Abstract
To test if Caenorhabditis elegans could be established as a model organism for prion study, we created transgenic C. elegans expressing the cytosolic form of the mouse prion .protein, MoPrP(23-231), which lacks the N-terminal signal sequence and the C-terminal glycosylphosphatidylinisotol (GPI) anchor site. We report here that transgenic worms expressing MoPrP(23-231)–CFP exhibited a wide range of distinct phenotypes: from normal growth and development, reduced mobility and development delay, complete paralysis and development arrest, to embryonic lethality. Similar levels of MoPrP (23-231)-CFP were produced in animals exhibiting these distinct phenotypes, suggesting that MoPrP (23-231)-CFP might have misfolded into distinct toxic species. In combining with the observation that mutations in PrP that affect prion pathogenesis also affect the toxic phenotypes in C. elegans, we conclude that the prion protein folding mechanism is similar in mammals and C. elegans. Thus, C. elegans can be a useful model organism for prion research.
Keywords: prion, C. elegans, toxicity, aggregation, protein misfolding, neurodegeneration
Introduction
Prion diseases are a group of mammalian neurodegenerative disorders with a complicated etiology that can appear in sporadic, inheritable, or infectious form [1]. It is generally accepted that the infectious agent of prion disease is a host protein (PrP) undergone conformational change from a normal cellular form, PrPC, to the altered pathogenic form, PrPSc [1; 2]. Encoded by a highly conserved single-copy gene, Prnp, PrP is a ubiquitously expressed glycosylphosphatidylinositol (GPI)-anchored glycoprotein [3]. Although studies from transgenic mammalian models have provided valuable information regarding the nature of PrPSc infectivity, the complexity of mammalian organisms, high cost of such studies, lengthy experimental periods, and numerous socio-ethical concerns have greatly limited our progress in understanding the devastating diseases. To date, the manner in which a normal host protein acquires the pathogenic conformation continues to evade our understanding, and the elucidation of the cellular mechanisms conferring PrP-mediated cellular toxicity remains a central problem in prion etiology. It is therefore of great importance to establish prion disease model in a genetically tractable organism with a nervous system.
The non-pathogenic nematode, C. elegans, offers several advantages as a model organism for prion research: small (1mm long), a short lifecycle (~3 days at 20°C), and multi-cellular organism containing a nervous system of 302 neurons in the hermaphrodite. Moreover, C. elegans is proven to be an ideal system for studying nerve function, behavior, aging, and neurodegenerative diseases [4; 5; 6; 7; 8]. Moreover, C. elegans does not have a direct PrP ortholog and thus any gain-of-function phenotype resulting from PrP production can be easily detected. Thus, C. elegans provides us the ideal compromise of complexity and tractability necessary to advance research in prion disease. In this study, we examine the ability of mouse PrP expression in C. elegans to induce a gain-of-function toxicity and the effects of PrP mutations that influence prion etiologies on this toxic phenotype.
Materials and methods
Strain and culture
The N2 Bristol strain of C. elegans and its transgenic derivatives were cultured and maintained according to standard methods in a 20°C incubator [9].
Plasmids and constructs
The DNA fragment of MoPrP(23-231) carrying the 3F4 epitope was amplified by PCR using the primers of 5’-GCGCGGCTAGCATGTCTAAAAAGCGGCCAAAGCCTG-3’ (forward), 5’-GCGCGCCGCGGGCTGGATCTTCTC CCGTC-3’ (reverse), and the template of PrP1-254-mPrP1 plasmid [10]. The resulting PCR product was digested with NheI/SacII and ligated to pECFP-N1 that was predigested with NheI/SacII to create pECFP- MoPrP(23-231). Following NcoI digestion and treatment with the Klenow, the MoPrP(23-231)-CFP fragment were ligated to pPD30.38 that was predigested with NheI and EcoRV to give the final C. elegans expression plasmid, pPD30.38- MoPrP(23-231)-CFP.
The Q167R and P101L mutations were created using a PCR-based site-directed mutagenesis. DNA fragments of MoPrP(23-231) containing these two mutations were ligated to pPD30.38 using the same procedure as described above.
Protein electrophoresis and Immunoblot analysis
Animals were frozen in liquid nitrogen and homogenized by bead-beater in lysis buffer, 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% Triton, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, 1 µg/ml leupeptin, and protease inhibitor cocktail (Roche). Crude protein extracts were resolved by SDS-PAGE, immunobloted with monoclonal PrP antibody (3F4), and detected with ECL kit (Amersham).
Phalloidin staining and fluorescence microscopy
The F-actin staining by Phalloidin and the following fluorescent detection were performed as described [11].
Behavioral assay
Liquid thrashing assays were performed in 20µl of M9 buffer as described [9].
