Abstract
Purpose
Elevated cellular proliferation and cell cycle abnormalities, which have been associated with premalignant lesions, may be caused by inactivation of tumor suppressor genes. We measured proliferative and cell cycle fractions of biopsies from a cohort of patients with Barrett’s esophagus to better understand the role of proliferation in early neoplastic progression and the association between cell cycle dysregulation and tumor suppressor gene inactivation.
Experimental Design
Cell proliferative fractions (determined by Ki67/DNA multiparameter flow cytometry) and cell cycle fractions (DNA content flow cytometry) were measured in 853 diploid biopsies from 362 patients with Barrett’s esophagus. The inactivation status of CDKN2A and TP53 was assessed in a subset of these biopsies in a cross-sectional study. A prospective study followed 276 of the patients without detectable aneuploidy for an average of 6.3 years with esophageal adenocarcinoma as an endpoint.
Results
Diploid S and 4N (G2/tetraploid) fractions were significantly higher in biopsies with TP53 mutation and LOH. CDKN2A inactivation was not associated with higher Ki67-positive, diploid S, G1, or 4N fractions. High Ki67-positive and G1 phase fractions were not associated with the future development of esophageal adenocarcinoma (p=0.13 and p=0.15, respectively), while high diploid S phase and 4N fractions were (p=0.03 and p<0.0001, respectively).
Conclusions
High Ki67-positive proliferative fractions were not associated with inactivation of CDKN2A and TP53 or future development of cancer in our cohort of patients with Barrett’s esophagus. Bi-allelic inactivation of TP53 was associated with elevated 4N fractions, which have been associated with the future development of esophageal adenocarcinoma.
Keywords: Barrett’s esophagus, esophageal adenocarcinoma, cell cycle, p16, p53
Introduction
Increased cellular proliferation and dysregulation of the cell cycle have been reported in advancing histologic grades of neoplastic progression in a large number of cross-sectional studies(1–4). Advances in basic science over several decades support the hypothesis that these changes are due to mutation, loss, or inactivation of cell cycle control genes.. The initial genetic model of the eukaryotic cell cycle reported that a regulatory element, called START in the yeast Saccharomyces cerevisiae, controlled the transition from G1 to S phase(5). This regulatory element was subsequently shown to be evolutionarily conserved and similar G1, S phase controls were identified in mammalian species, including humans(6). The potential importance of G1, S regulation in human neoplastic progression became clearer when tumor suppressor genes, such as CDKN2A (p16) and TP53 (p53), were identified, and well designed molecular biological studies in model systems and organisms elucidated mechanisms of tumor suppression that included control of the G1, S phase transition(7–10). These tumor suppressors could be inactivated by a two hit mechanism involving loss of heterozygosity (LOH) of one allele and mutation or methylation of the second(11–13). Abnormalities involving CDKN2A and TP53 are among the most commonly reported in human cancers and premalignant neoplasms(14, 15).
Barrett’s esophagus (BE) is a condition in which the normal esophageal squamous epithelium is replaced by an intestinal metaplasia associated with an increased risk of developing esophageal adenocarcinoma (EA)(16). Cell proliferation in Barrett’s epithelium is similar to the small intestine, but increased compared to normal esophageal squamous epithelium(17, 18). Proliferation has been measured by a variety of techniques in BE, including tritiated thymidine incorporation, immunohistochemical markers, such as Ki67, PCNA, cyclin A, and minichromosome maintenance proteins, DNA content flow cytometry and multiparameter flow cytometry(17–26). Several, but not all, cross-sectional studies using DNA content flow cytometry, multiparameter flow cytometry and immunohistochemistry have reported associations between abnormal proliferation/cell cycle fractions and advancing grades of dysplasia(18, 19, 21, 24–27). Similarly, in cross-sectional studies of individual BE crypts, the total number of proliferating cells appears to increase with progressive grades of dysplasia due to an expansion of the crypt proliferative compartment(22). Two previous studies, one in BE and one in colonic adenomas, have reported an association between TP53 abnormalities and elevated 4N fractions(28, 29). However the effects of loss of these genes on cell proliferation in human diploid biopsies in vivo are largely unknown.
The number of prospective studies of proliferative/cell cycle abnormalities as predictors of progression from BE to EA is much smaller. One study reported increased 4N fraction (and to a lesser extent, S phase) to be associated with progression to EA in persons with BE(23). However, no previous prospective cohort study has comprehensively evaluated increased proliferation and cell cycle fractions as candidate predictors of progression from BE to EA. Also, no previous cohort study has comprehensively evaluated associations between proliferation and cell cycle fractions and inactivation of CDKN2A and TP53 to determine whether abnormal proliferation is associated with loss of CDKN2A and TP53 regulation of the transition from G1 to S phase.
The Seattle Barrett’s Esophagus Study is designed around a dynamic prospective cohort whose research participants are being followed to identify risk and protective factors that are associated with progression or lack of progression to esophageal adenocarcinoma(23, 30–33). Here, we report for the first time results of a comprehensive study of proliferative and cell cycle abnormalities in diploid cells as predictors of progression from BE to EA using Ki67/DNA content multiparameter flow cytometry. Ki67 antibody labels nuclei in the G1/S/G2/M phases of the cell cycle, but not in G0 (quiescent cells)(34). By combining Ki67 labeling and DNA content flow cytometry, it is possible to measure total proliferative fractions as well as individual cell cycle phases, including G1, S, and G2/M(21). We tested the associations between total proliferative and cell cycle fractions and progression from BE to EA in 276 individuals. To our knowledge, this is the first prospective cohort study of cancer risk prediction based on cellular proliferation in cancer-free individuals. We were also able to compare the proliferative/cell cycle abnormalities directly with CDKN2A and TP53 status in the same biopsies by Ki67/DNA content multiparameter flow sorting for LOH, mutation and methylation detection. This cross-sectional analysis of biopsies from 362 individuals is, to our knowledge, the first to comprehensively investigate diploid cell proliferative/cell cycle fractions and CDKN2A and TP53 abnormalities in the same biopsies in humans.
