Abstract
Death receptors induce apoptosis through either the Type I or Type II pathway. In Type I cells, the initiator caspase-8 directly activates effector caspases such as caspase-3, whereas in Type II cells, the death signal is amplified through mitochondria thereby activating effector caspases causing cell death. Recently, there have been advances in elucidating the early events in the CD95 signaling pathways and how posttranslational modifications regulate CD95 signaling. This review will focus on recent insights into the mechanisms of the two different types of CD95 signaling pathways, and will introduce miRNAs as regulators of death receptor signaling.
Keywords: apoptosis, Fas, let-7, superclusters, TRAIL
1. Introduction
Apoptosis can be executed either through the extrinsic or intrinsic pathway [1]. The intrinsic pathway, which involves the release of a number of factors from mitochondria including cytochrome c, can be activated by a diverse set of stressors. Upon release, cytochrome c interacts with the adapter protein Apaf-1, dATP and caspase-9 to form the apoptosome complex, which subsequently activates caspase-3, leading to cell death [2]. Other factors that are released from the mitochondria that promote apoptosis include Apoptosis Inducing Factor (AIF) [3], Smac/Diablo [4, 5], endonuclease G [6] and Omi/HtrA [7].
The extrinsic pathway is activated when death receptors are engaged by their cognate ligands. Death receptors are members of the TNF superfamily and include CD95 (Fas/APO-1), TNF-R1 and the TRAIL receptors, DR4 and DR5. All death receptors carry a conserved cytoplasmic domain of about 80 amino acids called the death domain (DD) which is crucial for initiating apoptotic signals. The most well characterized death receptor with respect to the mechanism of apoptosis induction is CD95. Activation of CD95, either by its cognate ligand (CD95L) or by agonistic antibodies, initiates a process of receptor clustering and consequent triggering of apoptotic signals. In some cells CD95 induces apoptosis by activating the effector caspases directly (Type I cells). In other cells (Type II cells) CD95 induces apoptosis by amplifying the death signal through the mitochondrial, "intrinsic," pathway [8, 9]. Upon CD95 stimulation, sequential recruitment of the adapter molecule FADD (Mort1), pro-forms of caspase-8 and caspase-10, and c-FLIP to the CD95 DD result in the formation of the death-inducing signaling complex (DISC) [10]. In the DISC, aggregation of procaspase-8 leads to proteolytic cleavage and activation. In Type I cells, large amounts of active caspase-8 are released and directly activate caspase-3 without the involvement of the mitochondria. In Type II cells, very little active caspase-8 is produced at the DISC. The amount of caspase-8 produced, however, is sufficient to cleave the BH3 containing protein "BH3-interacting domain death agonist" (Bid) to generate truncated Bid (tBid) [11, 12]. tBid translocates to the mitochondria and induces the release of mitochondrial factors that lead to apoptosis. The mitochondrial pathway can be blocked by the expression of antiapoptotic members of the Bcl-2 family [13]. Therefore, the amount of DISC formed at the activated CD95 and the ability of Bcl-2/Bcl-xL to inhibit apoptosis induced through CD95 can be used to distinguish Type I from Type II cells [8, 14].
2. Early events in the Type I and Type II CD95 signaling pathways
Very early signaling events of CD95 when CD95L binds to the preassociated CD95 in Type I cells can be summarized as follows [15, 16]:
Formation of CD95 microaggregates with a low level of DISC formation are detected as SDS-stable CD95 aggregates by SDS-PAGE.
Recruitment of CD95 into lipid rafts to form higher order CD95 oligomers.
Receptor clustering or capping and formation of large lipid raft platforms.
Internalization of CD95 and migration of internalized CD95 into an endosomal compartment.
Recruitment of large amounts of DISC components to endosomal vesicles to form a high-molecular weight DISC (hiDISC).
The initial events in CD95-mediated signaling after CD95 stimulation differ between Type I and II cells. In Type I cells, upon CD95 stimulation FADD is efficiently recruited in an actin filament-dependent manner [15] and allows the recruitment of other components of the DISC such as caspase-8, caspase-10 and c-FLIP. Type II cells, by contrast, exhibit decreased FADD recruitment and decreased DISC formation [10] resulting in very little active caspase-8 generation, albeit sufficient for cleaving Bid. In Type I cells some CD95 has been shown to be present in lipid rafts. However, in Type II cells CD95 is not found in the rafts [17–19] until it is recruited to the raft membrane fractions following CD95 stimulation [17, 18, 20]. Redistribution of CD95 into lipid rafts also occurs after costimulation of lymphocytes through CD28 and CD95 [18] and by TCR restimulation in primary activated CD4+ T cells through the action of the Rho GTPases Rac1 and Rac2 [19, 21], which have been shown to sensitize Type II cells to CD95-induced apoptosis. A summary of the differences between Type I and Type II cells is shown in Table 1.
Table 1.