Digestion by proteinase K and solubility of PrP in sarkosyl
All proteinase-K digestions and solubility assays were performed in 1 × PBS buffer. Protein extracts were prepared from worms expressing CFP or MoPrP(23-231)-CFP using a bead-beater. After centrifugation at 11,000 × rpm for 2 min, the supernatant was digested with 50 µg/ml of proteinase K at 37 °C for 1 hour. For sarkosyl solubility assay, 20% sarkosyl was added to the protein extracts to give a final concentration of 0%, 0.5%, 1.0%, or 2%. After incubation at room temperature for 5 min, the extracts were centrifugated at 75,000 × rpm for 30 min. The resulting supernatants and pellets were precipitated with methanol. After vacuum-dried, the proteins were solubilized with 1 × SDS sample buffer and examined by SDS-PAGE and immunoblot analysis.
Semi-denaturing agarose gel electrophoresis
Crude protein extracts prepared from C. elegans expressing MoPrP(23-231)-CFP, MoPrP(Q167R)(23-231)-CFP, and MoPrP(23-231)-CFP and MoPrP(Q167R)(23-231)-YFP were treated with the Sarkosyl sample buffer (50 mM Tris–HCl (pH 6.8), 5% glycerol, 2% Sarkosyl, and 0.05% bromophenol blue) at room temperature for 7 min and separated on 1.5% agarose gels supplemented with 0.1% SDS as described [12]. After transferring to a polyvinylidene difluoride membrane (Millipore), membranes were probed with anti-PrP antibody (3F4) and detected with ECL kit (Amersham).
Results
Targeted expression of the cytoplasmic form of mouse PrP in C. elegans muscle cells caused severe impairment in mobility, growth, and development
We first examined if the mouse prion protein (MoPrP) could be ectopically expressed in C. elegans body wall muscle cells using the muscle cell specific promoter, unc-54. Unc-54 is a strong promoter, which usually results in a robust expression. Several protein-folding neurodegenerative disease models have been established using the unc-54 expression system, such as Huntington disease [13] and Alzheimer’s disease [14]. We constructed two C. elegans expression plasmids, unc54-MoPrP(23-231)-CFP and unc54-CFP as shown in Fig. 1A. MoPrP(23-231) lacks both the N-terminal signal sequence and the C-terminal GPI anchor and is thus localized in the cytoplasm [10]. CFP-fusion was used since its fluorescent signal would help us to identify transgenic worms and facilitate characterization of MoPrP(23-231) expression and localization in live worms. It has been shown that GFP-tagged prion protein can be correctly localized and functionally active in the brains of transgenic mice [15].
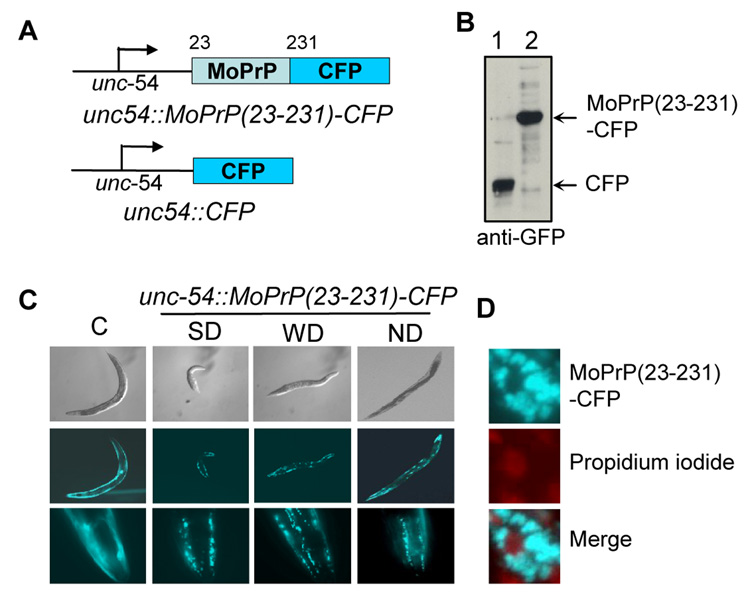
Fig. 1. Transgenic C. elegans expressing MoPrP(23-231)-CFP.
A.) Shown are CFP and MoPrP(23-231)-CFP expression constructs. B.) Immunoblot analysis of CFP or MoPrP(23-231)-CFP using a monoclonal GFP antibody. C.) A wide range of distinct phenotypes were observed for worms expressing MoPrP(23-231)-CFP. Pictures were taken at their young adult stage. C: control CFP worm; SD: strong-dumpy; WD: weak-dumpy; ND: non-dumpy. D.) Propidium iodide staining to show that MoPrP(23-231)-CFP aggregates were localized in the cytoplasm of the transgenic worms.