Materials and methods
Study Subjects and Tissue Acquisition
The 362 participants in the study (Table 1) were enrolled in the Seattle Barrett’s Esophagus Study between 1983 and 1999. Participants with EA at baseline were excluded from analysis. Baseline biopsies were obtained between 1989 and 2003. Biopsies from 214 of the participants were evaluated for CDKN2A mutation, 192 for TP53 mutation, 245 for 9p LOH, 241 for 17p LOH, and 96 for CDKN2A methylation. The 276 participants were followed from 89 days to 12.1 years. The Seattle Barrett’s Esophagus Study was approved by the Human Subjects Division of the University of Washington in 1983 and renewed annually thereafter with reciprocity from the Fred Hutchinson Cancer Research Center (FHCRC) Institutional Review Board from 1993 to 2001. Since 2001, the study has been approved by the FHCRC IRB with reciprocity from the University of Washington Human Subjects Division.
Table 1.
Patient information at the time of the baseline visit (362 patients).
| # persons | % of total | |
|---|---|---|
| Gender | ||
| Male | 293 | 80.9% |
| Female | 69 | 19.1% |
| Age | ||
| 26–55 | 108 | 29.8% |
| 55–70 | 142 | 39.2% |
| 70+ | 112 | 30.9% |
| Follow-up | ||
| None | 86 | 23.8% |
| 89 days-2 years | 40 | 11.0% |
| 2 years-5 years | 71 | 19.6% |
| 5 years-10 years | 114 | 31.5% |
| 10+ years | 51 | 14.1% |
Endoscopic biopsy protocols used in the Seattle Barrett’s Esophagus Study have been published previously(23, 30). Four quadrant biopsies for histology were taken every 1 cm (for patients with high-grade dysplasia) or every 2 cm (for patients without high-grade dysplasia) at intervals ranging from every 6 months (for high grade dysplasia) to 3 years, as described previously(23, 30). Endoscopic biopsies every 2-cm for flow cytometry and molecular studies were placed into media with 10% DMSO (dimethyl sulfoxide) within 15 seconds and held on ice until frozen and stored at −70C.
We included all cancers that developed subsequent to the baseline evaluation so that accurate risk stratification models can be developed based on findings at a single baseline endoscopy. All participants had at least one biopsy evaluated for biomarkers every two centimeters in the Barrett’s segment regardless of the histologic diagnosis. For the prospective study of cancer outcome, we included only the biopsies obtained for biomarker analysis by the standard one biopsy per 2 cm protocol in order to avoid sampling bias.
Ki67/DNA Content Multiparameter Flow Cytometry and Sorting
Frozen endoscopic biopsies (n=853) were prepared for flow cytometry as described previously(19, 35). To ensure accurate detection of 4N DNA content abnormalities, the suspension of unfixed nuclei from each biopsy was aliquoted into separate microfuge tubes for DNA content flow cytometric analysis and multiparameter Ki67/DNA content cell sorting. The aliquots with DAPI (10 μg/ml, Accurate Chemical, Westbury, NY) saturated nuclei for DNA content flow cytometry were never centrifuged and were triturated with a 25 gauge needle immediately before evaluation on the flow cytometer to prevent clumping of nuclei that will increase the 4N population as an artifact. DNA content analysis was performed using MultiCycle software with a standardized peak vs area gate to exclude residual doublets and with “sliced nucleus” background correction. In accordance with published guidelines(36), histograms containing less than 10,000 events or debris in excess of 20% were not considered adequate for S phase analysis. Two of the authors (CAS and PSR) interpreted flow cytometric histograms independently, with disagreements resolved by joint review of the histogram. See Figure 1C for a representative DNA content histogram.
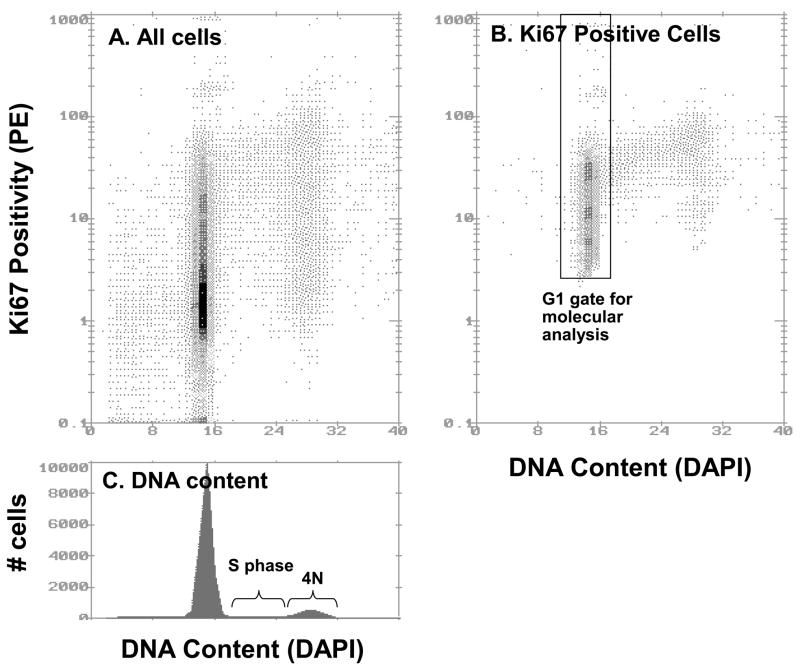
Figure 1.
Ki67-Phycoerthyrin (PE) and DNA content flow cytometry. Panel A: Bivariate histogram displaying DAPI fluorescence on the x-axis (linear) and Ki67-PE fluorescence on the y-axis (logarithmic; 4 decades) of diploid cells in a Barrett’s esophagus biopsy. Panel B: Ki67-positive proliferating diploid cells, as determined using the bivariate subtraction algorithm (Multicycle). The total Ki67-positive fraction of this sample was calculated to be 35.5%. A representation of the diploid G1 sorting gate is shown. Panel C: A single parameter histogram of DNA content, with S and 4N (G2/M) cells indicated. The S phase fraction is 6.5% and the 4N fraction is 9.9%. There was no evidence of a tetraploid cell cycle.
Ki67/DNA content multiparameter cell sorting was used to purify the proliferating BE epithelium from non-proliferating G0 cells into cell cycle fractions including G1, 4N (G2/tetraploid) for molecular studies as previously described(21, 33, 35). Briefly, the nuclei were incubated with directly conjugated Ki67–RPE (phycoerythrin) or isotype control–RPE (DAKO R0840, Carpenteria, CA (DAKO reagent no longer available, BD Pharmigen #556027 markets a similar reagent) and DAPI for DNA content. The antibody protocol includes a centrifugation step and is therefore inadequate for accurate assessment of the 4N fraction. Ki67/DNA content flow cytometry was also used to assess total Ki67-positive proliferative fractions and Ki67-positive G1 fractions. The Ki67-positive fraction is calculated by bivariate curve subtraction of the negative control from the Ki67 stained cytogram with the program Multicycle (Phoenix Flow Systems, San Diego, CA) as previously described(21). Samples were sorted on a Beckman Coulter (Fullerton, CA) Elite cell sorter. For the molecular analyses used in this study, the Ki67-positive diploid G1 cells were sorted (Figure 1B). Data from biopsies with aneuploid fractions were not used in this study.