Differences between Type I and Type II cells
| Phenotype | Type I | Type II |
|---|---|---|
| CD95 expression1 | High | High |
| Potential for CD95 apoptosis sensitivity2 | High | High |
| CD95 apoptosis kinetics2 | Fast | Fast |
| Constitutive localization of CD95 in lipid rafts3 | Yes | No |
| Recruitment of CD95 to lipid rafts upon stimulation3 | No | Yes |
| DISC formation2 | High | Low |
| hiDISC formation on endosomes5 | Yes | No |
| Cytochrome c release from mitochondria2 | Yes | Yes |
| Protection from apoptosis by Bcl-2/Bcl-xL2 | No | Yes |
| Apoptosis is actin-dependent6 | Yes | No |
| Apoptosis sensitivity to soluble CD95 ligand7 | No | Yes |
| Sensitivity to actin disrupting drugs7 | Low | Low |
| Sensitivity to Taxanes7 | Low | High |
| Sensitivity to C2-ceramide8 | No | High |
| Inhibition of apoptosis by activating PKC8 | Low | Yes |
| Expression of let-79 | High |
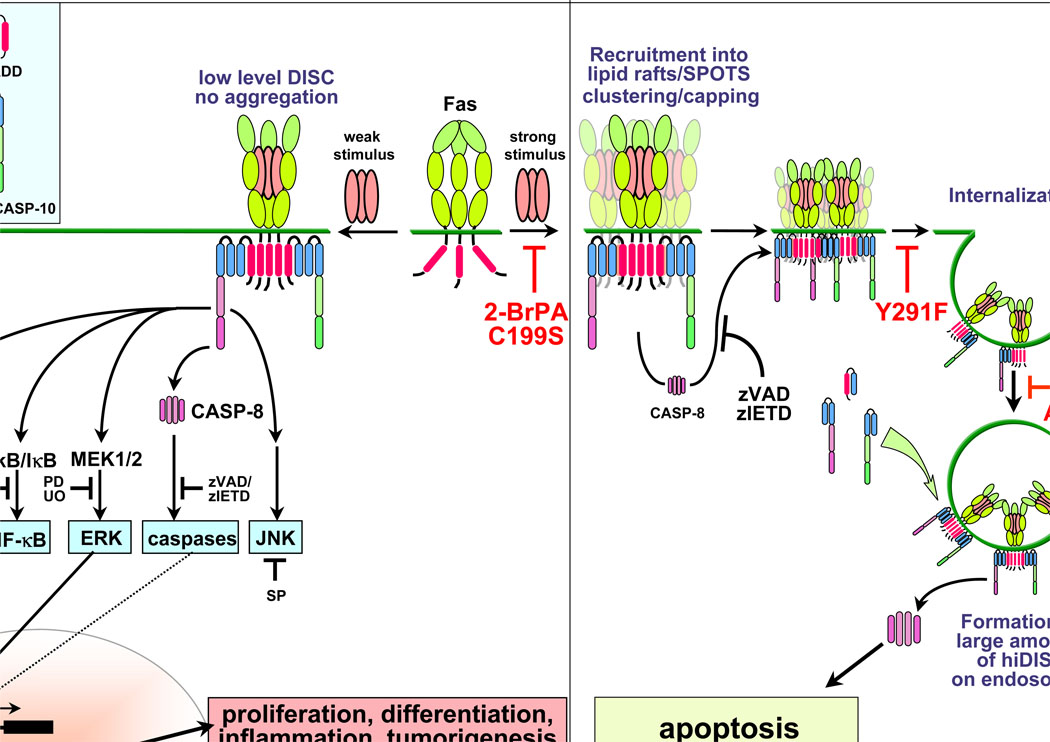
Activated CD95 forms higher order oligomers [22] and these forms can be visualized as SDS and β-mercaptoethanol stable aggregates [23, 24]. Recent studies have shown that formation of SDS-stable aggregates and recruitment to lipid rafts involves palmitoylation of the membrane proximal cysteine 199 of CD95. Treatment with the palmitoylation inhibitor 2-bromopalmitic acid prevented the formation of SDS-stable aggregates [25, 26] suggesting that palmitoylation allows CD95 to be efficiently recruited to the rafts. In Type I cells, aggregated CD95 is then internalized through an endosomal pathway that involves clathrin-coated pit-mediated endocytosis. Tyrosine 291 within the consensus AP-2 binding motif found in the intracellular domain of CD95 was found to be important for CD95 internalization. The internalization step is dependent on actin filaments as it can be inhibited by latrunculin A (Ltn A). In Type II cells, however, there is no indication that CD95 is internalized into endosomes, and consistent with this, treatment with Ltn A does not inhibit CD95-mediated apoptosis in Type II cells [10, 16].
In the past decade, many studies have shown that CD95 also activates nonapoptotic pathways [27] that link CD95 to various nonapoptotic functions including proliferation [28, 29], differentiation [30], inflammation [31, 32] and tumorigenesis [33, 34]. CD95 activates various nonapoptotic pathways including NF-κB and MAP kinases [30, 33]. CD95 utilizes the DISC components to engage non-apoptotic pathways and there is good evidence that CD95 activates the canonical NF-κB pathway through activation of the IκB kinase (IKK) complex [33, 35]. However, the mechanism by which the nonapoptotic and apoptotic pathways are differentially activated at the receptor level is still unknown. Recently, it was shown that if Y291 in the AP-2 binding motif is mutated to prevent CD95 from internalizing, CD95 engagement induces activation of NF-κB and ERK but not apoptosis [16]. Furthermore, when the palmitoylation site C199 of CD95 was mutated, CD95 was unable to form SDS-stable CD95 aggregates, internalization was strongly reduced, and apoptotic pathways were blocked. However, the nonapoptotic pathways were still intact [25]. These studies show that inhibition of receptor internalization enables the activated CD95 to enhance signaling pathways that activate nonapoptotic pathways. Figure 1 summarizes recent advances in our understanding of the proapoptotic and apoptosis-independent activities of CD95.
Figure 1.
Signaling pathways of CD95. When CD95 is strongly activated in Type I cells, CD95 forms clusters and palmitoylation at C199 allows efficient recruitment of CD95 into lipid rafts. In the rafts, further receptor clustering or capping occurs and large lipid raft platforms are formed. CD95 is then internalized through clathrin-mediated endocytosis, which can be inhibited by siRNAs against the AP-2 adaptor complex or the clathrin heavy chain (CHC). In the endosomal compartment, large amounts of DISC are recruited to CD95, thereby inducing apoptosis. When CD95 is weakly activated, or when recruitment of CD95 to lipid rafts to form higher order CD95 oligomers is impaired by inhibition of palmitoylation, or when internalization is blocked, only a low level of DISC with markedly reduced receptor aggregation is formed. This formation allows efficient activation of nonapoptotic pathways such as NF-κB and members of the MAPK family without apoptosis induction. Lnt A, latrunculin A; 2-BrPA, 2-bromopalmitic acid; CHC, clathrin heavy chain; hiDISC, high-molecular weight DISC; SB, SB203580; PD, PD98059; UO, UO126; SP, SP600125.
3. Different sensitivities of Type 1 and Type II cells to antitumor drugs
Previously, we determined apoptosis sensitivity to soluble CD95L and DISC formation among 58 cell lines from the National Cancer Institute anticancer drug screening panel of 60 cancer cell lines representing 9 different human cancers (NCI60), and found that 22 of these cells were CD95 sensitive. Of these 22 cell lines, half were classified as Type I and the other half as Type II [10]. A comprehensive microarray analysis of the gene expression profiles of the NCI60 cell lines had shown that the cells clustered into two distinct branches, one expressing an epithelial and the other a stromal/mesenchymal gene signature [36]. These two branches of the NCI60 cells may reflect two different stages of tumor development. Interestingly, most of the Type I cells fell into the stromal/mesenchymal branch (supercluster 1, SC1) and the Type II cells were found in the epithelial branch (supercluster 2, SC2). Gene array data on the expression of 8,000 distinct human genes in the NCI60 cell lines were obtained from the NCI and a COMPARE analysis was performed. Among the genes that were more abundant in Type I cells were CD95 and many actin-regulating genes, which is consistent with the requirement for actin filaments in apoptosis of Type I cells [10]. By using the public NCI Developmental Therapeutics Program anticancer drug screening database, which holds data on more than 42,000 publicly available compounds, Type I cells were found to have selective cell death sensitivity to actin-binding reagents. In contrast Type II cells were found to be much more sensitive to tubulin-binding compounds. Thus, Type I and Type II cells can be distinguished by their sensitivity to actin- or tubulin-binding drugs (Table 1), which suggests that the differences between Type I/SC1 and Type II/SC2 cells are not limited to CD95 signaling.
