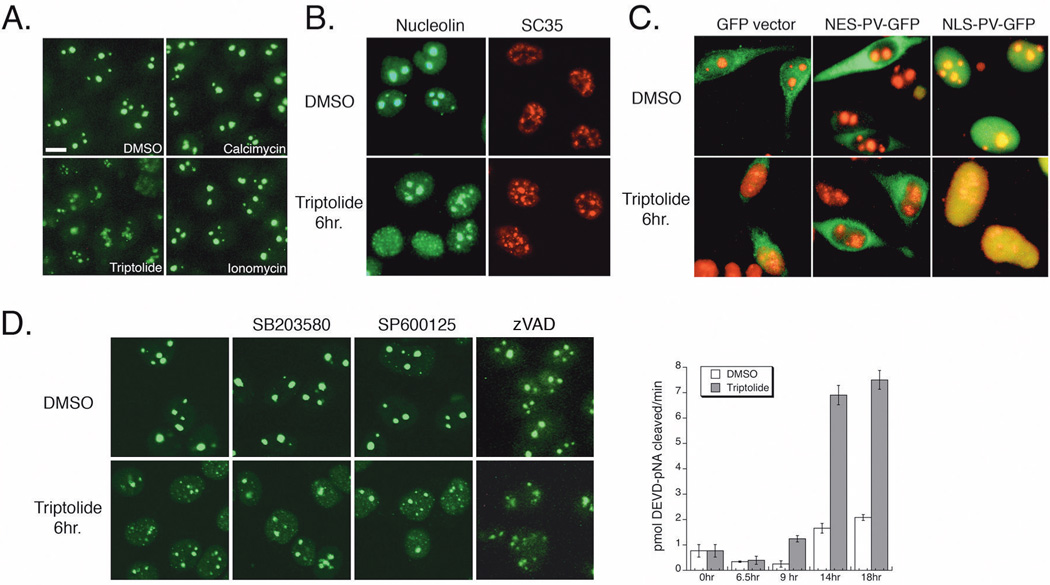
Nuclear Changes are Independent of Calcium, Stress Kinases, and Caspase Activation. A) HeLa cells were incubated for 6 hours with DMSO, 100 nM triptolide, 5 µM calcimycin, or 3 µM ionomycin before fixation and nucleolin detection by indirect immunofluorescence (25X magnification). Scale bar is 10 µm. B) HeLa cells were incubated in calcium free media for 16 hours before addition of DMSO or 100 nM triptolide for 6 hours. Cells were stained for SC35 (red) or nucleolin (green) by indirect immunofluorescence (confocal microscopy, 40X). C) HeLa cells were transiently transfected with GFP vector, nuclear excluded (NES) or nuclear localized (NLS) parvalbumin (PV) – GFP constructs for 24 hours before a 6-hour incubation with DMSO or 100 nM triptolide. GFP localization is in green, nucleolin is detected by indirect immunofluorescence (red). Images acquired separately under 40X magnification and artificially merged. D) HeLa cells were treated with DMSO, 5 µM SB203580, 10 µM SP600125, or 2 µM zVAD-fmk ± 100 nM triptolide for 6 hours. Nucleolar integrity was detected by indirect immunofluorescence of nucleolin (green). Graph represents caspase-3 activity in HeLa cells treated with DMSO or 100 nM triptolide over an 18-hour time course. Cell lysates were prepared and activity assessed by cleavage of the colorimetric substrate, DEVD-pNA. Data represent Mean±SE, n=3.
Nucleolar disassembly has also been utilized to detect activation of the cellular stress response through p38 kinase or JNK signaling (34). To determine if triptolide induced a general stress response that was then reflected on nucleolar integrity, we incubated cells with the p38 kinase inhibitor SB203580 or the JNK inhibitor SP600125 ± 100 nM triptolide for 6 hours. Treatment with DMSO or either inhibitor alone did not alter nucleolar structure and in addition, neither could rescue the effect of triptolide on nucleolar dispersal (Fig. 4D). Since caspase activation by triptolide has been well documented in the literature, we assessed whether the observed changes in nuclear structure could be due to proteolytic cleavage by active caspases. Incubation with the irreversible broad-spectrum caspase inhibitor, zVAD-fmk, failed to rescue triptolide induced nucleolar disassembly (Fig. 4D). Furthermore, triptolide induced caspase-3 activation was first evident after 9 hours of incubation, with strong activation beginning at 14 hours (Fig. 4D). Since nucleolar and speckle changes begin as early as 2 hours post-triptolide addition, it is unlikely that caspases are inducing these effects. We therefore continued to probe changes in proteins associated with either the nucleolus or nuclear speckles to determine a possible mechanism for triptolide induced transcriptional inhibition.