Microsatellite LOH, DNA Sequence, and p16 Methylation Analyses
Flow-sorted fractions from diploid biopsies were evaluated for 9p21 (CDKN2A/p16) and 17p13 (TP53/p53) LOH using polymorphic microsatellite markers, as described previously(35, 37–39). DNA was extracted from the Ki67/DNA content sorted cell populations and whole genome amplification using primer extension preamplification (PEP) was performed as described previously(37) for each sorted fraction and three constitutive controls per participant. LOH status for chromosome 9 was assessed in 526 samples from 245 participants and for chromosome 17 was assessed in 521 samples from 241 participants.
DNA was sequenced using BigDye or BigDyeV3 Terminator cycle sequencing (Applied Biosystems, Foster City, CA) on either an ABI 377, 3700, or 3730 DNA sequencer. Wild type sequences were confirmed using constitutive samples. All mutations were confirmed by at least two independent PCR and sequencing reactions. Evaluation of mutation of exons 5–9 of the TP53 gene was performed on 382 flow-purified fractions from 192 participants as described previously(11). Sequence analysis of exon 2 of the CDKN2A gene was performed on 424 flow-purified fractions from 214 participants as described previously(13).
One hundred and sixty five samples from 96 participants were evaluated for methylation of the CDKN2A CpG islands as described previously(13). A subset of these results were previously reported in a cross-sectional study(13). Samples were classified as either positive or negative for CDKN2A methylation on the basis of a positive result from methylation specific PCR performed on bisulfite treated DNA. The requirement for larger amounts of DNA for input into the bisulfite reaction limited the number of samples that could be analyzed by this technique.
Statistical Analysis
Univariate proportional-hazards models were used to calculate the hazard ratios (HR), confidence intervals (CI), and p values for the association of Ki67-positive and cell cycle fractions with cancer outcome. The Wilcoxon rank-sum test was used to compare Ki67-positive and cell cycle fractions between sets of biopsies. Linear regression was used to measure the correlation between Ki67 and S phase fractions. A chi-square test was used to determine the association among CDKN2A and between TP53 inactivation events. Receiver operating characteristic (ROC) curves(40) were used to plot the sensitivity and specificity of using various thresholds of S phase or 4N fractions to discriminate between biopsies with and without TP53 inactivation. The sensitivity and specificity using all S phase and 4N fractions observed in our samples were used as thresholds to create the ROC curves. Areas under the curves (AUC), a measure of discriminator accuracy with a range of 0.5 (no discrimination) to 1.0 (perfect discriminator), were computed using the trapezoidal rule. In standard ROC analyses, the output of the classifier is inverted if a classifier performs worse than random. However, because we hypothesize that inactivation of TP53 increases S and 4N fractions, all specificities and sensitivities are computed in terms of the number of TP53-inactivated samples with S or 4N fractions above a threshold. Therefore, we allow our analysis to include classifiers that perform worse than chance. In all analyses, neutral TP53 mutations were considered equivalent to TP53 wild type. The confidence intervals for the AUCs were estimated using the bootstrap (10,000 trials). Statistical analyses were carried out using the R statistical computing language version 2.5.1 (R Foundation for Statistical Computing, Vienna, Austria, 2007).
Results
Of the 276 participants with follow-up (average of 6.3 years, 89 days–12.1 years, total of 1752 person–years), 29 developed EA. High S and 4N fractions were significantly associated with future development of EA (p = 0.03 and p < 0.0001 modeling the fractions as continuous variables, respectively), while high Ki67-positive (the proliferative fraction that comprises cells in G1, S, and G2/M) and G1 fractions were not associated with subsequent progression to cancer (p = 0.13 and 0.15, respectively) (Table 2). Because cells with the largest cell cycle abnormalities may have the greatest contribution to neoplastic progression, the associations were computed using the biopsy with the maximum Ki67-positive, G1 phase, S phase, and 4N fractions for patients with more than one biopsy. However, estimating the associations using the average values did not affect the results (data not shown). Aneuploid biopsies were excluded from this analysis because aneuploidy is a late event in neoplastic progression and has already been associated with a high risk of progression to EA in this cohort(23). This allowed us to directly test the hypotheses that proliferative and cell cycle abnormalities are predictors of progression from BE to EA that develop as early events due to inactivation of the tumor suppressors CDKN2A and TP53. Further, the calculation of the diploid S phase is more accurate in diploid biopsies because the software algorithm that calculates the diploid S phase fractions does not have to account for the overlapping aneuploid cell population.
Table 2.
Associations between the future development of EA and Ki67-positive and cell cycle fractions. The Cox regressions were performed using the maximum values per patient obtained from their baseline endoscopies using a 2-cm sampling protocol.
| index | outcomes | # persons | HR | 95% CI | p value |
|---|---|---|---|---|---|
| Ki67-positive fraction | 29 | 276 | 1.02 | 0.99–1.05 | 0.13 |
| G1 | 29 | 276 | 1.02 | 0.99–1.06 | 0.15 |
| S phase fraction | 26 | 253b | 1.16 | 1.01–1.32 | 0.03 |
| 4N fraction | 29 | 269 | 1.36 | 1.26–1.47 | < 0.0001 |
HR and 95% CI are scaled to indicate increased risk of progression for each 1% increase in Ki67-positive, S phase, or 4N fraction.
S phase fractions in biopsies from 16 patients could not be accurately estimated
Although Ki67-positive fractions were not associated with cancer outcome and diploid S phase fractions were, these two frequently used measures of cellular proliferation were statistically significantly correlated but with a low correlation coefficient in the BE biopsies (p < 0.0001, adjusted r2 = 0.12).
