Abstract
Type 4 glutathione peroxidase (GPx4) is a widely expressed mammalian selenoenzyme known to play a vital role in cytoprotection against lipid hydroperoxide (LOOH)-mediated oxidative stress and regulation of oxidative signaling cascades. Since prokaryotes are not equipped to express mammalian selenoproteins, preparation of recombinant GPx4 via commonly used bacterial transformation is not feasible. A published procedure for isolating the enzyme from rat testis employs affinity chromatography on bromosulfophthalein-glutathione-linked agarose as the penultimate step in purification. Since this resin is no longer commercially available and preparing it in satisfactory operational form is tedious, we have developed an alternative purification approach based on sequential anion exchange, size exclusion, and cation exchange chromatography. Final preparations were found to be essentially homogeneous in GPx4 (Mr ∼20 kDa), as demonstrated by SDS-PAGE with protein staining and immunoblotting. Specific enzymatic activity was determined using a novel thin layer chromatographic approach in which the kinetics of phosphatidylcholine hydroperoxide loss or cholesterol-7α-hydroperoxide loss were monitored. A >400-fold purification of active enzyme has been attained. The relatively straightforward isolation procedure described should prove valuable for further functional studies on GPx4, e.g. how its ability to catalyze LOOH reduction compares with that of other LOOH detoxifying enzymes.
Introduction
Eukaryotic cells are equipped with a variety of primary and secondary enzymatic defenses against the damaging and potentially lethal effects of oxidative stress. Primary defenses are mainly preventative, e.g. scavenging of superoxide by superoxide dismutases or hydrogen peroxide by catalase and peroxidases. Secondary or back-up defenses, on the other hand, typically involve inactivation (detoxification) of oxidized groups at damage sites, followed by removal (or vice versa), and finally repair steps [1,2]. Reductive inactivation of fatty acid hydroperoxides or lipid hydroperoxides in membranes or lipoproteins is an important example of secondary antioxidant defense in which glutathione (GSH)-dependent enzymes in the selenoperoxidase family and certain non-selenoperoxidases in the peroxiredoxin family are known to play an important role [2-4]. Two major intracellular members of the first family have been studied extensively in terms of cytoprotection against oxidative stress, regulation of cyclooxygenase and lipoxygenase activity/expression, and regulation of redox signaling cascades: type-1 or “classical” glutathione peroxidase (GPx1) and type-4 glutathione peroxidase (GPx4), also known as phospholipid hydroperoxide glutathione peroxidase [3,5-7]. Both of these enzymes contain an active site selenocysteine (CySeH) residue, which participates in the two-electron reduction/detoxification of hydroperoxides to alcohols at the expense of GSH [3,5] (Eqs. 1-3)
| (1) |
| (2) |
| (3) |
However, the enzymes differ in functional molecular size (85 kDa homotetramer for GPx1 vs. 20 kDa monomer for GPx4), amino acid sequence, and average level of protein expressed in any given cell (GPx1 >> GPx4) [5,6]. They also exhibit striking differences in peroxide reactivity. GPx4 can catalyze the direct reduction of phospholipid hydroperoxides (PLOOHs) in biomembranes, whereas GPx1 lacks this ability and acts primarily on H2O2 or fatty acid hydroperoxides liberated by sn-2 acyl bond hydrolysis of PLOOHs [2,8,9]. GPx4 can also catalyze the reduction of peroxidized cholesterol and cholesteryl esters in membranes or lipoproteins and, again, GPx1 is incapable of this [10,11]. For mice, homozygous GPx1-null mutations have no obvious effects on viability, whereas such mutations in GPx4 are embryonic lethal [12]. Although heterozygous knockout mice (Gpx4+/−) survive, their fibroblasts are substantially more sensitive to oxidative challenges than wild types [13]. Moreover, GPx4 transgenic mice have been found to be hyperresistant to oxidative insults, including neurotoxic insults [14,15].
Studies involving isolated GPx4 require that the enzyme be prepared from natural sources, since bacterial systems typically cannot express recombinant eukaryotic selenoproteins [16]. GPx4 was originally purified from porcine liver [17] and more recently from rat testis [18], which is not only rich in the enzyme, but readily available from commercial sources. One of the resins previously used for column purification of the rat enzyme was bromosulfophthalein-glutathione (BSP-GS)-linked agarose [18]. Although an approximately 10-fold increase in purification was reported with this material (presumably functioning as an affinity resin), the underlying molecular basis for this is not completely clear. In any event, BSP-GS-agarose is no longer commercially available, necessitating either de novo preparation of the resin in suitable form, which is not straightforward [19], or development of a new purification strategy that omits it. In this report we describe such a new approach, which routinely affords active GPx4 of at least 95% final purity based on electrophoretic analysis.