We microinjected the unc54-MoPrP(23-231)-CFP and unc54-CFP constructs into healthy young adult worms according to an established protocol [9]. As shown in Fig. 1B, the resulting transgenic worms were able to express MoPrP(23-231)-CFP and CFP with the expected sizes, respectively. Although a diffused fluorescent pattern was seen for the CFP control worms, numerous fluorescent foci were observed for the MoPrP(23-231)-CFP worms, suggesting that MoPrP(23-231)-CFP was aggregated (Fig. 1C). MoPrP(23-231)-CFP was cytoplasmically localized since the signals of CFP and propidium iodide, a fluorescent dye that specifically binds to DNA, did not overlap (Fig. 1D).
Although worms expressing CFP showed normal growth and development, a significant portion (~10 to 30%, varying upon different injections) of transgenic worms expressing MoPrP(23-231)-CFP exhibited a striking altered phenotype: short and chunky-looking, with a body shape similar to dumpy, a previously described phenotype mainly caused by mutations in collagen or collagen modifying enzyme [16]. Although some dumpy mutations can cause mobility defects, most dumpy worms move freely and show normal growth and reproduction [17; 18]. However, the dumpy-like worms expressing unc54-MoPrP(23-231)-CFP showed a severe mobility defect. Interestingly, a wide range of distinct phenotypes were seen, from normal growth and development, partial paralysis, to embryonic lethality. For clarity, we categorized them into three groups: strong dumpy-like, weak dumpy-like, and non-dumpy (Fig. 1C). The non-dumpy worms had normal body size and shape and moved freely. The strong dumpy-like worms were very short and nearly paralyzed. The weak dumpy-like worms had intermediate body lengths and moved slower than the non-dumpy worms.
Fig 2A shows pictures of representative tracks left by strong dumpy-like, weak dumpy-like, and non-dumpy worms expressing MoPrP(23-231)-CFP. Thirty minutes after the worms were placed on the marked positions, the N2 non-transgenic worm had moved out of sight and the non-dumpy worm also left a long track, indicating that it could move freely. The weak dumpy-like worm left a short track, indicating that it was defective in mobility. Remarkably, the strong dumpy-like worm remained in the same position where it was placed (Fig 2A). In fact, the majority of strong dumpy-like worms were virtually paralyzed. They stayed where they hatched and exhibited the bag-of-worm phenotype, larvae hatching inside their mother's body (data not shown). Both weak dumpy-like and non-dumpy worms were able to lay eggs and give rise to similar numbers of progenies as control worms. Although the strong dumpy-like, weak dumpy-like, and non-dumpy worms exhibited striking differences in phenotype, they expressed similar amount of MoPrP(23-231)-CFP fusion proteins and there were no significant differences in their fluorescence patterns (Fig. 1C). All dumpy-like worms were able to produce healthy N2 progenies upon the loss of the extra-chromosomal arrays of MoPrP(23-231)-CFP (data not shown), indicating that the observed dumpy-like phenotype was solely due to MoPrP(23-231)- CFP expression. Apparently, this dumpy-like trait was acquired de novo because dumpy-like and non-dumpy worms were able to produce progenies exhibiting a wide range of dumpiness (data not shown).
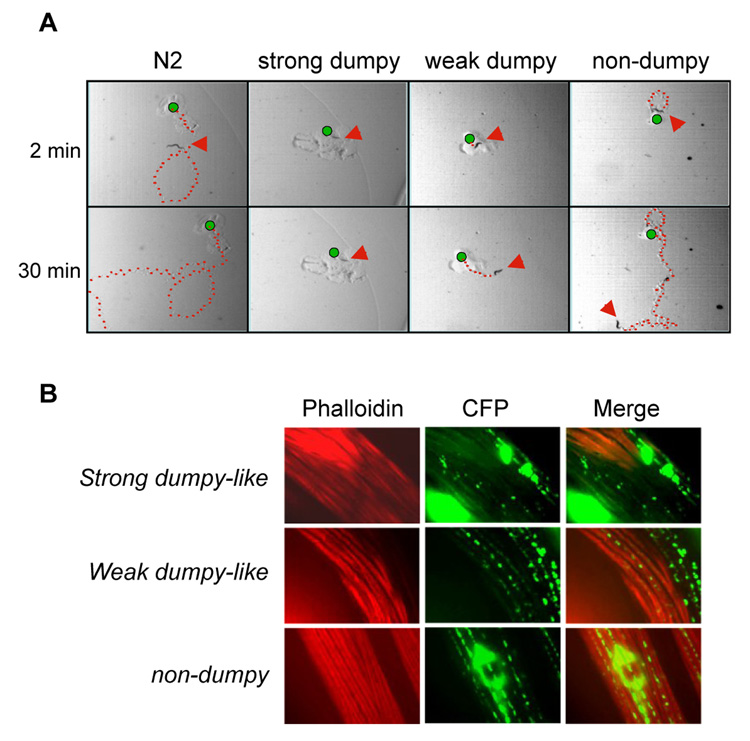
Fig. 2. Dumpy-like worms were defective in mobility.