Nucleolar Disassembly is Characterized by Changes in Nucleolin, Nucleophosmin, and UBF Localization
Since we had previously characterized the loss of nucleolar integrity only by nucleolin immunofluorescent localization, we wanted to determine if NPM and UBF were also connected with this observed effect. Following triptolide incubation, nucleolin, NPM and UBF dispersed into the nucleoplasm leaving only a small remnant of the nucleolus in some cells (Fig. 5A). To examine if there were any protein stability or modification changes, nuclear lysates from an 8-hour triptolide time course were separated by SDS-PAGE, however all protein levels remained constant and there was no apparent cleavage or shift in mobility (Fig. 5B).
Figure 5.
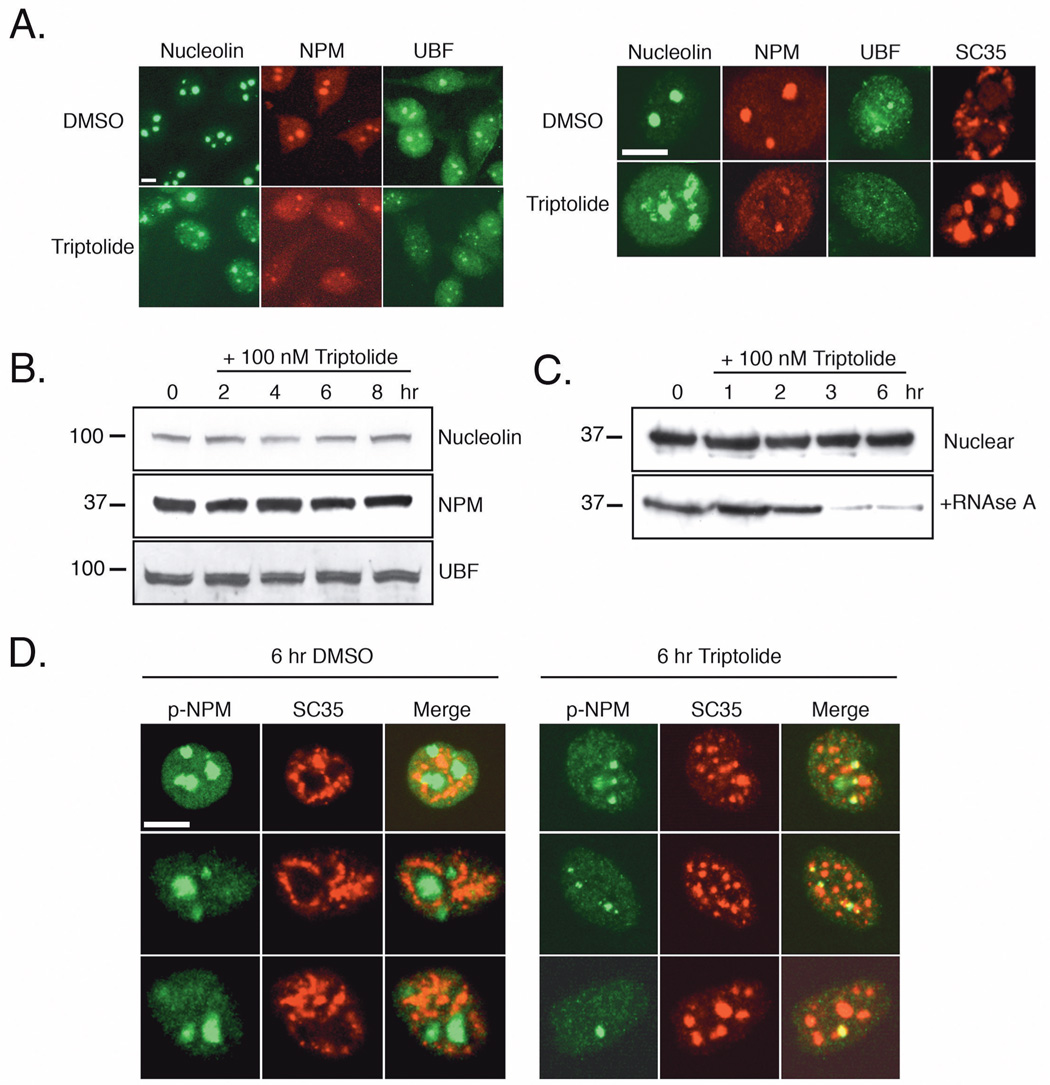
Nucleolar Disassembly is Characterized by Nucleolin, Nucleophosmin, and UBF Localization. A) HeLa cells were incubated with DMSO or 100 nM triptolide for 6 hours before fixation and immunofluorescent detection of nucleolin, nucleophosmin (NPM), upstream binding factor (UBF), or SC35. Left panel images were acquired under 25X magnification, right panel images were acquired by confocal microscopy at 63X. B) Nuclear cell lysates were prepared from cells treated with 100 nM triptolide over an 8-hour time course and separated by SDS-PAGE. Shown are representative western blots of nucleolin, NPM, and UBF protein levels. Molecular weight markers in kD are represented on the left. C) Following a 6-hour time course with 100 nM triptolide, total nuclear lysates were first prepared (nuclear). The remaining pellet was resolubilized with nuclear lysis buffer plus 0.1 mg/ml RNAse A (+RNAse A). All samples were separated by SDS-PAGE and NPM expression was detected in each fraction. D) HeLa cells were treated with DMSO vehicle or 100 nM triptolide for six hours and cells were co-stained with p-NPM (green) and SC35 (red). Representative images were acquired in separate channels and artificially merged. All scale bars represent 10 µm.