4. Let-7, a miRNA that separates Type I and Type II cells
Micro (mi)RNAs are a class of noncoding RNAs of 18–24 nucleotides (nt) that posttranscriptionally regulate protein expression. The first miRNA to be identified, lin-4, was discovered in 1993 by Victor Ambros and colleagues in a genetic screen for genes that control developmental timing in Caenorhabditis elegans [37]. Subsequently, it was shown that lin-4 encodes a 22 nt small RNA that negatively regulates lin-14, a gene that is necessary for differentiation of specific cell lineages [37]. In recent years, the known roles of miRNAs have expanded from their initially identified functions in development to various biological activities including proliferation, cell death, and differentiation (reviewed in [38–40]). The dysregulation of various miRNAs is associated with many diseases including cancer. Currently, more than 500 human miRNAs are known to exist and computational analysis predicts up to 30% of human protein coding genes to be regulated by miRNAs [41], making miRNAs one of the largest classes of gene regulators.
MiRNAs are initially transcribed by RNA polymerase II (pol II) as long primary transcripts known as primary-miRNAs (pri-miRNAs). Pri-RNA transcripts are generally several kilobases in length, and are capped and polyadenylated like other pol II transcripts [42]. A significant number of miRNAs are located within introns of protein-coding RNAs [43] as well as in the exons and introns of non-coding RNAs [44]. The pri-miRNA contains one or more stem loop structures of 60–80 bases within which the miRNA resides. The stem loop is a double stranded RNA structure with imperfect base-pairing. This structure is recognized and cleaved in the nucleus by an RNase III enzyme, Drosha and its cofactor DGCR8 [45]. Pri-miRNAs are cleaved to form precursor-miRNAs (pre-miRNA) which contain a two nt 3’ overhang that is recognized by the nuclear export factor, exportin-5, allowing rapid export to the cytoplasm [46]. Further processing of the pre-miRNA by Dicer, another RNase III enzyme, generates the mature miRNA that is incorporated into the RNA-induced silencing complex (RISC). MiRNAs direct the RISC to target specific mRNAs, which are either subsequently cleaved or translationally silenced. The mode of negative regulation of the mRNA is determined by the extend of complementarity between the miRNA and target mRNA. When the miRNA and target site in the 3’UTR of the target mRNA exhibit imperfect base-pairing, which is common with mammalian miRNAs, regulation is by translational repression [47]. However, in case of perfect complementarity between miRNA and mRNA, the target mRNA is degraded by RISC [48].
Because of the role miRNAs play in differentiation processes we recently tested whether miRNAs could be differentially expressed between Type I and Type II cells. We subjected 10 Type I and 10 Type II cell lines to a miRNA gene array analysis and found that members of the let-7 family were preferentially expressed in Type II cells [49]. Further analysis showed that let-7 was generally expressed more highly in SC2 cells when compared to SC1 cells, irrespective of CD95 sensitivity. Since let-7 plays an important role during embryonic development, this suggests that let-7 expression could influence the differentiation state of tumor cells and their sensitivity to CD95L and chemotherapeutic drugs.
Let-7 is one of the first identified miRNA families [50, 51] originally discovered in C. elegans [52]. Let-7 is highly conserved in animals from worm to humans and is important for embryonic development [53]. Let-7 expression is low in early embryonic stages, upregulated late during embryonic development and highly expressed in differentiated tissues [54, 55]. Downregulation of let-7 has been shown in various cancers including head and neck, lung, colon and ovarian cancer. Its expression levels serve as a prognostic marker for various cancers [49, 56–58]. Let-7 is considered to be a tumor suppressor in lung cancer through regulation of the oncogene, Ras [57]. Recently, four groups have shown that high mobility group A2 (HMGA2) is another direct target of let-7 [49, 59–61]. HMGA2 is an architectural transcription factor that binds to DNA and is involved in chromatin remodeling. HMGA2 is an early embryonic gene that is undetectable in most differentiated tissues but is highly expressed in various cancers including neuroblastoma, thyroid, pancreatic and lung cancer [62–65]. Because 1) let-7 is expressed late in embryonic development and its direct target, HMGA2, is an embryonic gene, and 2) let-7 is preferentially expressed in SC2 of the NCI60 cells which represent more mesenchymal/stromal cells, and 3) let-7 is downregulated in cancer in which dedifferentiation is an essential process, it is likely that the physiological targets of let-7 are embryonic genes. We recently hypothesized that during carcinogenesis embryonic genes in adult tissues that are normally repressed by let-7 are relieved from let-7 control and can be re-expressed, promoting dedifferentiation of tissues leading to cancer progression [66]. Consistent with that hypothesis are data that demonstrated that the most undifferentiated stage in breast cancer, the cancer stem cell, is devoid of let-7 expression [67].
5. miRNAs regulating apoptosis sensitivity
The discovery of miRNAs that mediate post-transcriptional silencing of specific target genes has shed light on how noncoding RNAs can play important roles in essential processes such as proliferation, differentiation, and also in apoptosis. Readers are referred to several recent reviews for detailed descriptions of the function and biogenesis of miRNAs [39, 48, 51, 68, 69]. In the following we will review selected miRNAs that regulate pro- and anti-apoptotic genes involved in the apoptotic pathways.
5.1 miRNA with anti-apoptotic functions
First observations on regulation of cell death by miRNAs were made in Drosophila. Bantam, which was originally identified as a gene that causes overgrowth of wing and eye tissue, was demonstrated to be a miRNA [70]. Bantam was shown to promote proliferation and inhibit apoptosis by targeting the proapoptotic gene, hid [71]. Another Drosophila miRNA, miR-14, which was identified in a screen for genes that alter cell death in the eye, was demonstrated to inhibit apoptosis, possibly by regulating an effector caspase, Drice, which carries a putative miR-14 target site in its 3'UTR [72]. In addition inactivation of members of the miR-2 family, miR-2/6/11/13/308, in Drosophila embryos by injection of antisense oligoribonucleotides complementary to the miRNAs caused widespread cell death. These miRNAs were demonstrated to post-transcriptionally repress the proapoptotic proteins Reaper, Grim, Hid and Sickle [73].