We tested the associations between CDKN2A inactivation and cell proliferative and cell cycle fractions. Ki67-positive, S phase, and 4N fractions were measured in diploid BE biopsies at the same time as the Ki67-positive G1 cells were sorted for subsequent examination of the mutation and LOH statuses of CDKN2A and TP53 and the CpG island methylation status of CDKN2A. Eighty-six biopsies without detectable TP53 inactivation were evaluated for all three mechanisms of CDKN2A inactivation. Among these biopsies, there were no significant differences in Ki67 proliferative, G1, diploid, S phase, or 4N fractions between biopsies with no detectable CDKN2A inactivation and those with at least one form of inactivation (p > 0.1 for all fractions) or those with two or all three forms (p > 0.1). In addition, none of the CDKN2A inactivation mechanisms were associated with statistically significantly higher Ki67-positive, G1 phase, S phase, or 4N fractions when the mechanisms were analyzed univariately in TP53+/+ biopsies. CDKN2A methylation and 9p LOH co-occurred in biopsies less frequently than expected by chance (p = 0.0001, chi-square test).
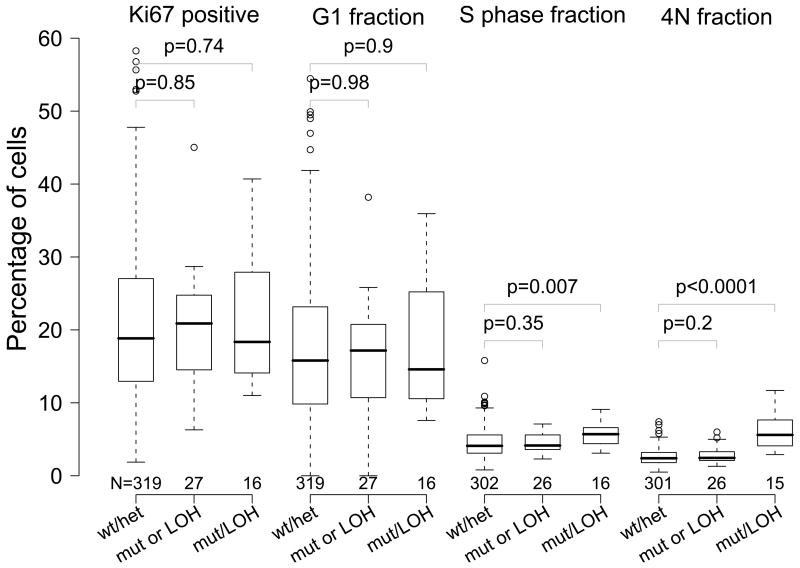
S phase and 4N fractions were significantly higher in diploid biopsies with both TP53 mutation and 17p LOH than those with no detectable TP53 alterations (+/+) (p = 0.007 and p < 0.0001, respectively), while they were not significantly different between TP53 +/+ samples and those with only one form of TP53 abnormality (Figure 2). TP53 mutation and 17p LOH co-occurred in biopsies more frequently than expected by chance (p = 0.0005, chi-square test). Analyses of the effects of CDKN2A inactivation excluded biopsies with detectable TP53 alterations in order to determine the effects of CDKN2A inactivation in isolation. Biopsies without detectable CDKN2A inactivation could not be excluded in the analyses of TP53 inactivation because TP53 inactivation usually occurs in a background of CDKN2A inactivation(41).
Figure 2.
Ki67-positive, G1 phase, S phase, and 4N fractions of biopsies by TP53 inactivation status. Measurements from biopsies no TP53 inactivation (TP53 wt/17p het), with either TP53 mutation or 17p LOH (TP53 mutation or 17p LOH), and both mutation and LOH (TP53 mutation and 17p LOH) are summarized. The boxes indicate median and middle two quartiles, the whiskers the 95th percentiles, and the outliers are plotted as circles. The number of samples in each group is printed below each box. The biopsies with no TP53 inactivation (TP53+/+) were compared with the other two groups (TP53+/− and TP53−/−) using the Wilcoxon rank sum test, and the p values are shown on the plot.
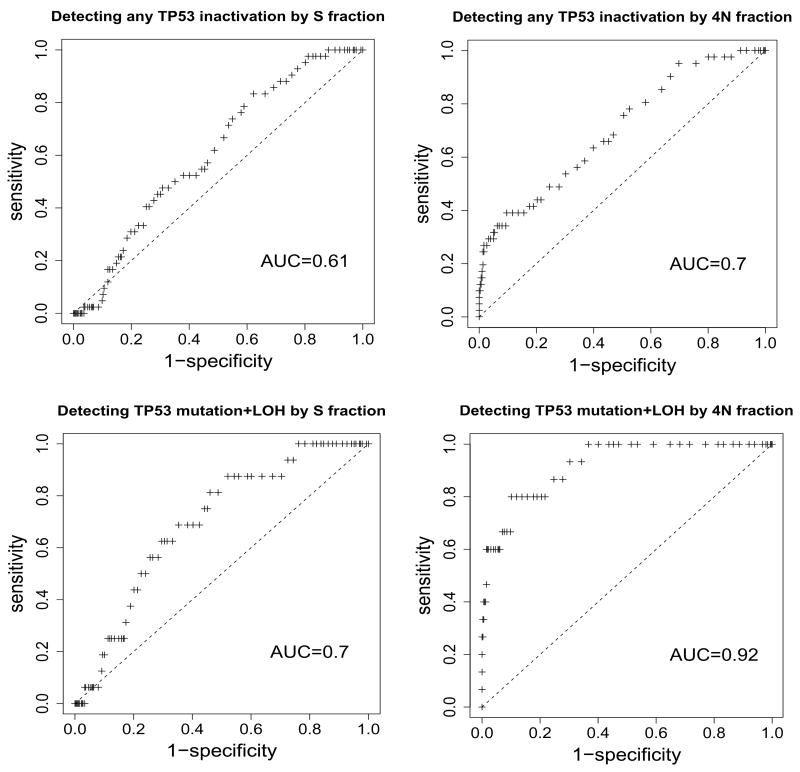
Because TP53 abnormalities were significantly associated with high S phase and 4N fractions, we estimated the sensitivity and specificity of these fractions for detecting biopsies with TP53 mutation and LOH (Figure 3). Diploid S phase fraction was a poor discriminator of biopsies with one or more TP53 inactivation events (AUC=0.61, 95% CI=0.54–0.68), and 4N fraction was better but still not strong (AUC=0.70, 95% CI=0.63–0.77). 4N fraction was a good discriminator of biopsies with both TP53 mutation and LOH (AUC=0.92, 95% CI=0.87–0.97), but diploid S phase fraction was not (AUC=0.70, 95% CI=0.60–0.79).
Figure 3.