Since NPM has been found to bind RNA and aid in processing, we assessed if triptolide could alter this interaction. Following nuclear protein extraction, the remaining pellet was treated with RNAse A to release any bound NPM, as Triton X-100 extraction alone has been shown to be insufficient to solubilize this population of NPM (35). While total nuclear levels of NPM before RNAse treatment again remained stable following triptolide incubation, it was evident that the functionally active NPM population bound to RNA decreased following 3 hours (Fig. 5C). This finding suggests that RNA processing and transcription is altered in the presence of triptolide.
NPM phosphorylation at Thr199 (p-NPM) has previously been reported to target NPM to nuclear speckles and repress pre-mRNA processing in the presence of the RNA Pol II inhibitor, α-amanitin (36). Since we observed both a change in nucleolar integrity and nuclear speckle rounding following triptolide addition, we next examined p-NPM expression and localization in relation to nuclear speckles. In control treated HeLa cells, p-NPM expression was nucleolar with little or no overlap with the nuclear speckle marker SC35 (Fig. 5D). In contrast, immunofluorescent staining of triptolide treated cells showed co-localization of punctate p-NPM regions with nuclear speckles (Fig. 5D). It remained to be elucidated whether nucleolar dispersal was due to an inhibition of RNA Pol I or was an indirect effect following RNA Pol II inhibition. To address this, we next studied key phosphorylation sites on the CTD of the large subunit of RNA Pol II and its subsequent transcriptional activity.
RNA Pol II Phosphorylation and Transcriptional Activity Decreases Upon Triptolide Addition
The CTD in mammalian cells is comprised of more than 50 repeats of a heptapeptide sequence in which Ser5 and Ser2 are required for initiation and elongation of transcription, respectively (25). To determine if triptolide could affect general transcription by influencing either Ser2 or Ser5 phosphorylation, we examined nuclear lysates prepared from a triptolide time course. Western blot analysis indicated that Ser2 phosphorylation levels significantly decreased between 2–3 hours of incubation, while Ser5 levels remained largely elevated although some fluctuation was evident (Fig. 6A). It is also noteworthy that a higher molecular weight band is detected in the 3- and 6- hour time points and may be indicative of a sustained or increased level of Ser5 phosphorylation over the length of the CTD. As an additional method to evaluate changes in CTD phosphorylation, we examined Ser5 and Ser2 by immunofluorescence microscopy, and again observed only a significant decrease in Ser2 phosphorylation that was not dependent on calcium (Fig. 6A). To examine the effect of triptolide concentration on RNA Pol II phosphorylation, we incubated HeLa cells with 10–100 nM triptolide for 6-hours. Beginning at 50 nM, a mild inhibition of Ser2 phosphorylation was evident and this decrease became prominent at the 75–100 nM concentrations (Fig. 6A). This regulation of RNA Pol II seemed to be relevant to the pro-apoptotic or transcriptional inhibitory effects of triptolide as Clone-2 was not Ser2 responsive (Fig. 6B).
Figure 6.