In mammalian cells, several miRNAs were recently reported to be involved in cell death. MiR-21 is a miRNA that was described as having anti-apoptotic function. MiR-21 was initially found during a study screening for miRNAs involved in cell growth and apoptosis in which it was found that inhibition of miR-21 led to a profound increase in cell growth [74]. Up-regulation of miR-21 is frequently associated with solid cancers including breast and pancreatic cancer [60, 75, 76]. Involvement of miR-21 in apoptosis was shown in glioblastoma in which inhibition of miR-21 caused activation of effector caspases and increased cell death [77]. Furthermore, suppression of miR-21 together with TRAIL treatment was shown to synergistically kill gliomas in vitro and in vivo [78]. Studies have shown that PTEN, tropomyosin and most recently, Programmed Cell Death 4 (PDCD4) are direct targets of miR-21 [79–81].
Additional miRNAs with anti-apoptotic function include miR-17-5p and miR-20a both of which are part of the miR-17–92 cluster that is often overexpressed in cancer including B cell lymphoma [82–85]. The miR-17–92 cluster, which consists of 7 miRNAs, is transcribed as a polycistronic unit driven by c-myc expression [84, 86]. In lung cancer cells that overexpress miR-17–92, inhibition of these two miRNAs with antisense oligonucleotides was shown to induce apoptosis [83], which may be partly due to the relief of suppression of its target, E2F1 [87]. Although E2F2 and E2F3 are also negatively regulated by miR-17-5p and miR-20a, E2F1 is more strongly affected [88]. The antiapoptotic activity of miR-17–92 miRNAs has been proposed to reflect the distinct physiological roles of different E2Fs in apoptosis [89]. Unlike E2F2 or E2F3, E2F1 has been particularly associated with apoptosis in addition to its function as an activator of cell cycle [90]. E2F1−/− mice exhibit defects in apoptosis [91], and forced expression of E2F1 in vitro causes apoptosis in various cell types [92]. E2F1 is activated by the ATM/ATR DNA damage signaling pathway [93] and regulates the expression of various caspases [94] and other pro-apoptotic proteins [95] through direct and indirect transcriptional mechanisms. Although E2F2 and E2F3 are also downregulated by miR-17–92 based on reporter assays, the effect on E2F1 is much stronger [88]. Therefore, the miR-17–92 cluster has been proposed to inhibit apoptosis mostly by decreasing E2F1 levels [88, 96].
5.2 miRNAs with proapoptotic functions
Due to the way that miRNAs work, a particular miRNA is likely to have multiple targets. This allows miRNAs to regulate distinct, but functionally connected activities such as apoptotic and antiproliferative functions, or survival and proliferation. MiRNAs with proapoptotic function are often linked to antiproliferative activity. MiR-34 represents a clear example of a miRNA with such a dual function. A key player in the intrinsic apoptotic pathway is p53, a transcription factor activated by diverse forms of cellular stress [97]. Earlier studies indicated that the primary action of p53 in apoptosis is to regulate the activity of the Bcl-2 family proteins through transcription-dependent and independent functions [98]. Recently, five studies correlating miRNA expression profiles with p53 status in various systems independently identified the miR-34 family (miR-34a,b,c) as transcriptional targets of p53 [76, 99–102]. Studies were focused on determining if miR-34 was sufficient or necessary for the two main p53 functions, growth arrest and apoptosis. Ectopic expression of miR-34 in primary and tumor-derived cell lines led to significant cell cycle arrest, recapitulating the p53 growth arresting effect [100]. This likely occurs through the regulation by miR-34 of p53 cell cycle targets which include cyclin-E2 (CCNE), cyclin dependent kinase 4 (CDK4), hepatocyte growth factor receptor (c-Met), cyclin dependent kinase 6 (CDK6), and E2F3 [99, 100]. Furthermore, overexpression of miR-34a increased apoptosis in several tumor cell lines [76, 101, 102]. Consistent with this, inactivation of miR-34 by high affinity locked nucleic acid (LNA) antisense miRNA oligonucleotides was shown to inhibit p53-mediated apoptosis in cells exposed to genotoxic stress [99, 101]. Bommer et al identified Bcl-2 as a miR-34 target involved in apoptosis that is regulated by targeting of its 3‘UTR [99].
MiR-15a and miR-16-1 represent another class of miRNAs that regulate apoptosis. These miRNAs are located on chromosome 13q14, which is a region frequently deleted in B-cell chronic lymphocytic leukemia [103]. Cimmeno et al showed that the expression of these two miRNAs inversely correlated with Bcl-2 protein levels in samples from chronic lymphocytic leukemia patients, and identified a conserved target site for miR-15a and miR-16-1 in the 3’UTR of Bcl-2 [104]. Furthermore, overexpression of these miRNAs reduced Bcl-2 protein levels, activated the intrinsic apoptosis pathway, and caused cell death.
Another Bcl-2 family member that is regulated by miRNAs is Mcl-1. Mcl-1 exerts its protective role by binding to the proapoptotic proteins Bim, Bid, Bik, Noxa and Puma [105], as well as Bak [106]. Mcl-1 overexpression results in resistance to TRAIL-induced apoptosis, whereas Mcl-1 suppression leads to sensitization of cells to apoptosis [107]. Mcl-1 is upregulated in some cancers [108] including cholangiocarcinoma [109], but is downregulated in aggressive chronic lymphocytic leukemia, colon and breast cancer [110–112]. MiR-29 expression reduces Mcl-1 protein levels and thereby sensitizes cells to TRAIL-induced apoptosis. Conversely, inhibition of miR-29 by LNA-modified antisense oligonucleotides increased Mcl-1 levels and reduced the apoptosis inducing activity of TRAIL, demonstrating that miR-29 is an endogenous regulator of Mcl-1 [113].
6. miRNAs that modulate death receptor-induced apoptosis
Recently, a screen for modulators of the TRAIL apoptotic pathway was performed on a miRNA library containing about 200 synthetic miRNAs [114]. A breast cancer cell line, MDA-MB-453, was transfected with these miRNAs, and forty-eight hours post-transfection, cells were treated with TRAIL. Relevant miRNAs were identified based on their ability to alter susceptibility to TRAIL-induced apoptosis, which was assessed by monitoring caspase-3 activity. Thirty-four of these miRNAs caused a change in caspase activity. Using the miRBase Target database (http://microrna.sanger.ac.uk/cgibin/targets/v3/download.pl) a subset of these 34 miRNAs was selected and several interesting candidates were found. Among them, miR-144, miR-182 and miR-155 were predicted to target caspase-3. MiR-145 and miR-216 had DR4 and DR5 as predicted targets. MiR-182 and miR-96 were predicted to target FADD, the adaptor molecule that binds to the death receptors. MiR-7 was predicted to target BAD, and let-7c was predicted to target CD95L. However, most of these target identifications are based on predictions and await functional validation.