A.) Time-lapse moving tracks (marked with red dots). Young adult worms were placed onto soft agar plates with the original positions marked (green dots). Their moving tracks were photographed after 2 and 30 min. The red arrows indicate the positions of animals when the pictures were taken. B.) Muscle structures of MoPrP(23-231)-CFP transgenic worms. Shown are Phalloidin staining (red) and MoPrP(23-231)-CFP (green).
We next examined if dumpy-like worms had altered muscle morphology. We stained the worms with phalloidin, a dye specifically binding to F-actins in permeable cells [19]. As shown in Fig. 2B, the F-actin organization was severely disrupted in muscle cells of strong dumpy-like worms. The weak dumpy-like worms also showed a noticeable disorganization of F-actins but less severe than that of strong dumpy-like worms (Fig. 2B). In contrast, non-dumpy worms showed a well-organized muscle architecture (Fig. 2B). Overall, there were more overlapping signals of MoPrP(23-231)-CFP and F-actins in the non-dumpy animals than that of dumpy-like animals, indicating that the presence of MoPrP(23-231)-CFP could affect the architecture of F-actins (Fig. 2B).
Mutations in PrP that affect prion pathogenesis also affect PrP mediated cellular toxicities in C. elegans
We next examined if mutations in PrP that reduce or enhance TSE pathogenesis would also affect PrP mediated dumpy-like phenotype accordingly in C. elegans. It was previously reported that transgenic mice co-expressing the wild type MoPrP and a mutant PrP with a missense mutation of glutamine to arginine at the codon 167, MoPrP(Q167R), were resistant to scrapie inoculation [20]. MoPrP(Q167R) was thus considered as a dominant negative mutant [20]. To test if the Q167R mutation would have similar effect in C. elegans, we created transgenic worms expressing MoPrP(Q167R)(23-231)-CFP alone or co-expressing MoPrP(Q167R)(23-231)-YFP and MoPrP(23-231)-CFP and found both of them could move freely and no dumpy-like phenotypes were developed (Fig. 3A and data not shown). Moreover, the developmental delay caused by MoPrP(23-231) expression was also relieved to some extent when MoPrP(Q167R)(23-231)-YFP was co-expressed (Supplemental Table 1). The similar intensities of the YFP- and CFP-fluorescent signals suggest that similar amounts of the wild type and the mutant fusion proteins were produced in the co-expressors (Fig. 3B). The two fusion proteins appeared to co-localize, as the -CFP and -YFP fluorescent signals were almost completely overlapped (Fig. 3B).
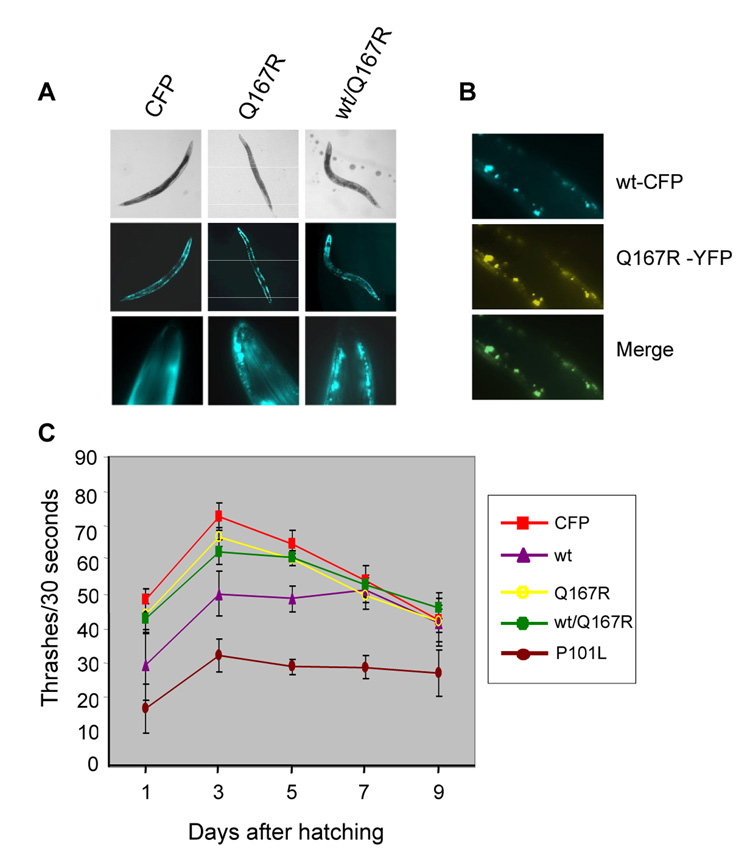
Fig. 3. Transgenic worms expressing PrP mutants.