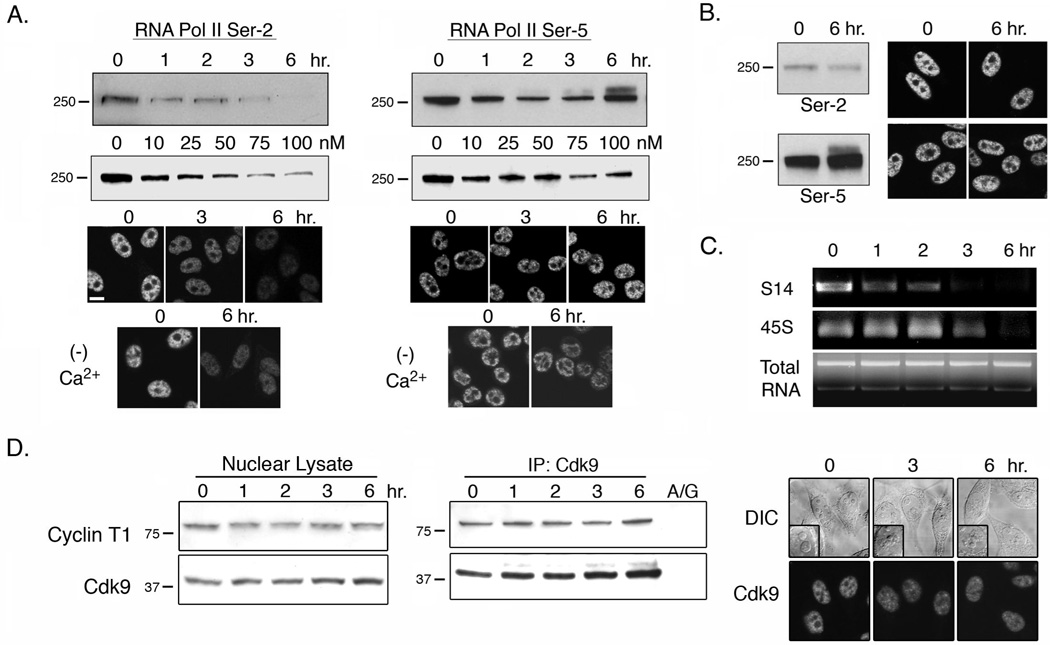
Triptolide Inhibits RNA Pol II Activity. A) HeLa cells were treated with 100 nM triptolide over a 6-hour time course or increasing triptolide concentrations for 6 hours before nuclear lysates were prepeared. RNA Pol II phosphorylation at Ser2 or Ser5 of the CTD was assessed by western blot analysis (molecular weight markers in kD are indicated to the left of each panel). Immunofluorescence localization of RNA Pol II Ser2 or Ser5 phosphorylation in the presence or absence (− Ca2+) of calcium containing media was also completed. Results are representative of four separate experiments. Scale bar is 10 µm. B) Clone-2 cells were treated with 100 nM triptolide for 6 hours and assessed for differences in Ser2 and Ser5 phosphorylation by western blot analysis and immunofluorescence localization. Results are representative of three separate experiments. C) HeLa cells were treated with 100 nM triptolide over a 6-hour time course and total RNA was extracted from each sample. RNA was normalized per sample and RT-PCR was performed to assess Pol I and Pol II activity. The ribosomal protein S14 transcript is transcribed by RNA Pol II and the 5’ ETS of 45S pre-ribosomal RNA is transcribed by RNA Pol I. Results shown are representative of three separate experiments. D) HeLa cells were incubated in the presence of 100 nM triptolide during a 6-hour time course. Total Cyclin T1 and Cdk9 protein levels (left panels), Cdk9/ Cyclin T1 complex interaction by Cdk9 IP (center panels), and Cdk9 immunolocalization (right panels) were assessed. A/G represents IP with Protein A/G agarose beads and DIC inset shows nucleolar integrity. Results are representative of three separate experiments.
All of our data collected thus far linked nucleolar disruption, nuclear speckle rounding, and a decrease in Ser2 phosphorylation of the CTD of RNA Pol II to the same time frame (6 hours) and triptolide concentrations (≥50 nM) that are associated with NFκB inhibition in HeLa cells. We next wanted to determine if RNA Pol I and/ or Pol II transcriptional activity was inhibited and if so, was there a chronological order to this effect. To do this we assessed RNA Pol II activity by transcript levels of ribosomal subunit S14 and RNA Pol I by the 5’ ETS (external transcribed spacer) region of the pre-ribosomal 45S transcript. Total RNA from an equal number of cells per time point was extracted following incubation with 100 nM triptolide over 6 hours. Regardless of triptolide addition, total RNA levels appeared similar in all samples (Fig. 6C). Following RT-PCR analysis, we observed a dramatic reduction in RNA Pol II transcriptional activity that was evident as early as 1-hour post-triptolide addition (Fig. 6C). This effect was most prominent following 3 hours of incubation that corresponds with a clear rounding of nuclear speckles (Fig. 2A). Since the 5’ ETS of the 45S rRNA is rapidly cleaved and processed following transcription, we used this sequence as a target for RT-PCR amplification to assess RNA Pol I activity. Levels of this transcript did not start to decline until 3 hours after triptolide addition, which is concomitant with the onset of nucleolar disassembly (Fig. 6C). Since the Cdk9/Cyclin T1 complex (P-TEFb) is known to phosphorylate Ser2 of the RNA Pol II CTD, we next examined the expression of these two proteins. Over a 6 hour, 100 nM triptolide time course, total nuclear levels of Cdk9 or Cyclin T1 were constant (Fig. 6D). Complex formation was tested by co-immunoprecipitation experiments and again Cdk9/Cyclin T1 interaction remained stable (Fig. 6D). Additionally, Cdk9 localization remained nuclear even as nucleolar disassembly became evident (Fig. 6D).
Taken together, our data support a model in which high pro-apoptotic concentrations of triptolide cause RNA Pol II transcriptional inhibition through a decrease in Ser2 phosphorylation. Subsequent to RNA Pol II inhibition, RNA Pol I transcriptional activity is also attenuated impacting RNA ribosome biogenesis as early as 3 hours following triptolide addition. Therefore, we conclude that triptolide affects global transcription in a concentration dependent manner as evidenced by: 1) attenuation of transcripts associated with ribosome biogenesis, 2) RNA Pol II activity and its regulatory phosphorylation decreases, 3) nuclear speckle rounding is evident indicating mRNA splicing has ceased, and 4) normal nucleolar structure is lost and RNA Pol I activity decreases.