7. Conclusions
It is now well established that death receptor-sensitive cells respond to death ligands either through the Type I or Type II pathway. Recent studies advanced the understanding of the early events in these two pathways. Studies demonstrated the mechanism by which recruitment of activated CD95 receptor from the plasma membrane to lipid rafts can lead to efficient clustering of receptor aggregates and internalization leading to the generation of strong apoptotic signals. These events, however, are not needed for triggering nonapoptotic pathways. Therefore, the dynamic of CD95 membrane localization and internalization is critical for balancing apoptotic and nonapoptotic pathways in driving cells either to undergo cell death or to perform other functions. MiRNAs are a new class of regulators which function in various essential processes including apoptosis. Future studies will be directed at elucidating how miRNAs affect death receptor signaling, and determining the mechanism of how let-7 causes changes in the differentiation of cancer.
Acknowledgements
We would like to thank E. Milner for proofreading the manuscript. S.M.P. was supported by a University of Chicago Women's Board fellowship. This work was supported by NIH grant R01 GM61712.
Abbreviations used
- AIF
Apoptosis Inducing Factor
- DISC
death-inducing signaling complex
- HMGA2
high mobility group A2
- Ltn A
latrunculin A
- RISC
RNA-induced silencing complex
- tBid
truncated Bid
Biographies
Marcus E. Peter is a Professor in the Ben May Department for Cancer Research at the University of Chicago, Illinois. He received his PhD degree from the University of Baureuth, Germany in 1988 and completed a postdoctoral training at the Dana Farber Cancer Institute in Boston before joining the German Cancer Research Center in Heidelberg, Germany in 1992 where he published a number of seminal papers on the Fas signaling pathway including the description of the death-inducing signaling complex, the cloning of caspase-8 and the description of the two signaling pathways of Fas. He received the Walther and Christine Richtzenhain Prize for Experimental Cancer Research 1999. In that year he joined the faculty of the University of Chicago. His research interests include signaling pathways of death receptors. Recently he has begun to study the role of microRNAs in cancer progression. Dr. Peter has been a member of the Editorial boards of Cell Death and Differentiation and Apoptosis.

Sun Mi Park received her PhD degree from Yonsei University, Seoul (South Korea) in 2004 where she studied signaling pathways of members of the TNFR family. For her postdoctoral training, she joined the lab of Dr. Marcus Peter at the University of Chicago. In Dr. Peterís lab she has been working on the non-apoptotic roles of CD95 in several cancer models. She has published papers on the role of the microRNA let-7 in the two differentiation stages of cancer referred to as Type I and Type II cells, the protective role of CD95 against liver cancer and the role of miR-200 in maintaining the epithelial phenotype of cancer cells. Her current research interests are to understand the regulation of let-7 in Type I and Type II cells, and the role of CD95 in tumor promotion. In 2006 she was awarded a fellowship from the University of Chicago Women's Board.

Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Sun-Mi Park, Email: smpark@uchicago.edu.
Marcus E. Peter, Email: mpeter@uchicago.edu.
References
- 1.Hengartner MO. The biochemistry of apoptosis. Nature. 2000;407:770–776. doi: 10.1038/35037710. [DOI] [PubMed] [Google Scholar]
- 2.Li P, Nijhawan D, Budihardjo I, Srinivasula SM, Ahmad M, Alnemri ES, et al. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell. 1997;91:479–489. doi: 10.1016/s0092-8674(00)80434-1. [DOI] [PubMed] [Google Scholar]
- 3.Susin SA, Lorenzo HK, Zamzami N, Marzo I, Snow BE, Brothers GM, et al. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature. 1999;397:441–446. doi: 10.1038/17135. [DOI] [PubMed] [Google Scholar]
- 4.Du C, Fang M, Li Y, Li L, Wang X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell. 2000;102:33–42. doi: 10.1016/s0092-8674(00)00008-8. [DOI] [PubMed] [Google Scholar]
- 5.Verhagen AM, Ekert PG, Pakusch M, Silke J, Connolly LM, Reid GE, et al. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell. 2000;102:43–53. doi: 10.1016/s0092-8674(00)00009-x. [DOI] [PubMed] [Google Scholar]
- 6.Li LY, Luo X, Wang X. Endonuclease G is an apoptotic DNase when released from mitochondria. Nature. 2001;412:95–99. doi: 10.1038/35083620. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki Y, Imai Y, Nakayama H, Takahashi K, Takio K, Takahashi R. A serine protease, HtrA2, is released from the mitochondria and interacts with XIAP, inducing cell death. Mol Cell. 2001;8:613–621. doi: 10.1016/s1097-2765(01)00341-0. [DOI] [PubMed] [Google Scholar]
- 8.Scaffidi C, Fulda S, Srinivasan A, Friesen C, Li F, Tomaselli KJ, et al. Two CD95 (APO-1/Fas) signaling pathways. Embo J. 1998;17:1675–1687. doi: 10.1093/emboj/17.6.1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barnhart BC, Alappat EC, Peter ME. The CD95 type I/type II model. Semin Immunol. 2003;15:185–193. doi: 10.1016/s1044-5323(03)00031-9. [DOI] [PubMed] [Google Scholar]
- 10.Algeciras-Schimnich A, Pietras EM, Barnhart BC, Legembre P, Vijayan S, Holbeck SL, et al. Two CD95 tumor classes with different sensitivities to antitumor drugs. Proc Natl Acad Sci U S A. 2003;100:11445–11450. doi: 10.1073/pnas.2034995100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Zhu H, Xu CJ, Yuan J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell. 1998;94:491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- 12.Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell. 1998;94:481–490. doi: 10.1016/s0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- 13.Green DR, Reed JC. Mitochondria and apoptosis. Science. 1998;281:1309–1312. doi: 10.1126/science.281.5381.1309. [DOI] [PubMed] [Google Scholar]
- 14.Scaffidi C, Schmitz I, Zha J, Korsmeyer SJ, Krammer PH, Peter ME. Differential modulation of apoptosis sensitivity in CD95 type I and type II cells. J Biol Chem. 1999;274:22532–22538. doi: 10.1074/jbc.274.32.22532. [DOI] [PubMed] [Google Scholar]