A.) Q167R expression did not result in dumpy-like phenotypes. B.) Colocalization of MoPrP(23-231)-CFP and PrP(23-231)(Q167R)-YFP in a young adult animal co-expressing these two fusion proteins. C.) Thrashing assay. Each point represents the mean value of thrashing scores of 5 individual animals. CFP: control worm; wt: MoPrP(23-231)-CFP expressor; Q167R: MoPrP(23-231)(Q167R)-CFP expressor; wt/Q167R: co-expressors of MoPrP(23-231)-CFP and MoPrP(Q167R)(23-231)-YFP; P101L: MoPrP(23-231)(P101L)-CFP expressor. Note, only non-dumpy worms were used in this experiment.
We next examined if mutations in PrP that promote mammalian prion pathogenesis would also enhance cellular toxicity in C. elegans. We generated transgenic worms expressing MoPrP(P101L)(23-231)-CFP, which contains a point mutation from proline to leucine at codon 101 (P101L). The corresponding mutation in human PrP (P102L) is linked to a familial prion disease known as Gerstmann-Straüssler-Scheinker (GSS) disease [1]. Comparing to worms expressing the wild-type allele, a larger portion of the MoPrP(P101L)(23-231)-CFP worms showed dumpy-like phenotype (data not shown) and exhibited a more severe mobility defect. As shown in Fig. 3C, the MoPrP(P101L)(23-231)-CFP non-dumpy worms were scored very poorly in a thrash assay, a measurement quantifying the head movement of C. elegans whereas the non-dumpy worms expressing the wild-type allele obtained significantly higher scores. Moreover, worms expressing MoPrP(Q167R)(23-231)-CFP or co-expressing MoPrP(Q167R)(23-231)-YFP and wild type MoPrP(23-231)-CFP had thrash scores comparable to the CFP control worms, confirming that Q167R mutation can reduce wild-type PrP medicated toxicity. As shown in Supplemental Table 1, MoPrP(P101L)(23-231)-CFP worms also suffered longer developmental delays. Seventy-two hours after hatching, there were only ~ 19% of MoPrP(P101L)(23-231)-CFP worms entered the L4/adult stage and 46% of them remained in the L1/L2 stage. However, ~44% of wild-type MoPrP(23-231) worms had already entered L4/adult stage and only 29% of them were in L1/L2 stage. Thus, the P101L mutation augmented the PrP-mediated cellular toxicity in C. elegans.
The cytoplasmic form of MoPrP(23-231), produced in transgenic C. elegans is sarkosyl insoluble but proteinase K sensitive
Accumulation of PrPSc is the hallmark of TSE etiology [21]. Due to its unique compact folding and β-rich amyloid structure, PrPSc is significantly more resistant to detergent treatment and proteolysis than its native isomer, PrPC. To investigate if MoPrP(23-231)-CFP worms produced PrPSc-like species, we examined the detergent solubility of MoPrP(23-231)-CFP. As shown in Fig. 4A, both MoPrP(23-231)-CFP and MoPrP(Q167R)(23-231)-CFP were not extractable in buffers containing 0 - 2% sarkosyl, while CFP produced in the control worms was soluble in all tested conditions (data not shown). To examine if MoPrP(23-231)-CFP produced in C. elegans is proteinase K resistant, we prepared total lysates from worms expressing CFP or MoPrP(23-231)-CFP and treated them with 50 µg/ml proteinase K. As shown in Fig 4B, there were no detectable bands with slower mobility than CFP were detected when GFP antibody was used for immunoblot analysis. When the 3F4 antibody was used, there were no detectable bands in any samples (data not shown). Our results indicate that PrPSc was not formed in transgenic worms expressing MoPrP(23-231)-CFP.
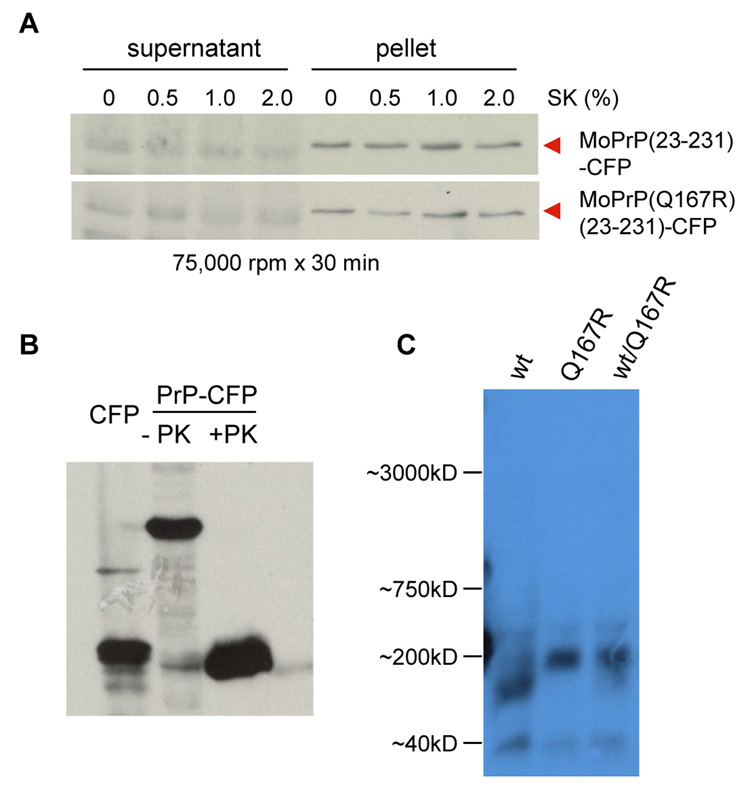
Fig. 4. Biochemical analyses of MoPrP(23-231)-CFP and MoPrP(23-231)(Q167)- CFP.