Discussion
Triptolide, a natural product isolated from a Chinese medicinal plant, may be therapeutically relevant for a variety of proliferative disorders such as cancer (37, 38) and Autosomal Dominant Polycystic Kidney Disease (7), or autoimmune diseases and inflammation such as systemic lupus erythematosus and arthritis (39, 40). Its cellular effects and mechanisms of action are highly complex and undoubtedly involve multiple biological pathways. Ongoing studies in several laboratories have elucidated key points in triptolide’s mode of action such as concentration and cell type dependent effects (37), a partial dependence on calcium (6), modulation of apoptosis activating proteins (3–5), and inhibition of the transcription factor NFκB at a step following DNA binding (8). While the emphasis of triptolide-induced transcriptional inhibition has focused on NFκB and its pro-inflammatory gene targets, triptolide has also been reported to suppress AP-1 (41), NFAT (8) and HSF1 (42) transactivation as well. In contrast to the hypothesis that triptolide targets transcription factors with specificity, triptolide has also been reported to suppress total RNA synthesis (9), however a mechanism has not been described. We began a careful analysis of nuclear sub-structure changes indicative of global transcriptional arrest at triptolide concentrations and time-points known to inhibit NFκB mediated transcription (6). Furthermore, we were able to characterize changes in RNA polymerase activity describing a potential mechanistic action.
Known transcriptional inhibitors that affect RNA Pol II such as actinomycin D, DRB, and α-amanitin have all been previously shown to cause nuclear speckle rounding and/ or nucleolar disruption. It should, however, be noted that coordination of RNA Pol I, II, and III activity is tightly regulated as inhibition of one will affect the efficiency of the others (43, 44). This coordinated action of all RNA Polymerases ensures sufficient protein subunits and rRNA to complete ribosome assembly. We first observed a reversible nucleolar disassembly induced by triptolide and further explored if it was an indication of global transcriptional changes in the cell as assessed by nuclear speckle morphology. While speckle size, shape, and number can vary per cell, RNA Pol II transcriptional inhibition is marked by an accumulation of the splicing machinery and a concomitant increase in speckle size. Although 100 nM triptolide incubation resulted in a decrease in RNA Pol II transcription after only one hour, changes in speckle morphology and a decrease in Ser2 RNA Pol II phosphorylation were readily observed after two hours. Additionally, residual p-NPM was found associated with nuclear speckles, an event previously correlated with transcriptional inactivity (36). Ser5 phosphorylation was relatively constant throughout our time course, however we did observe minor fluctuations. This could be due to cell cycle dependent effects or may reflect differences in the number of Ser5 phosphorylations along the full-length of the CTD. While we believe that Ser2 is the site of primary regulation by triptolide, we cannot rule out an additional or indirect effect on Ser5 phosphorylation as well.
If we examine the time-line of data stemming from these experiments, a model emerges where triptolide (100 nM) has an effect on RNA Pol II efficiency in just one hour as assessed by transcript levels of the S14 ribosomal subunit. After two hours, nuclear speckles become rounded, Pol II transcriptional efficiency continues to decline, and RNA Pol II Ser2 phosphorylation decreases. Following three hours, the nucleolus begins to unravel, and RNA Pol I activity now begins to decrease. It is of note that at this time, these effects are still reversible and cells may fully recover provided that triptolide is removed from culture. However, after six hours of continual culture, RNA Pol I and II activities are absent and nucleolar organization has dissolved, which may therefore be the turning point towards the commitment to cell death. We should also note that this time line of cellular effects may be shifted by triptolide concentration. For example, our data with 25 nM triptolide (previously shown to induce cell death in HeLa) does not show any effect on transcription, nucleolar disassembly, or speckle rounding at the six-hour time point. However, after 16 hours in culture, 25 nM causes prominent enlargement and rounding of speckles ultimately leading to cell death.
We additionally generated a triptolide resistant clonal population of HeLa (Clone-2) to test if these nuclear changes were specific to both transcription and cell death. While Clone-2 cells exhibited a very minor fluctuation in nucleolar integrity and transcriptional inhibition, cells were able to escape from death and dissolution of the nucleolus during an extended incubation with 100 nM triptolide. The cells from this clonal line retain the majority of their transcriptional activity, and do not undergo nuclear speckle rounding or inhibition of Ser2 phosphorylation. We have yet to discover why these cells are relatively resistant to triptolide induced nucleolar disruption or RNA Pol II inactivation, but we are currently investigating these questions as we further dissect triptolide’s mechanism of action.