- 15.Algeciras-Schimnich A, Shen L, Barnhart BC, Murmann AE, Burkhardt JK, Peter ME. Molecular ordering of the initial signaling events of CD95. Mol Cell Biol. 2002;22:207–220. doi: 10.1128/MCB.22.1.207-220.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee KH, Feig C, Tchikov V, Schickel R, Hallas C, Schutze S, et al. The role of receptor internalization in CD95 signaling. EMBO J. 2006;25:1009–1023. doi: 10.1038/sj.emboj.7601016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hueber AO, Bernard AM, Herincs Z, Couzinet A, He HT. An essential role for membrane rafts in the initiation of Fas/CD95-triggered cell death in mouse thymocytes. EMBO Rep. 2002;3:190–196. doi: 10.1093/embo-reports/kvf022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Legembre P, Daburon S, Moreau P, Moreau JF, Taupin JL. Modulation of Fas-mediated apoptosis by lipid rafts in T lymphocytes. J Immunol. 2006;176:716–720. doi: 10.4049/jimmunol.176.2.716. [DOI] [PubMed] [Google Scholar]
- 19.Muppidi JR, Siegel RM. Ligand-independent redistribution of Fas (CD95) into lipid rafts mediates clonotypic T cell death. Nat Immunol. 2004;5:182–189. doi: 10.1038/ni1024. [DOI] [PubMed] [Google Scholar]
- 20.Eramo A, Sargiacomo M, Ricci-Vitiani L, Todaro M, Stassi G, Messina CG, et al. CD95 death-inducing signaling complex formation and internalization occur in lipid rafts of type I and type II cells. Eur J Immunol. 2004;34:1930–1940. doi: 10.1002/eji.200324786. [DOI] [PubMed] [Google Scholar]
- 21.Ramaswamy M, Dumont C, Cruz AC, Muppidi JR, Gomez TS, Billadeau DD, et al. Cutting edge: Rac GTPases sensitize activated T cells to die via Fas. J Immunol. 2007;179:6384–6388. doi: 10.4049/jimmunol.179.10.6384. [DOI] [PubMed] [Google Scholar]
- 22.Henkler F, Behrle E, Dennehy KM, Wicovsky A, Peters N, Warnke C, et al. The extracellular domains of FasL and Fas are sufficient for the formation of supramolecular FasL-Fas clusters of high stability. J Cell Biol. 2005;168:1087–1098. doi: 10.1083/jcb.200501048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kischkel FC, Hellbardt S, Behrmann I, Germer M, Pawlita M, Krammer PH, et al. Cytotoxicity-dependent APO-1 (Fas/CD95)-associated proteins form a death-inducing signaling complex (DISC) with the receptor. Embo J. 1995;14:5579–5588. doi: 10.1002/j.1460-2075.1995.tb00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kamitani T, Nguyen HP, Yeh ET. Activation-induced aggregation and processing of the human Fas antigen. Detection with cytoplasmic domain-specific antibodies. J Biol Chem. 1997;272:22307–22314. doi: 10.1074/jbc.272.35.22307. [DOI] [PubMed] [Google Scholar]
- 25.Feig C, Tchikov V, Schutze S, Peter ME. Palmitoylation of CD95 facilitates formation of SDS-stable receptor aggregates that initiate apoptosis signaling. Embo J. 2007;26:221–231. doi: 10.1038/sj.emboj.7601460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chakrabandhu K, Herincs Z, Huault S, Dost B, Peng L, Conchonaud F, et al. Palmitoylation is required for efficient Fas cell death signaling. Embo J. 2007;26:209–220. doi: 10.1038/sj.emboj.7601456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peter ME, Budd RC, Desbarats J, Hedrick SM, Hueber A-O, Newell MK, et al. The CD95 receptor: Apoptosis revisited. Cell. 2007;129:447–450. doi: 10.1016/j.cell.2007.04.031. [DOI] [PubMed] [Google Scholar]
- 28.Alderson MR, Armitage RJ, Maraskovsky E, Tough TW, Roux E, Schooley K, et al. Fas transduces activation signals in normal human T lymphocytes. J Exp Med. 1993;178:2231–2235. doi: 10.1084/jem.178.6.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freiberg RA, Spencer DM, Choate KA, Duh HJ, Schreiber SL, Crabtree GR, et al. Fas signal transduction triggers either proliferation or apoptosis in human fibroblasts. J Invest Dermatol. 1997;108:215–219. doi: 10.1111/1523-1747.ep12334273. [DOI] [PubMed] [Google Scholar]
- 30.Desbarats J, Birge RB, Mimouni-Rongy M, Weinstein DE, Palerme JS, Newell MK. Fas engagement induces neurite growth through ERK activation and p35 upregulation. Nat Cell Biol. 2003;5:118–125. doi: 10.1038/ncb916. [DOI] [PubMed] [Google Scholar]
- 31.Park DR, Thomsen AR, Frevert CW, Pham U, Skerrett SJ, Kiener PA, et al. Fas (CD95) induces proinflammatory cytokine responses by human monocytes and monocyte-derived macrophages. J Immunol. 2003;170:6209–6216. doi: 10.4049/jimmunol.170.12.6209. [DOI] [PubMed] [Google Scholar]
- 32.Ahn JH, Park SM, Cho HS, Lee MS, Yoon JB, Vilcek J, et al. Non-apoptotic signaling pathways activated by soluble Fas ligand in serum-starved human fibroblasts. Mitogen-activated protein kinases and NF-kappaB-dependent gene expression. J Biol Chem. 2001;276:47100–47106. doi: 10.1074/jbc.M107385200. [DOI] [PubMed] [Google Scholar]
- 33.Barnhart BC, Legembre P, Pietras E, Bubici C, Franzoso G, Peter ME. CD95 ligand induces motility and invasiveness of apoptosis-resistant tumor cells. Embo J. 2004;23:3175–3185. doi: 10.1038/sj.emboj.7600325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee JK, Sayers TJ, Back TC, Wigginton JM, Wiltrout RH. Lack of FasL-mediated killing leads to in vivo tumor promotion in mouse Lewis lung cancer. Apoptosis. 2003;8:151–160. doi: 10.1023/a:1022918625509. [DOI] [PubMed] [Google Scholar]
- 35.Imamura R, Konaka K, Matsumoto N, Hasegawa M, Fukui M, Mukaida N, et al. Fas ligand induces cell-autonomous NF-kappaB activation and interleukin-8 production by a mechanism distinct from that of tumor necrosis factor-alpha. J Biol Chem. 2004;279:46415–46423. doi: 10.1074/jbc.M403226200. [DOI] [PubMed] [Google Scholar]
- 36.Ross DT, Scherf U, Eisen MB, Perou CM, Rees C, Spellman P, et al. Systematic variation in gene expression patterns in human cancer cell lines. Nat Genet. 2000;24:227–235. doi: 10.1038/73432. [DOI] [PubMed] [Google Scholar]
- 37.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 38.Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 39.Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Lett. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- 40.Carthew RW. Gene regulation by microRNAs. Curr Opin Genet Dev. 2006;16:203–208. doi: 10.1016/j.gde.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 41.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 42.Cullen BR. Transcription and processing of human microRNA precursors. Mol Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 43.Rodriguez A, Griffiths-Jones S, Ashurst JL, Bradley A. Identification of mammalian microRNA host genes and transcription units. Genome Res. 2004;14:1902–1910. doi: 10.1101/gr.2722704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weber MJ. New human and mouse microRNA genes found by homology search. Febs J. 2005;272:59–73. doi: 10.1111/j.1432-1033.2004.04389.x. [DOI] [PubMed] [Google Scholar]