Receiver operating characteristic (ROC) curves for detecting TP53 inactivation by mutation or LOH in diploid samples using S or 4N fractions. The upper plots are ROC curves for discriminating between biopsies with no detectable TP53 inactivation and those with any inactivation. 42/344 biopsies in which TP53 inactivation and S phase fractions were ascertained had detectable TP53 mutation or LOH, and 41/342 biopsies in which TP53 inactivation and 4N fractions were ascertained had detectable TP53 mutation or LOH. The associated areas under the curves (AUC) are indicated. The lower plots are ROC curves for discriminating between biopsies with both forms of TP53 inactivation (mutation and LOH) and all other biopsies. 16/344 biopsies had detectable TP53 mutation and LOH in the S phase plot, and 15/342 biopsies had detectable TP53 mutation and LOH for the 4N plot.
Discussion
Increased cellular proliferation has been reported to be associated with BE neoplastic progression in cross-sectional studies that used a variety of methods to evaluate proliferative or S phase fractions, including tritiated thymidine incorporation, monoclonal antibodies such as Ki67, PCNA, cyclin A, and minichromosome maintenance proteins, as well as flow cytometry(17–26). However, to our knowledge, no previous studies have been conducted in which patients were characterized by an unbiased sampling protocol for total proliferative and cell cycle fractions and followed prospectively for the development of cancer as an endpoint. In this prospective cohort study (a phase 4 study, as defined by the Early Detection Research Network of the National Cancer Institute), we report that an elevated 4N fraction in biopsies from BE is a strong and significant predictor of progression from BE to EA (p < 0.0001), and diploid S phase fractions had a smaller but significant (p = 0.03) association with the future development of EA. These results are consistent with an earlier analysis of our cohort, which found that a 4N fraction of 6% is the optimal threshold for defining elevated 4N for cancer risk prediction, indicating that 4N fraction should not be treated as a continuous value(23). However, total Ki67-positive proliferative fractions and G1 fractions in diploid biopsies were not associated with progression to EA (p = 0.13 and 0.15, respectively). There was a low correlation coefficient between the Ki67-positive and S phase fractions in diploid biopsies, indicating that progression to EA is more closely associated with S phase (an increased G1 to S phase transition) than total proliferative fraction (G0 to G1 transition).
In cross-sectional studies, elevated cellular proliferation has been associated with risk factors of neoplastic progression in colonic epithelium, which, like metaplastic Barrett’s epithelium, is formed of crypts. High S phase fractions measured using 3H-thymidine have been observed in normal-appearing mucosa in patients with adenomas(42), and high proliferating cell nuclear antigen (PCNA) stained fractions in colorectal epithelial biopsies have been associated with risk factors for colorectal cancer(43). However, no association was found between increased proliferative fractions based on PCNA expression and future development of adenomas in a large prospective study of colon cancer patients(44). In another study, the unusual distribution of Ki67-stained cells in colonic crypts, but not the total fraction of Ki67-positive epithelial cells, was associated with an increased risk of development of secondary tumors in participants who had early colorectal tumors(45). Our study of Barrett’s mucosal diploid biopsies agrees with these observations that overall cell proliferation has not been found to predict cancer outcome in the colon.
We also evaluated inactivation of the tumor suppressor genes CDKN2A and TP53, which develop in diploid cells prior to aneuploidy and EA, and the cell cycle to determine possible associations between proliferation and somatic genetic and epigenetic changes during neoplastic progression(35, 41, 46). TP53 is involved in the G1/S checkpoint(8), thus, inactivation of this tumor suppressor would be expected to increase the numbers of cells in S and G2/M phases of the cell cycle. Here, we report that elevated diploid S and 4N fractions, but not diploid Ki67-positive or G1 fractions, were associated with TP53 inactivation in BE biopsies. These results are consistent with the known negative regulatory functions of TP53 in the G1/S transition in model systems. TP53 mutation and LOH were found in the same biopsies more frequently than expected by chance, suggesting that bi-allelic inactivation of TP53 is selected, which is consistent with Knudson’s two hit hypothesis for inactivating tumor suppressor genes and previous studies of our cohort(41, 47). We found that high 4N fractions were a sensitive and specific indicator of bi-allelic TP53 inactivation (Figure 3). Elevated 4N fractions in a TP53-deficient background can consist of diploid G2/M cells and tetraploid G0 cells that develop from cells that do not complete chromosome segregation(48). Studies in rodents and humans have observed 4N populations in neoplasia(49). Further, studies have reported that tetraploid cell populations are genetically unstable intermediates that develop aneuploidy during neoplastic progression to cancer(10, 28, 46, 50–54). However, murine models appear to differ from humans in the stringency of mitotic checkpoints, and cycling tetraploids are observed more frequently in rodent models(46, 49–54). 4N populations in our study of BE rarely have detectable tetraploid S phase or G2/M fractions by flow cytometry. In vitro, TP53-deficient BE cell cultures express a number of transcripts associated with G2/M and appear to be heterogeneous by FISH analysis, suggesting that they may have an extremely prolonged G2/M delay or the cells with genetic damage are arresting at a variety of G2/M checkpoints(48). Since TP53 abnormalities can be pleiotropic and could be selected for loss of proliferative control, decreased apoptosis or gain of function, it will be difficult to determine the exact mechanism in human biopsies in vivo, given heterogeneity in both constitutive and neoplastic genetic backgrounds and uncontrolled environment exposures. This may be a case in which a translational research study identifies an important biological problem that can then be addressed in model systems.
CDKN2A negatively regulates the transition from G1 to S phase(7), so we had expected to observe higher S phase and 4N fractions in samples with CDKN2A inactivation. However, since at least one allele of CDKN2A was inactivated in most of our samples, it was difficult to evaluate the effects of CDKN2A inactivation on proliferation and cell cycle in the absence of sufficient wild-type samples with BE to use as a comparison group. CDKN2A methylation and LOH were detected in the same biopsies less frequently than expected by chance, which suggests that they are alternate pathways of p16 inactivation or that the methylated alleles of CDKN2A can be lost. We did not find an association between inactivation of CDKN2A or TP53 and increased Ki67-positive or G1 fractions, a result consistent with their known biological properties in studies in model systems.