Taken together we conclude that NFκB transcriptional inhibition in HeLa cells correlates with global transcriptional and structural changes in the cell. Since the RelA subunit of NFκB has been shown to recruit P-TEFb to stimulate RNA Pol II transcriptional elongation in response to cytokine stimulation (45), it is possible that this explains why the NFκB pathway is exquisitely sensitive to triptolide regulation in multiple cell lines. In fact, treatment of cells with the P-TEFb inhibitor, DRB, sensitizes cells to TNF-α induced apoptosis, similar to the effect observed with triptolide. While we found no obvious changes in Cdk9/Cyclin T1 interaction and no changes in protein levels or localization in Cdk9 it is likely that other mechanisms of regulation exist such as co-factor recruitment to the RNA Pol II transcriptional complex, acetylation events, or regulation of Cdk9 binding to HEXIM1 and 7SK snRNA (46, 47).
Triptolide is a small molecule with multiple physiological effects such as immune modulation and chemotherapeutic. Its biological effects on cell growth and transcription vary by concentration, duration of exposure, and cell type. Here, we have demonstrated that triptolide induces nuclear sub-structural changes in both the nucleolus and nuclear speckles that are associated with a rapid decrease in RNA Pol II transcriptional efficiency. Use of these nuclear sub-structures as surrogate markers for transcriptional arrest and possibly cytotoxicity will be of great utility in delineating the separate biological functions of triptolide and its derivatives. As triptolide’s mode of action continues to unfold it may eventually be possible to understand its multiple regulatory actions on the cell as well as its therapeutic potentials.
Acknowledgments
Financial Support: This work was funded by the NIH (AI055914) and a Postdoctoral Fellowship from the American Cancer Society (SJL).
References
- 1.Wu Y, Cui J, Bao X, et al. Triptolide attenuates oxidative stress, NF-kappaB activation and multiple cytokine gene expression in murine peritoneal macrophage. Int J Mol Med. 2006;17:141–150. [PubMed] [Google Scholar]
- 2.Zhao G, Vaszar LT, Qiu D, Shi L, Kao PN. Anti-inflammatory effects of triptolide in human bronchial epithelial cells. Am J Physiol Lung Cell Mol Physiol. 2000;279:L958–L966. doi: 10.1152/ajplung.2000.279.5.L958. [DOI] [PubMed] [Google Scholar]
- 3.Carter BZ, Mak DH, Schober WD, et al. Triptolide induces caspase-dependent cell death mediated via the mitochondrial pathway in leukemic cells. Blood. 2006;108:630–637. doi: 10.1182/blood-2005-09-3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi YJ, Kim TG, Kim YH, et al. Immunosuppressant PG490 (triptolide) induces apoptosis through the activation of caspase-3 and down-regulation of XIAP in U937 cells. Biochem Pharmacol. 2003;66:273–280. doi: 10.1016/s0006-2952(03)00282-x. [DOI] [PubMed] [Google Scholar]
- 5.Wan CK, Wang C, Cheung HY, Yang M, Fong WF. Triptolide induces Bcl-2 cleavage and mitochondria dependent apoptosis in p53-deficient HL-60 cells. Cancer Lett. 2006;241:31–41. doi: 10.1016/j.canlet.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 6.Leuenroth SJ, Crews CM. Studies on calcium dependence reveal multiple modes of action for triptolide. Chem Biol. 2005;12:1259–1268. doi: 10.1016/j.chembiol.2005.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leuenroth SJ, Okuhara D, Shotwell JD, et al. Triptolide is a traditional Chinese medicine-derived inhibitor of polycystic kidney disease. Proc Natl Acad Sci U S A. 2007;104:4389–4394. doi: 10.1073/pnas.0700499104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Qiu D, Zhao G, Aoki Y, et al. Immunosuppressant PG490 (triptolide) inhibits T-cell interleukin-2 expression at the level of purine-box/nuclear factor of activated T- cells and NF-kappaB transcriptional activation. J Biol Chem. 1999;274:13443–13450. doi: 10.1074/jbc.274.19.13443. [DOI] [PubMed] [Google Scholar]
- 9.McCallum C, Kwon S, Leavitt P, Shen DM, Liu W, Gurnett A. Triptolide binds covalently to a 90 kDa nuclear protein. Role of epoxides in binding and activity. Immunobiology. 2007;212:549–556. doi: 10.1016/j.imbio.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Boisvert FM, van Koningsbruggen S, Navascues J, Lamond AI. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- 11.Haaf T, Ward DC. Inhibition of RNA polymerase II transcription causes chromatin decondensation, loss of nucleolar structure, and dispersion of chromosomal domains. Exp Cell Res. 1996;224:163–173. doi: 10.1006/excr.1996.0124. [DOI] [PubMed] [Google Scholar]