- 45.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 46.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17:3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 48.Engels BM, Hutvagner G. Principles and effects of microRNA-mediated post-transcriptional gene regulation. Oncogene. 2006;25:6163–6169. doi: 10.1038/sj.onc.1209909. [DOI] [PubMed] [Google Scholar]
- 49.Shell S, Park SM, Radjabi AR, Schickel R, Kistner EO, Jewell DA, et al. Let-7 expression defines two differentiation stages of cancer. Proc Natl Acad Sci U S A. 2007;104:11400–11405. doi: 10.1073/pnas.0704372104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 51.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 52.Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 53.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 54.Miska EA, Alvarez-Saavedra E, Townsend M, Yoshii A, Sestan N, Rakic P, et al. Microarray analysis of microRNA expression in the developing mammalian brain. Genome Biol. 2004;5:R68. doi: 10.1186/gb-2004-5-9-r68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomson JM, Parker J, Perou CM, Hammond SM. A custom microarray platform for analysis of microRNA gene expression. Nat Methods. 2004;1:47–53. doi: 10.1038/nmeth704. [DOI] [PubMed] [Google Scholar]
- 56.Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29:903–906. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- 57.Johnson SM, Grosshans H, Shingara J, Byrom M, Jarvis R, Cheng A, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 58.Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 59.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee YS, Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hebert C, Norris K, Scheper MA, Nikitakis N, Sauk JJ. High mobility group A2 is a target for miRNA-98 in head and neck squamous cell carcinoma. Mol Cancer. 2007;6:5. doi: 10.1186/1476-4598-6-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Giannini G, Kim CJ, Di Marcotullio L, Manfioletti G, Cardinali B, Cerignoli F, et al. Expression of the HMGI(Y) gene products in human neuroblastic tumours correlates with differentiation status. Br J Cancer. 2000;83:1503–1509. doi: 10.1054/bjoc.2000.1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Chiappetta G, Bandiera A, Berlingieri MT, Visconti R, Manfioletti G, Battista S, et al. The expression of the high mobility group HMGI (Y) proteins correlates with the malignant phenotype of human thyroid neoplasias. Oncogene. 1995;10:1307–1314. [PubMed] [Google Scholar]
- 64.Abe N, Watanabe T, Suzuki Y, Matsumoto N, Masaki T, Mori T, et al. An increased high-mobility group A2 expression level is associated with malignant phenotype in pancreatic exocrine tissue. Br J Cancer. 2003;89:2104–2109. doi: 10.1038/sj.bjc.6601391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sarhadi VK, Wikman H, Salmenkivi K, Kuosma E, Sioris T, Salo J, et al. Increased expression of high mobility group A proteins in lung cancer. J Pathol. 2006;209:206–212. doi: 10.1002/path.1960. [DOI] [PubMed] [Google Scholar]
- 66.Park SM, Shell S, Radjabi AR, Schickel R, Feig C, Boyerinas B, et al. Let-7 Prevents Early Cancer Progression by Suppressing Expression of the Embryonic Gene HMGA2. Cell Cycle. 2007;6:2585–2590. doi: 10.4161/cc.6.21.4845. [DOI] [PubMed] [Google Scholar]
- 67.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, et al. let-7 Regulates Self Renewal and Tumorigenicity of Breast Cancer Cells. Cell. 2007;131:1109–1123. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 68.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 69.Schickel R, Boyerinas B, Park SM, Peter ME. Oncogene. doi: 10.1038/onc.2008.274. in press. [DOI] [PubMed] [Google Scholar]
- 70.Hipfner DR, Weigmann K, Cohen SM. The bantam gene regulates Drosophila growth. Genetics. 2002;161:1527–1537. doi: 10.1093/genetics/161.4.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 72.Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13:790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- 73.Leaman D, Chen PY, Fak J, Yalcin A, Pearce M, Unnerstall U, et al. Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell. 2005;121:1097–1108. doi: 10.1016/j.cell.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 74.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci U S A. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chang TC, Wentzel EA, Kent OA, Ramachandran K, Mullendore M, Lee KH, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chan JA, Krichevsky AM, Kosik KS. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65:6029–6033. doi: 10.1158/0008-5472.CAN-05-0137. [DOI] [PubMed] [Google Scholar]
- 78.Corsten MF, Miranda R, Kasmieh R, Krichevsky AM, Weissleder R, Shah K. MicroRNA-21 knockdown disrupts glioma growth in vivo and displays synergistic cytotoxicity with neural precursor cell delivered S-TRAIL in human gliomas. Cancer Res. 2007;67:8994–9000. doi: 10.1158/0008-5472.CAN-07-1045. [DOI] [PubMed] [Google Scholar]
- 79.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133:647–658. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhu S, Si ML, Wu H, Mo YY. MicroRNA-21 targets the tumor suppressor gene tropomyosin 1 (TPM1) J Biol Chem. 2007;282:14328–14336. doi: 10.1074/jbc.M611393200. [DOI] [PubMed] [Google Scholar]
- 81.Frankel LB, Christoffersen NR, Jacobsen A, Lindow M, Krogh A, Lund AH. Programmed cell death 4 (PDCD4) is an important functional target of the microRNA miR-21 in breast cancer cells. J Biol Chem. 2007 doi: 10.1074/jbc.M707224200. [DOI] [PubMed] [Google Scholar]
- 82.Hayashita Y, Osada H, Tatematsu Y, Yamada H, Yanagisawa K, Tomida S, et al. A polycistronic microRNA cluster, miR-17–92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005;65:9628–9632. doi: 10.1158/0008-5472.CAN-05-2352. [DOI] [PubMed] [Google Scholar]