Our study has several strengths: a) the prospective cohort study design and long-term follow-up, b) a defined, unbiased protocol for biopsy collection, c) use of primary cancer rather than a surrogate such as incident high-grade dysplasia as an endpoint(30, 55, 56), d) the measurement of total proliferation, cell cycle fractions, CDKN2A and TP53 inactivation in the same cells, and e) the use of flow cytometry instead of immunohistochemistry in order to obtain more precise estimates of Ki67-positive and cell cycle fractions based on an average of 15,000 cells per sample. However, immunohistochemistry can be used to determine the location of the proliferating cells within Barrett’s epithelium, which may expand towards the luminal surface during neoplastic progression(22). There is also the concern that Ki67 labeling might not be specific for epithelial cells in BE biopsies. However, we rarely observed non-epithelial Ki67-positive cells in BE biopsy slides in earlier studies(57, 58). In addition, a previous study reported that an average of 90% of proliferating cells in whole endoscopic biopsies from Barrett’s metaplasia were epithelial in origin(21). Finally, although proton pump inhibitor (PPI) use has been associated with reduced proliferative fractions in BE(59, 60), our study did not examine their effects because most patients in the cohort were managed with PPIs and we did not have a comparison group. PPI use can alleviate the symptoms of gastroesophageal reflux, but its effects on the development of cancer are not known, although dose can be studied in a randomized trial such as AspECT (61).
In a recent study of the same cohort, the best predictor of cancer outcome was a combination of chromosomal instability biomarkers that included early (9p LOH), intermediate (17p LOH), and late events (tetraploidy, aneuploidy) in neoplastic progression(33). Detection of CDKN2A and TP53 loss are promising biomarkers for predicting neoplastic progression in many premalignant conditions because they are among the most common somatic genetic abnormalities in human cancers(14, 15). However, LOH at TP53 and DNA content abnormalities (high 4N, aneuploidy) have independent contributions to cancer risk prediction(33), indicating that, despite the strong association between genetic (TP53 inactivation) and cell cycle abnormalities (high 4N fractions) found in the present study, the characterization of both can improve cancer risk stratification in the esophagus and very likely other organs.
Acknowledgments
Financial support: NIH P01 CA91955 (DLC, CAS, PCG, PLB, DSC, KA, RDO, PSR, BJR), NIH R01 CA61202 (CAS, PCG, PLB, DSC, RDO, PSR, BJR), NIH T32 CA80416 (DLC), NIH K07 CA089147 (TGP), Ryan Hill Foundation (TGP), and interim funding from the Fred Hutchinson Cancer Research Center
We thank Mike Shen for developing software to analyze the flow cytometry data; Xiaohong Li for statistical advice; Christine Karlsen for coordination of patient care; Valerie Cerera for coordination of research biopsies and flow cytometry; Jessica Arnaudo, Heather Kissel, and David Wong for molecular assays; and the study participants in the Seattle Barrett’s Esophagus Program for making this research possible.
Footnotes
Statement of Clinical Relevance
Increased cellular proliferation has long been hypothesized to be associated with progression to cancer. In this prospective cohort study, we found no association between cellular proliferation (measured using Ki67) and the future development of esophageal adenocarcinoma (EA) in cancer-free individuals with Barrett’s esophagus (BE). However, we found evidence that high 4N fractions, which had previously been shown to be strongly predictive of the development of EA, are a result of bi-allelic inactivation of TP53. Cell cycle abnormalities and loss of tumor suppressor genes are common somatic genetic abnormalities in human cancers, and are therefore promising biomarkers for predicting neoplastic progression in many premalignant conditions. In this study, we have shown a connection between these two types of biomarkers in human clinical samples.
References
- 1.Preston-Martin S, Pike MC, Ross RK, Jones PA, Henderson BE. Increased cell division as a cause of human cancer. Cancer Res. 1990;50:7415–21. [PubMed] [Google Scholar]
- 2.Gerdes J. Ki-67 and other proliferation markers useful for immunohistological diagnostic and prognostic evaluations in human malignancies. Semin Cancer Biol. 1990;1:199–206. [PubMed] [Google Scholar]
- 3.Martin B, Paesmans M, Mascaux C, et al. Ki-67 expression and patients survival in lung cancer: systematic review of the literature with meta-analysis. Br J Cancer. 2004;91:2018–25. doi: 10.1038/sj.bjc.6602233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Azambuja E, Cardoso F, de Castro G, Jr, et al. Ki-67 as prognostic marker in early breast cancer: a meta-analysis of published studies involving 12,155 patients. Br J Cancer. 2007;96:1504–13. doi: 10.1038/sj.bjc.6603756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartwell LH, Culotti J, Pringle JR, Reid BJ. Genetic control of the cell division cycle in yeast. Science. 1974;183:46–51. doi: 10.1126/science.183.4120.46. [DOI] [PubMed] [Google Scholar]
- 6.Lee MG, Nurse P. Complementation used to clone a human homologue of the fission yeast cell cycle control gene cdc2. Nature. 1987;327:31–5. doi: 10.1038/327031a0. [DOI] [PubMed] [Google Scholar]
- 7.Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–7. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 8.Kastan MB, Zhan Q, el-Deiry WS, et al. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–97. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- 9.Yin Y, Tainsky MA, Bischoff FZ, Strong LC, Wahl GM. Wild-type p53 restores cell cycle control and inhibits gene amplification in cells with mutant p53 alleles. Cell. 1992;70:937–48. doi: 10.1016/0092-8674(92)90244-7. [DOI] [PubMed] [Google Scholar]