- 12.Perry RP, Kelley DE. Inhibition of RNA synthesis by actinomycin D: characteristic dose-response of different RNA species. J Cell Physiol. 1970;76:127–139. doi: 10.1002/jcp.1040760202. [DOI] [PubMed] [Google Scholar]
- 13.Pinol-Roma S, Dreyfuss G. Transcription-dependent and transcription-independent nuclear transport of hnRNP proteins. Science. 1991;253:312–314. doi: 10.1126/science.1857966. [DOI] [PubMed] [Google Scholar]
- 14.Andersen JS, Lam YW, Leung AK, et al. Nucleolar proteome dynamics. Nature. 2005;433:77–83. doi: 10.1038/nature03207. [DOI] [PubMed] [Google Scholar]
- 15.Scherl A, Coute Y, Deon C, et al. Functional proteomic analysis of human nucleolus. Mol Biol Cell. 2002;13:4100–4109. doi: 10.1091/mbc.E02-05-0271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ginisty H, Sicard H, Roger B, Bouvet P. Structure and functions of nucleolin. J Cell Sci. 1999;112:761–772. doi: 10.1242/jcs.112.6.761. [DOI] [PubMed] [Google Scholar]
- 17.Okuda M, Horn HF, Tarapore P, et al. Nucleophosmin/B23 is a target of CDK2/cyclin E in centrosome duplication. Cell. 2000;103:127–140. doi: 10.1016/s0092-8674(00)00093-3. [DOI] [PubMed] [Google Scholar]
- 18.Szebeni A, Olson MO. Nucleolar protein B23 has molecular chaperone activities. Protein Sci. 1999;8:905–912. doi: 10.1110/ps.8.4.905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Herrera JE, Savkur R, Olson MO. The ribonuclease activity of nucleolar protein B23. Nucleic Acids Res. 1995;23:3974–3979. doi: 10.1093/nar/23.19.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Falini B, Nicoletti I, Bolli N, et al. Translocations and mutations involving the nucleophosmin (NPM1) gene in lymphomas and leukemias. Haematologica. 2007;92:519–532. doi: 10.3324/haematol.11007. [DOI] [PubMed] [Google Scholar]
- 21.Beckmann H, Chen JL, O'Brien T, Tjian R. Coactivator and promoter-selective properties of RNA polymerase I TAFs. Science. 1995;270:1506–1509. doi: 10.1126/science.270.5241.1506. [DOI] [PubMed] [Google Scholar]
- 22.Cavanaugh AH, Hempel WM, Taylor LJ, Rogalsky V, Todorov G, Rothblum LI. Activity of RNA polymerase I transcription factor UBF blocked by Rb gene product. Nature. 1995;374:177–180. doi: 10.1038/374177a0. [DOI] [PubMed] [Google Scholar]
- 23.Lamond AI, Spector DL. Nuclear speckles: a model for nuclear organelles. Nat Rev Mol Cell Biol. 2003;4:605–612. doi: 10.1038/nrm1172. [DOI] [PubMed] [Google Scholar]
- 24.Herrmann CH, Mancini MA. The Cdk9 and cyclin T subunits of TAK/P-TEFb localize to splicing factor-rich nuclear speckle regions. J Cell Sci. 2001;114:1491–1503. doi: 10.1242/jcs.114.8.1491. [DOI] [PubMed] [Google Scholar]
- 25.Hirose Y, Ohkuma Y. Phosphorylation of the C-terminal domain of RNA polymerase II plays central roles in the integrated events of eucaryotic gene expression. J Biochem (Tokyo) 2007;141:601–608. doi: 10.1093/jb/mvm090. [DOI] [PubMed] [Google Scholar]
- 26.Komarnitsky P, Cho EJ, Buratowski S. Different phosphorylated forms of RNA polymerase II and associated mRNA processing factors during transcription. Genes Dev. 2000;14:2452–2460. doi: 10.1101/gad.824700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marshall NF, Peng J, Xie Z, Price DH. Control of RNA polymerase II elongation potential by a novel carboxyl-terminal domain kinase. J Biol Chem. 1996;271:27176–27183. doi: 10.1074/jbc.271.43.27176. [DOI] [PubMed] [Google Scholar]
- 28.Peterlin BM, Price DH. Controlling the elongation phase of transcription with P-TEFb. Mol Cell. 2006;23:297–305. doi: 10.1016/j.molcel.2006.06.014. [DOI] [PubMed] [Google Scholar]
- 29.O'Keefe RT, Mayeda A, Sadowski CL, Krainer AR, Spector DL. Disruption of pre-mRNA splicing in vivo results in reorganization of splicing factors. J Cell Biol. 1994;124:249–260. doi: 10.1083/jcb.124.3.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spector DL, Schrier WH, Busch H. Immunoelectron microscopic localization of snRNPs. Biol Cell. 1983;49:1–10. doi: 10.1111/j.1768-322x.1984.tb00215.x. [DOI] [PubMed] [Google Scholar]