- 83.Matsubara H, Takeuchi T, Nishikawa E, Yanagisawa K, Hayashita Y, Ebi H, et al. Apoptosis induction by antisense oligonucleotides against miR-17-5p and miR-20a in lung cancers overexpressing miR-17–92. Oncogene. 2007;26:6099–6105. doi: 10.1038/sj.onc.1210425. [DOI] [PubMed] [Google Scholar]
- 84.He L, Thomson JM, Hemann MT, Hernando-Monge E, Mu D, Goodson S, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tagawa H, Karube K, Tsuzuki S, Ohshima K, Seto M. Synergistic action of the microRNA-17 polycistron and Myc in aggressive cancer development. Cancer Sci. 2007;98:1482–1490. doi: 10.1111/j.1349-7006.2007.00531.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dews M, Homayouni A, Yu D, Murphy D, Sevignani C, Wentzel E, et al. Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat Genet. 2006;38:1060–1065. doi: 10.1038/ng1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 88.Sylvestre Y, De Guire V, Querido E, Mukhopadhyay UK, Bourdeau V, Major F, et al. An E2F/miR-20a autoregulatory feedback loop. J Biol Chem. 2007;282:2135–2143. doi: 10.1074/jbc.M608939200. [DOI] [PubMed] [Google Scholar]
- 89.Coller HA, Forman JJ, Legesse-Miller A. "Myc'ed messages": myc induces transcription of E2F1 while inhibiting its translation via a microRNA polycistron. PLoS Genet. 2007;3:e146. doi: 10.1371/journal.pgen.0030146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Stanelle J, Putzer BM. E2F1-induced apoptosis: turning killers into therapeutics. Trends Mol Med. 2006;12:177–185. doi: 10.1016/j.molmed.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 91.Yamasaki L, Jacks T, Bronson R, Goillot E, Harlow E, Dyson NJ. Tumor induction and tissue atrophy in mice lacking E2F-1. Cell. 1996;85:537–548. doi: 10.1016/s0092-8674(00)81254-4. [DOI] [PubMed] [Google Scholar]
- 92.Wu X, Levine AJ. p53 and E2F-1 cooperate to mediate apoptosis. Proc Natl Acad Sci U S A. 1994;91:3602–3606. doi: 10.1073/pnas.91.9.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lin WC, Lin FT, Nevins JR. Selective induction of E2F1 in response to DNA damage, mediated by ATM-dependent phosphorylation. Genes Dev. 2001;15:1833–1844. [PMC free article] [PubMed] [Google Scholar]
- 94.Nahle Z, Polakoff J, Davuluri RV, McCurrach ME, Jacobson MD, Narita M, et al. Direct coupling of the cell cycle and cell death machinery by E2F. Nat Cell Biol. 2002;4:859–864. doi: 10.1038/ncb868. [DOI] [PubMed] [Google Scholar]
- 95.Hershko T, Ginsberg D. Up-regulation of Bcl-2 homology 3 (BH3)-only proteins by E2F1 mediates apoptosis. J Biol Chem. 2004;279:8627–8634. doi: 10.1074/jbc.M312866200. [DOI] [PubMed] [Google Scholar]
- 96.Woods K, Thomson JM, Hammond SM. Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J Biol Chem. 2007;282:2130–2134. doi: 10.1074/jbc.C600252200. [DOI] [PubMed] [Google Scholar]
- 97.Ko LJ, Prives C. p53: puzzle and paradigm. Genes Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- 98.Hemann MT, Lowe SW. The p53-Bcl-2 connection. Cell Death Differ. 2006;13:1256–1259. doi: 10.1038/sj.cdd.4401962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Bommer GT, Gerin I, Feng Y, Kaczorowski AJ, Kuick R, Love RE, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 100.He L, He X, Lim LP, de Stanchina E, Xuan Z, Liang Y, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Raver-Shapira N, Marciano E, Meiri E, Spector Y, Rosenfeld N, Moskovits N, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 102.Tarasov V, Jung P, Verdoodt B, Lodygin D, Epanchintsev A, Menssen A, et al. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 103.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Cimmino A, Calin GA, Fabbri M, Iorio MV, Ferracin M, Shimizu M, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci U S A. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, et al. Differential targeting of prosurvival Bcl-2 proteins by their BH3-only ligands allows complementary apoptotic function. Mol Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 106.Cuconati A, Mukherjee C, Perez D, White E. DNA damage response and MCL-1 destruction initiate apoptosis in adenovirus-infected cells. Genes Dev. 2003;17:2922–2932. doi: 10.1101/gad.1156903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Taniai M, Grambihler A, Higuchi H, Werneburg N, Bronk SF, Farrugia DJ, et al. Mcl-1 mediates tumor necrosis factor-related apoptosis-inducing ligand resistance in human cholangiocarcinoma cells. Cancer Res. 2004;64:3517–3524. doi: 10.1158/0008-5472.CAN-03-2770. [DOI] [PubMed] [Google Scholar]
- 108.Konopleva M, Contractor R, Tsao T, Samudio I, Ruvolo PP, Kitada S, et al. Mechanisms of apoptosis sensitivity and resistance to the BH3 mimetic ABT-737 in acute myeloid leukemia. Cancer Cell. 2006;10:375–388. doi: 10.1016/j.ccr.2006.10.006. [DOI] [PubMed] [Google Scholar]
- 109.Kobayashi S, Werneburg NW, Bronk SF, Kaufmann SH, Gores GJ. Interleukin-6 contributes to Mcl-1 up-regulation and TRAIL resistance via an Akt-signaling pathway in cholangiocarcinoma cells. Gastroenterology. 2005;128:2054–2065. doi: 10.1053/j.gastro.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 110.Calin GA, Ferracin M, Cimmino A, Di Leva G, Shimizu M, Wojcik SE, et al. A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N Engl J Med. 2005;353:1793–1801. doi: 10.1056/NEJMoa050995. [DOI] [PubMed] [Google Scholar]
- 111.Cummins JM, He Y, Leary RJ, Pagliarini R, Diaz LA, Jr, Sjoblom T, et al. The colorectal microRNAome. Proc Natl Acad Sci U S A. 2006;103:3687–3692. doi: 10.1073/pnas.0511155103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yanaihara N, Caplen N, Bowman E, Seike M, Kumamoto K, Yi M, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 113.Mott JL, Kobayashi S, Bronk SF, Gores GJ. mir-29 regulates Mcl-1 protein expression and apoptosis. Oncogene. 2007;26:6133–6140. doi: 10.1038/sj.onc.1210436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Ovcharenko D, Kelnar K, Johnson C, Leng N, Brown D. Genome-scale microRNA and small interfering RNA screens identify small RNA modulators of TRAIL-induced apoptosis pathway. Cancer Res. 2007;67:10782–10788. doi: 10.1158/0008-5472.CAN-07-1484. [DOI] [PubMed] [Google Scholar]
- 115.Algeciras-Schimnich A, Peter ME. Actin dependent CD95 internalization is specific for Type I cells. FEBS Lett. 2003;546:185–188. doi: 10.1016/s0014-5793(03)00558-1. [DOI] [PubMed] [Google Scholar]