- 10.Livingstone LR, White A, Sprouse J, et al. Altered cell cycle arrest and gene amplification potential accompany loss of wild-type p53. Cell. 1992;70:923–35. doi: 10.1016/0092-8674(92)90243-6. [DOI] [PubMed] [Google Scholar]
- 11.Prevo LJ, Sanchez CA, Galipeau PC, Reid BJ. p53-mutant clones and field effects in Barrett’s esophagus. Cancer Res. 1999;59:4784–7. [PubMed] [Google Scholar]
- 12.Sherr CJ. Principles of Tumor Suppression. Cell. 2004;116:235–46. doi: 10.1016/s0092-8674(03)01075-4. [DOI] [PubMed] [Google Scholar]
- 13.Wong DJ, Paulson TG, Prevo LJ, et al. p16 INK4a lesions are common, early abnormalities that undergo clonal expansion in Barrett’s metaplastic epithelium. Cancer Res. 2001;61:8284–9. [PubMed] [Google Scholar]
- 14.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 15.Sharpless NE. INK4a/ARF: a multifunctional tumor suppressor locus. Mutat Res. 2005;576:22–38. doi: 10.1016/j.mrfmmm.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 16.Thomas T, Abrams KR, De Caestecker JS, Robinson RJ. Meta analysis: Cancer risk in Barrett’s oesophagus. Aliment Pharmacol Ther. 2007;26:1465–77. doi: 10.1111/j.1365-2036.2007.03528.x. [DOI] [PubMed] [Google Scholar]
- 17.Herbst JJ, Berenson MM, McCloskey DW, Wiser WC. Cell proliferation in esophageal columnar epithelium (Barrett’s esophagus) Gastroenterology. 1978;75:683–7. [PubMed] [Google Scholar]
- 18.Pellish LJ, Hermos JA, Eastwood GL. Cell proliferation in three types of Barrett’s epithelium. Gut. 1980;21:26–31. doi: 10.1136/gut.21.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reid BJ, Haggitt RC, Rubin CE, Rabinovitch PS. Barrett’s esophagus. Correlation between flow cytometry and histology in detection of patients at risk for adenocarcinoma. Gastroenterology. 1987;93:1–11. [PubMed] [Google Scholar]
- 20.Gray MR, Hall PA, Nash J, et al. Epithelial proliferation in Barrett’s esophagus by proliferating cell nuclear antigen immunolocalization. Gastroenterology. 1992;103:1769–76. doi: 10.1016/0016-5085(92)91433-5. [DOI] [PubMed] [Google Scholar]
- 21.Reid BJ, Sanchez CA, Blount PL, Levine DS. Barrett’s esophagus: cell cycle abnormalities in advancing stages of neoplastic progression. Gastroenterology. 1993;105:119–29. doi: 10.1016/0016-5085(93)90017-7. [DOI] [PubMed] [Google Scholar]
- 22.Hong MK, Laskin WB, Herman BE, et al. Expansion of the Ki-67 proliferative compartment correlates with degree of dysplasia in Barrett’s esophagus. Cancer. 1995;75:423–9. doi: 10.1002/1097-0142(19950115)75:2<423::aid-cncr2820750202>3.0.co;2-5. [DOI] [PubMed] [Google Scholar]
- 23.Rabinovitch PS, Longton G, Blount PL, Levine DS, Reid BJ. Predictors of progression in Barrett’s esophagus III: baseline flow cytometric variables. Am J Gastroenterol. 2001;96:3071–83. doi: 10.1111/j.1572-0241.2001.05261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sirieix PS, O’Donovan M, Brown J, et al. Surface expression of minichromosome maintenance proteins provides a novel method for detecting patients at risk for developing adenocarcinoma in Barrett’s esophagus. Clin Cancer Res. 2003;9:2560–6. [PubMed] [Google Scholar]
- 25.Lao-Sirieix P, Lovat L, Fitzgerald RC. Cyclin A immunocytology as a risk stratification tool for Barrett’s esophagus surveillance. Clin Cancer Res. 2007;13:659–65. doi: 10.1158/1078-0432.CCR-06-1385. [DOI] [PubMed] [Google Scholar]
- 26.Kerkhof M, Steyerberg EW, Kusters JG, et al. Aneuploidy and high expression of p53 and Ki67 is associated with neoplastic progression in Barrett esophagus. Cancer Biomarkers. 2008;4:1–10. doi: 10.3233/cbm-2008-4101. [DOI] [PubMed] [Google Scholar]
- 27.Lao-Sirieix P, Brais R, Lovat L, Coleman N, Fitzgerald RC. Cell cycle phase abnormalities do not account for disordered proliferation in Barrett’s carcinogenesis. Neoplasia. 2004;6:751–60. doi: 10.1593/neo.04280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galipeau PC, Cowan DS, Sanchez CA, et al. 17p (p53) allelic losses, 4N (G2/tetraploid) populations, and progression to aneuploidy in Barrett’s esophagus. Proc Natl Acad Sci U S A. 1996;93:7081–4. doi: 10.1073/pnas.93.14.7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Carder P, Wyllie AH, Purdie CA, et al. Stabilised p53 facilitates aneuploid clonal divergence in colorectal cancer. Oncogene. 1993;8:1397–401. [PubMed] [Google Scholar]
- 30.Reid BJ, Levine DS, Longton G, Blount PL, Rabinovitch PS. Predictors of progression to cancer in Barrett’s esophagus: baseline histology and flow cytometry identify low- and high-risk patient subsets. Am J Gastroenterol. 2000;95:1669–76. doi: 10.1111/j.1572-0241.2000.02196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reid BJ, Prevo LJ, Galipeau PC, et al. Predictors of progression in Barrett’s esophagus II: baseline 17p (p53) loss of heterozygosity identifies a patient subset at increased risk for neoplastic progression. Am J Gastroenterol. 2001;96:2839–48. doi: 10.1111/j.1572-0241.2001.04236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vaughan TL, Dong LM, Blount PL, et al. Non-steroidal anti-inflammatory drugs and risk of neoplastic progression in Barrett’s oesophagus: a prospective study. Lancet Oncol. 2005;6:945–52. doi: 10.1016/S1470-2045(05)70431-9. [DOI] [PubMed] [Google Scholar]
- 33.Galipeau PC, Li X, Blount PL, et al. NSAIDs modulate CDKN2A, TP53, and DNA content risk for future esophageal adenocarcinoma. PLoS Med. 2007;4:e67. doi: 10.1371/journal.pmed.0040067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerdes J, Lemke H, Baisch H, et al. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol. 1984;133:1710–5. [PubMed] [Google Scholar]
- 35.Blount PL, Galipeau PC, Sanchez CA, et al. 17p allelic losses in diploid cells of patients with Barrett’s esophagus who develop aneuploidy. Cancer Res. 1994;54:2292–5. [PubMed] [Google Scholar]
- 36.Shankey TV, Rabinovitch PS, Bagwell B, et al. Guidelines for implementation of clinical DNA cytometry. International Society for Analytical Cytology [published erratum appears in Cytometry 1993 Oct;14(7):842] Cytometry. 1993;14:472–7. doi: 10.1002/cyto.990140503. [DOI] [PubMed] [Google Scholar]