- 31.Pusl T, Wu JJ, Zimmerman TL, et al. Epidermal growth factor-mediated activation of the ETS domain transcription factor Elk-1 requires nuclear calcium. J Biol Chem. 2002;277:27517–27527. doi: 10.1074/jbc.M203002200. [DOI] [PubMed] [Google Scholar]
- 32.Ferrari S, Tagliafico E, Manfredini R, et al. Abundance of the primary transcript and its processed product of growth-related genes in normal and leukemic cells during proliferation and differentiation. Cancer Res. 1992;52:11–16. [PubMed] [Google Scholar]
- 33.Grandori C, Gomez-Roman N, Felton-Edkins ZA, et al. c-Myc binds to human ribosomal DNA and stimulates transcription of rRNA genes by RNA polymerase I. Nat Cell Biol. 2005;7:311–318. doi: 10.1038/ncb1224. [DOI] [PubMed] [Google Scholar]
- 34.Mayer C, Bierhoff H, Grummt I. The nucleolus as a stress sensor: JNK2 inactivates the transcription factor TIF-IA and down-regulates rRNA synthesis. Genes Dev. 2005;19:933–941. doi: 10.1101/gad.333205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okuwaki M, Tsujimoto M, Nagata K. The RNA binding activity of a ribosome biogenesis factor, nucleophosmin/B23, is modulated by phosphorylation with a cell cycle-dependent kinase and by association with its subtype. Mol Biol Cell. 2002;13:2016–2030. doi: 10.1091/mbc.02-03-0036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tarapore P, Shinmura K, Suzuki H, et al. Thr199 phosphorylation targets nucleophosmin to nuclear speckles and represses pre-mRNA processing. FEBS Lett. 2006;580:399–409. doi: 10.1016/j.febslet.2005.12.022. [DOI] [PubMed] [Google Scholar]
- 37.Kiviharju TM, Lecane PS, Sellers RG, Peehl DM. Antiproliferative and proapoptotic activities of triptolide (PG490), a natural product entering clinical trials, on primary cultures of human prostatic epithelial cells. Clin Cancer Res. 2002;8:2666–2674. [PubMed] [Google Scholar]
- 38.Yang S, Chen J, Guo Z, et al. Triptolide inhibits the growth and metastasis of solid tumors. Mol Cancer Ther. 2003;2:65–72. [PubMed] [Google Scholar]
- 39.Qiu D, Kao PN. Immunosuppressive and anti-inflammatory mechanisms of triptolide, the principal active diterpenoid from the Chinese medicinal herb Tripterygium wilfordii Hook. f. Drugs R D. 2003;4:1–18. doi: 10.2165/00126839-200304010-00001. [DOI] [PubMed] [Google Scholar]
- 40.Tao X, Cush JJ, Garret M, Lipsky PE. A phase I study of ethyl acetate extract of the chinese antirheumatic herb Tripterygium wilfordii hook F in rheumatoid arthritis. J Rheumatol. 2001;28:2160–2167. [PubMed] [Google Scholar]
- 41.Jiang XH, Wong BC, Lin MC, et al. Functional p53 is required for triptolide-induced apoptosis and AP-1 and nuclear factor-kappaB activation in gastric cancer cells. Oncogene. 2001;20:8009–8018. doi: 10.1038/sj.onc.1204981. [DOI] [PubMed] [Google Scholar]
- 42.Westerheide SD, Kawahara TL, Orton K, Morimoto RI. Triptolide, an inhibitor of the human heat shock response that enhances stress-induced cell death. J Biol Chem. 2006;281:9619–9622. doi: 10.1074/jbc.M512044200. [DOI] [PubMed] [Google Scholar]
- 43.Laferte A, Favry E, Sentenac A, Riva M, Carles C, Chedin S. The transcriptional activity of RNA polymerase I is a key determinant for the level of all ribosome components. Genes Dev. 2006;20:2030–2040. doi: 10.1101/gad.386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mayer C, Grummt I. Ribosome biogenesis and cell growth: mTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene. 2006;25:6384–6391. doi: 10.1038/sj.onc.1209883. [DOI] [PubMed] [Google Scholar]
- 45.Barboric M, Nissen RM, Kanazawa S, Jabrane-Ferrat N, Peterlin BM. NF-kappaB binds P-TEFb to stimulate transcriptional elongation by RNA polymerase II. Mol Cell. 2001;8:327–337. doi: 10.1016/s1097-2765(01)00314-8. [DOI] [PubMed] [Google Scholar]
- 46.Michels AA, Nguyen VT, Fraldi A, et al. MAQ1 and 7SK RNA interact with CDK9/cyclin T complexes in a transcription-dependent manner. Mol Cell Biol. 2003;23:4859–4869. doi: 10.1128/MCB.23.14.4859-4869.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yik JH, Chen R, Nishimura R, Jennings JL, Link AJ, Zhou Q. Inhibition of P-TEFb (CDK9/Cyclin T) kinase and RNA polymerase II transcription by the coordinated actions of HEXIM1 and 7SK snRNA. Mol Cell. 2003;12:971–982. doi: 10.1016/s1097-2765(03)00388-5. [DOI] [PubMed] [Google Scholar]