A.) Sarkosyl solubility assay. Soluble and pellet fractions of protein extracts containing different sarkosyl concentrations were prepared and analyzed as described in the materials and methods. A monoclonal PrP antibody (3F4) was used for immunoblot analysis. B.) Proteinase K digestion was performed as described in the Materials and Methods. A monoclonal GFP antibody was used for the immunoblot analysis. PrP-CFP: MoPrP(23-231)-CFP expressor. C.) Semi-denaturing agarose gel electrophoresis (SDAGE) assay. wt: MoPrP(23-231)-CFP expressor; Q167R: MoPrP(23-231)(Q167R)-CFP expressor; wt/Q167R: co-expressors of MoPrP(23-231)-CFP and MoPrP(Q167R)(23-231)-YFP.
Analysis of the polymer sizes of the wild-type MoPrP(23-231)-CFP and the dominant negative mutant, MoPrP(Q167R)(23-231)-CFP, in C. elegans
To test if MoPrP(23-231)-CFP and MoPrP(Q167R)(23-231)-CFP had folded into different conformations, we prepared crude protein extracts in the presence of 2% sarkosyl and analyzed their polymer sizes using a modified semi-denaturing agarose gel electrophoresis (SDAGE), a useful technique that has been successfully applied to examine the polymer sizes of several detergent stable protein aggregates, as described [22]. As shown in Fig 4C, there were no detectable large polymers formed for both MoPrP(23-231)-CFP and MoPrP(Q167R)(23-231)-CFP. The average size of MoPrP(23-231)-CFP oligomers was estimated ~120kDa, which is smaller than that of MoPrP(Q167R)(23-231)-CFP, ~200kDa (Fig 4C). Interestingly, although similar expression levels of MoPrP(23-231)-CFP and MoPrP(Q167R)(23-231)-YFP were observed in the co-localization experiment (Fig 3B), the majority fusion proteins were found in the higher molecular weight band corresponding to the MoPrP(Q167R)(23-231)-CFP conformer.
Discussion
In this study, we have explored the possibility of establishing the non-pathogenic nematode, C. elegans, as a model organism for prion study. We expressed the cytoplasmic form of mouse PrP as CFP or YFP fusion proteins in the body wall muscle cells of C. elegans. It has been shown previously that PrP accumulation in the cytoplasm of neuronal cells leads to the formation of PrPSc-like molecules and to a significant apoptosis [10; 23]. Moreover, elevated PrP levels were detected in muscle cells of patients with muscle related disease, such as inclusion-body myositis, polymyositis, dermatomyositis, and neurogenic muscle atrophy [24]. However, contradictive observations were also reported to suggest cytosolic expression of PrP was not toxic to N2a cells or primary neuron culture [25; 26]. We showed in this study that some transgenic worms expressing the cytoplasmic form of MoPrP displayed a striking dumpy-like phenotype, short and chunky-looking. Although the underlying mechanism of this dumpy-like phenotype is unclear, the abnormal muscle structures and severe mobility defects of the dumpy-like worms suggest that accumulation of cytosolic MoPrP can be toxic to C. elegans. The fact that MoPrP(23-231) expression in the body wall muscle cells of C. elegans can mimic certain unc-54 mutants to result in alteration of muscle morphology and function demonstrates that this gain-of-function phenotype can be a useful system to study prion mediated toxicity. Interestingly, transgenic worms expressing similar levels of MoPrP(23-231) could exhibit a wide range of phenotypes varying in different degrees of dumpiness and muscle dysfunction, indicating that multiple toxic species of MoPrP(23-231) can be formed in C. elegans’ muscle cells, perhaps due to the formation of different conformers. Since no proteinase K resistant species of MoPrP(23-231) were detected, MoPrP(23-231) produced in C. elegans muscle cells was not PrPSc-like. Our results support the view that PrPtoxic species are conformationally different from PrPSc [27].