- 37.Barrett MT, Reid BJ, Joslyn G. Genotypic analysis of multiple loci in somatic cells by whole genome amplification. Nucleic Acids Res. 1995;23:3488–92. doi: 10.1093/nar/23.17.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barrett MT, Galipeau PC, Sanchez CA, Emond MJ, Reid BJ. Determination of the frequency of loss of heterozygosity in esophageal adenocarcinoma by cell sorting, whole genome amplification and microsatellite polymorphisms. Oncogene. 1996;12:1873–8. [PubMed] [Google Scholar]
- 39.Galipeau PC, Prevo LJ, Sanchez CA, Longton GM, Reid BJ. Clonal expansion and loss of heterozygosity at chromosomes 9p and 17p in premalignant esophageal (Barrett’s) tissue. J Natl Cancer Inst. 1999;91:2087–95. doi: 10.1093/jnci/91.24.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hanley JA, McNeil BJ. The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology. 1982;143:29–36. doi: 10.1148/radiology.143.1.7063747. [DOI] [PubMed] [Google Scholar]
- 41.Maley CC, Galipeau PC, Li X, et al. Selectively advantageous mutations and hitchhikers in neoplasms: p16 lesions are selected in Barrett’s esophagus. Cancer Res. 2004;64:3414–27. doi: 10.1158/0008-5472.CAN-03-3249. [DOI] [PubMed] [Google Scholar]
- 42.Bleiberg H, Buyse M, Galand P. Cell kinetic indicators of premalignant stages of colorectal cancer. Cancer. 1985;56:124–9. doi: 10.1002/1097-0142(19850701)56:1<124::aid-cncr2820560119>3.0.co;2-y. [DOI] [PubMed] [Google Scholar]
- 43.Bostick RM, Fosdick L, Grandits GA, et al. Colorectal epithelial cell proliferative kinetics and risk factors for colon cancer in sporadic adenoma patients. Cancer Epidemiol Biomarkers Prev. 1997;6:1011–9. [PubMed] [Google Scholar]
- 44.Sandler RS, Baron JA, Tosteson TD, Mandel JS, Haile RW. Rectal mucosal proliferation and risk of colorectal adenomas: results from a randomized controlled trial. Cancer Epidemiol Biomarkers Prev. 2000;9:653–6. [PubMed] [Google Scholar]
- 45.Akedo I, Ishikawa H, Ioka T, et al. Evaluation of epithelial cell proliferation rate in normal-appearing colonic mucosa as a high-risk marker for colorectal cancer. Cancer Epidemiol Biomarkers Prev. 2001;10:925–30. [PubMed] [Google Scholar]
- 46.Barrett MT, Sanchez CA, Prevo LJ, et al. Evolution of neoplastic cell lineages in Barrett oesophagus. Nat Genet. 1999;22:106–9. doi: 10.1038/8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Knudson AG., Jr Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A. 1971;68:820–3. doi: 10.1073/pnas.68.4.820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barrett MT, Pritchard D, Palanca-Wessels C, et al. Molecular phenotype of spontaneously arising 4N (G2-tetraploid) intermediates of neoplastic progression in Barrett’s esophagus. Cancer Res. 2003;63:4211–7. [PubMed] [Google Scholar]
- 49.Schimke RT, Kung AL, Rush DF, Sherwood SW. Differences in mitotic control among mammalian cells. Cold Spring Harb Symp Quant Biol. 1991;56:417–25. doi: 10.1101/sqb.1991.056.01.049. [DOI] [PubMed] [Google Scholar]
- 50.Shackney SE, Smith CA, Miller BW, et al. Model for the genetic evolution of human solid tumors. Cancer Res. 1989;49:3344–54. [PubMed] [Google Scholar]
- 51.Levine DS, Sanchez CA, Rabinovitch PS, Reid BJ. Formation of the tetraploid intermediate is associated with the development of cells with more than four centrioles in the elastase-simian virus 40 tumor antigen transgenic mouse model of pancreatic cancer [published erratum appears in Proc Natl Acad Sci U S A 1991 Sep 15;88(18):8282] Proc Natl Acad Sci U S A. 1991;88:6427–31. doi: 10.1073/pnas.88.15.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cross SM, Sanchez CA, Morgan CA, et al. A p53-dependent mouse spindle checkpoint. Science. 1995;267:1353–6. doi: 10.1126/science.7871434. [DOI] [PubMed] [Google Scholar]
- 53.Wongsurawat VJ, Finley JC, Galipeau PC, et al. Genetic mechanisms of TP53 loss of heterozygosity in Barrett’s esophagus: implications for biomarker validation. Cancer Epidemiol Biomarkers Prev. 2006;15:509–16. doi: 10.1158/1055-9965.EPI-05-0246. [DOI] [PubMed] [Google Scholar]
- 54.Ganem NJ, Pellman D. Limiting the proliferation of polyploid cells. Cell. 2007;131:437–40. doi: 10.1016/j.cell.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 55.Fleming TR, Prentice RL, Pepe MS, Glidden D. Surrogate and auxiliary endpoints in clinical trials, with potential applications in cancer and AIDS research. Stat Med. 1994;13:955–68. doi: 10.1002/sim.4780130906. [DOI] [PubMed] [Google Scholar]
- 56.Schnell T, Sontag SJ, Chejfec G, et al. High grade dysplasia still is not an indication for surgery in patients with Barrett’s esophagus: An update. Gastroenterology. 1998;114:A280. [Google Scholar]
- 57.Hornick JL, Blount PL, Sanchez CA, et al. Biologic properties of columnar epithelium underneath reepithelialized squamous mucosa in Barrett’s esophagus. Am J Surg Pathol. 2005;29:372–80. doi: 10.1097/01.pas.0000147403.33509.de. [DOI] [PubMed] [Google Scholar]
- 58.Lomo LC, Blount PL, Sanchez CA, et al. Crypt dysplasia with surface maturation: a clinical, pathologic, and molecular study of a Barrett’s esophagus cohort. Am J Surg Pathol. 2006;30:423–35. doi: 10.1097/00000478-200604000-00001. [DOI] [PubMed] [Google Scholar]
- 59.Peters FT, Ganesh S, Kuipers EJ, et al. Epithelial cell proliferative activity of Barrett’s esophagus: methodology and correlation with traditional cancer risk markers. Dig Dis Sci. 1998;43:1501–6. doi: 10.1023/a:1018858713965. [DOI] [PubMed] [Google Scholar]
- 60.Lao-Sirieix P, Roy A, Worrall C, et al. Effect of acid suppression on molecular predictors for esophageal cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:288–93. doi: 10.1158/1055-9965.EPI-05-0528. [DOI] [PubMed] [Google Scholar]
- 61.Jankowski J, Barr H. Improving surveillance for Barrett’s oesophagus: AspECT and BOSS trials provide an evidence base. BMJ. 2006;332:1512. doi: 10.1136/bmj.332.7556.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]