It has been shown that the PrP(P101L) mutation is tightly linked to a genetic prion disease, Gerstmann-Sträussler-Scheinker syndrome. In flies, PrP(P101L) expression in cholinergic neurons resulted in severe locomotor dysfunction and premature death of larvae and adults [28]. We also found MoPrP(P101L)(23-231)-CFP expression in C. elegans augmented PrP mediated toxicity in C. elegans (Fig 3C and Supplemental Table 1). In contrast, Q167R, a PrP mutation conferring resistance to scrapie inoculation in the presence of the wild-type PrP expression [20], reduced PrP mediated toxic phenotypes in C. elegans. Although the underlying mechanism is unclear, our finding that MoPrP(23-231)-CFP and MoPrP(Q167R)(23-231)-YFP could exist in oligomers with different sizes strongly suggests that they had different conformations in the muscle cells of C. elegans and such a difference is likely responsible for the observed difference in cellular toxicity. These data, combined with the observation that MoPrP(Q167R)(23-231)-YFP and MoPrP(23-231)-CFP were co-localized (Fig 3B), strongly suggest that PrP(Q167R) was able to influence the folding of wild-type PrP to adopt its non-toxic conformation. As a consequence, the wild-type PrP is co-aggregated with the non-toxic PrP(Q167R) conformer(s) to result in a dominant negative phenotype.
Taken together, our results suggest that the cellular mechanism required for prion protein folding is similar in C. elegans and mammals. Thus it is possible to use C. elegans as a model organism to dissect the underlying cellular machinery. To date, the mechanism by which prions kill neurons remains a mystery. Establishing simple prion models, which are genetically amenable, such as C. elegans and D. melanogaster, will likely provide us better opportunities to identify cellular factors responsible for prion mediated infectivity and toxicity.
Supplementary Material
Acknowledgement
We thank R. Morimoto for the pPD30.38 plasmid; K. Iwasaki and T. Teramoto for the technical advices; J. Kramer and H. Epstein for helpful discussions; G.E. Kim for technical assistance; Z. Du and E. Crow for critical comments on the manuscript. This work was partially supported by grants from the United States Army (0850-370-R744), the Ellison Medical Foundation, and the US National Institutes of Health (R01NS056086) to L.L.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Prusiner SB. Prions. PNAS. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aguzzi A, Sigurdson C, Heikenwaelder M. Molecular Mechanisms of Prion Pathogenesis. Annual Review of Pathology: Mechanisms of Disease. 2008;3:11–40. doi: 10.1146/annurev.pathmechdis.3.121806.154326. [DOI] [PubMed] [Google Scholar]
- 3.Oesch B. A cellular gene encodes scrapie PrP 27–30 protein. Cell. 1985;40:735–746. doi: 10.1016/0092-8674(85)90333-2. [DOI] [PubMed] [Google Scholar]
- 4.Carter CS, Ramsey MM, Sonntag WE. A critical analysis of the role of growth hormone and IGF-1 in aging and lifespan. Trends Genet. 2002;18:295–301. doi: 10.1016/S0168-9525(02)02696-3. [DOI] [PubMed] [Google Scholar]
- 5.Goedert M. Neurodegenerative tauopathy in the worm. Proc Natl Acad Sci U S A. 2003;100:9653–9655. doi: 10.1073/pnas.1834191100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsu AL, Murphy CT, Kenyon C. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science. 2003;300:1142–1145. doi: 10.1126/science.1083701. [DOI] [PubMed] [Google Scholar]
- 7.Morley JF, Morimoto RI. Regulation of longevity in Caenorhabditis elegans by heat shock factor and molecular chaperones. Mol Biol Cell. 2004;15:657–664. doi: 10.1091/mbc.E03-07-0532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thomas JH. Nematodes are smarter than you think. Neuron. 2001;30:7–8. doi: 10.1016/s0896-6273(01)00256-2. [DOI] [PubMed] [Google Scholar]
- 9.Teramoto T, Lambie EJ, Iwasaki K. Differential regulation of TRPM channels governs electrolyte homeostasis in the C. elegans intestine. Cell Metab. 2005;1:343–354. doi: 10.1016/j.cmet.2005.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma J, Lindquist S. De novo generation of a PrPSc-like conformation in living cells. Nat Cell Biol. 1999;1:358–361. doi: 10.1038/14053. [DOI] [PubMed] [Google Scholar]
- 11.Ono S. The Caenorhabditis elegans unc-78 Gene Encodes a Homologue of Actin-Interacting Protein 1 Required for Organized Assembly of Muscle Actin Filaments. The Journal of Cell Biology. 2001;152:1313–1319. doi: 10.1083/jcb.152.6.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fan Q, Park KW, Du Z, Morano KA, Li L. The role of Sse1 in the de novo formation and variant determination of the [PSI+] prion. Genetics. 2007;177:1583–1593. doi: 10.1534/genetics.107.077982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Satyal SH, Schmidt E, Kitagawa K, Sondheimer N, Lindquist S, Kramer JM, Morimoto RI. Polyglutamine aggregates alter protein folding homeostasis in Caenorhabditis elegans. Proc Natl Acad Sci U S A. 2000;97:5750–5755. doi: 10.1073/pnas.100107297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Link CD. Expression of human beta-amyloid peptide in transgenic Caenorhabditis elegans. Proc Natl Acad Sci U S A. 1995;92:9368–9372. doi: 10.1073/pnas.92.20.9368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barmada S, Piccardo P, Yamaguchi K, Ghetti B, Harris DA. GFP-tagged prion protein is correctly localized and functionally active in the brains of transgenic mice. Neurobiol Dis. 2004;16:527–537. doi: 10.1016/j.nbd.2004.05.005. [DOI] [PubMed] [Google Scholar]
- 16.Kenning C, Kipping I, Sommer RJ. Isolation of mutations with dumpy-like phenotypes and of collagen genes in the nematode Pristionchus pacificus. Genesis. 2004;40:176–183. doi: 10.1002/gene.20084. [DOI] [PubMed] [Google Scholar]
- 17.von Mende N, Bird DM, Albert PS, Riddle DL. dpy-13: a nematode collagen gene that affects body shape. Cell. 1988;55:567–576. doi: 10.1016/0092-8674(88)90215-2. [DOI] [PubMed] [Google Scholar]
- 18.Page AP, Johnstone IL. The cuticle. WormBook. 2007:1–15. doi: 10.1895/wormbook.1.138.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wulf E, Deboben A, Bautz FA, Faulstich H, Wieland T. Fluorescent phallotoxin, a tool for the visualization of cellular actin. Proc Natl Acad Sci U S A. 1979;76:4498–4502. doi: 10.1073/pnas.76.9.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Perrier V, Kaneko K, Safar J, Vergara J, Tremblay P, DeArmond SJ, Cohen FE, Prusiner SB, Wallace AC. Dominant-negative inhibition of prion replication in transgenic mice. Proc Natl Acad Sci U S A. 2002;99:13079–13084. doi: 10.1073/pnas.182425299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Aguzzi A, Heikenwalder M. Prion diseases: cannibals and garbage piles. Nature. 2003;423:127–129. doi: 10.1038/423127a. [DOI] [PubMed] [Google Scholar]
- 22.Bagriantsev SN, Kushnirov VV, Liebman SW. Analysis of amyloid aggregates using agarose gel electrophoresis. Methods Enzymol. 2006;412:33–48. doi: 10.1016/S0076-6879(06)12003-0. [DOI] [PubMed] [Google Scholar]
- 23.Ma J, Wollmann R, Lindquist S. Neurotoxicity and neurodegeneration when PrP accumulates in the cytosol. Science. 2002;298:1781–1785. doi: 10.1126/science.1073725. [DOI] [PubMed] [Google Scholar]
- 24.Zanusso G, Vattemi G, Ferrari S, Tabaton M, Pecini E, Cavallaro T, Tomelleri G, Filosto M, Tonin P, Nardelli E, Rizzuto N, Monaco S. Increased expression of the normal cellular isoform of prion protein in inclusion-body myositis, inflammatory myopathies and denervation atrophy. Brain Pathol. 2001;11:182–189. doi: 10.1111/j.1750-3639.2001.tb00390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fioriti L, Dossena S, Stewart LR, Stewart RS, Harris DA, Forloni G, Chiesa R. Cytosolic prion protein (PrP) is not toxic in N2a cells and primary neurons expressing pathogenic PrP mutations. J Biol Chem. 2005;280:11320–11328. doi: 10.1074/jbc.M412441200. [DOI] [PubMed] [Google Scholar]
- 26.Roucou X, Guo Q, Zhang Y, Goodyer CG, LeBlanc AC. Cytosolic prion protein is not toxic and protects against Bax-mediated cell death in human primary neurons. J. Biol. Chem. 2003;278:40877–40881. doi: 10.1074/jbc.M306177200. [DOI] [PubMed] [Google Scholar]
- 27.Chiesa R, Pestronk A, Schmidt RE, Tourtellotte WG, Ghetti B, Piccardo P, Harris DA. Primary myopathy and accumulation of PrPSc-like molecules in peripheral tissues of transgenic mice expressing a prion protein insertional mutation. Neurobiol Dis. 2001;8:279–288. doi: 10.1006/nbdi.2001.0400. [DOI] [PubMed] [Google Scholar]
- 28.Gavin BA, Dolph MJ, Deleault NR, Geoghegan JC, Khurana V, Feany MB, Dolph PJ, Supattapone S. Accelerated accumulation of misfolded prion protein and spongiform degeneration in a Drosophila model of Gerstmann-Straussler-Scheinker syndrome. J Neurosci. 2006;26:12408–12414. doi: 10.1523/JNEUROSCI.3372-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.