Materials and Methods
Materials
Sigma-Aldrich (St. Louis, MO) supplied most of the chemicals and reagents, including cholesterol, cumene hydroperoxide (CuOOH), reduced glutathione (GSH), and desferrioxamine (DFO), along with DEAE-Sepharose CL-6B anion exchange resin and Sephadex G-50 (20-80 μm particles) for size exclusion. HiTrap SP cation exchange resin was obtained from GE Healthcare (Fairfield, CT). Frozen rat testes as a source of GPx4 were obtained from Pel-Freez (Rogers, AR). N,N,N′,N′-tetramethyl-p-phenylenediamine (TMPD) was from Alfa Aesar (Ward Hill, MA). The rabbit anti-human GPx4 polyclonal antibody was from Gene Tex (San Antonio, TX). 1-Palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (PC) was obtained from Avanti Polar Lipids (Birmingham, AL). PC in chloroform was peroxidized by dye-sensitized photooxidation as described [20]. After irradiation, the material was dried under nitrogen, dissolved in 2-propanol, and subjected to isocratic normal-phase HPLC, using a silica column with 2-propanol/hexane/water (51/39/10, v/v/v) as the mobile phase. Absorbance at 205 and 234 nm was monitored, the latter reflecting hydroperoxide (PCOOH) conjugated diene bonds. A PCOOH peak exhibiting maximal A234/A205 ratio was isolated and subjected to orthophosphate analysis for determination of total phospholipid content and iodometric analysis for determination of total peroxide content [20]. The PCOOH used for assessing GPx4 activity had a mole ratio of ∼1.0 - OOH group per Pi group, as previously observed [21]. HPLC-ESI-MS analysis revealed that the PCOOH consisted of equal amounts of PC with an -OOH group at either the 9- or 10-position of the sn-2 oleoyl moiety [21], as expected for attack of photogenerated singlet oxygen on PC. The cholesterol hydroperoxide 3β-hydroxycholest-5-ene-7α-hydroperoxide (7α-OOH) was generated by dye-sensitized photooxidation of cholesterol in liposomal form [22]. 7α-OOH was separated from parent cholesterol and other peroxides using sequential reversed-phase and normal-phase HPLC, and was determined by iodometric analysis [22]. Stock solutions of PCOOH and 7α-OOH in 2-propanol were stored at −20 °C.
Isolation of GPx4
Tissue Disruption
Frozen rat testes (10 g) from Pel-Freez were slowly thawed on ice and cut into small pieces using a razor blade. The minced tissue was suspended in 0.1 M Tris-HCl/0.5 mM phenylmethylsulphonyl fluoride (PMSF)/5 mM 2-mercaptoethanol (2-ME)/0.2 mM EDTA/0.6 % (w/v) CHAPS at pH 7.4 (3 mL/g of tissue) and disrupted for 1 min on ice, using a Polytron homogenizer.
Crude purification steps
All of the following steps were carried out on ice or at 4 °C. The tissue lysate was coarse-filtered through cheesecloth to remove large particulate material and then centrifuged for 15 min at 15000 × g to remove cellular debris. The supernatant fluid was subjected to a second centrifugation step at 100000 × g for 45 min. The recovered supernatant was brought to 50% saturation with ammonium sulfate over a 5 min interval with stirring, then centrifuged at 20000 × g for 20 min and the pellet discarded. The ammonium sulfate concentration was then taken to 70% saturation, followed by centrifugation at 20000 × g for 20 min and recovery of the pellet. The pellet was carefully rinsed with water to remove residual salt, then dissolved in 10 mL of KME Buffer A [10 mM potassium phosphate/5 mM 2-ME/50 μM EDTA (pH 7.3)] and dialyzed against this buffer using a 12-14 kDa cut-off dialysis membrane. All subsequent chromatographic separations were carried out in a cold room at 4 °C.
DEAE-Sepharose chromatography
The DEAE-Sepharose CL-6B column (1.5 × 16 cm) was first equilibrated with KME Buffer A. Dialyzed protein from the preceding step was centrifuged at 10000 × g for 20 min to remove any undissolved material. The supernatant solution was loaded onto the DEAE column at a flow rate of 1 mL/min, using a peristaltic pump and collecting 10-mL fractions. The column was washed with 5 column volumes of buffer A to remove unbound or weakly bound proteins. GPx4 was then eluted using a linear gradient from 0% to 100% KME Buffer B [10 mM potassium phosphate/500 mM KCl/5 mM 2-ME/50 μM EDTA (pH 7.3)] over 6 column volumes, followed by an isocratic wash with 50 mL of the same buffer. Regeneration of the column was accomplished by washing with 5 column volumes of 1.0 M NaCl. Protein levels were determined by measuring absorbance at 280 nm for each fraction, followed by measurement of GPx4 activity via coupled enzymatic assay (see below).
Size-exclusion chromatography
A Sephadex G-50 column (1.5 × 57 cm) was used for size-exclusion separation. Fractions from the DEAE-Sepharose column that had significant GPx4 activity based on CuOOH reduction were pooled, brought to 95% ammonium sulfate saturation, and centrifuged at 20000 × g for 20 min, the supernatant being discarded. The pellet was dissolved in 2 mL of KME Buffer C [10 mM potassium phosphate/100 mM KSCN/5 mM 2-ME/50 μM EDTA (pH 7.3)] and loaded onto the Sephadex G-50 column following equilibration with the same buffer. Proteins were separated using this buffer for gravity flow elution (0.8 mL/min), 3.2-mL fractions being collected. Elution was terminated after 100 mL of buffer had passed through the column. Protein concentration of each fraction was determined by A280 measurement. Each fraction after the void volume (∼32 mL) was also assessed for GPx4 activity using a CuOOH-based coupled assay.
HiTrap SP chromatography
Size-exclusion fractions containing significant peroxidatic activity were pooled, dialyzed against KME Buffer D [10 mM potassium phosphate/5 mM 2-ME/50 μM EDTA (pH 6.5)], concentrated to ∼0.5 mL by centrifugation-filtration using a 10 kDa cutoff filter, and loaded onto a 1-mL HiTrap SP column. An AKTA FPLC (GE Healthcare, Fairfield, CT) with Unicorn 5.11 software was used for elution. The column was first washed with 3 column volumes of KME Buffer D at a flow rate of 1.0 mL/min, 1-mL fractions being collected. GPx4 was then eluted using a linear gradient from 0% to 100% KME Buffer E [10 mM potassium phosphate/0.3 M KCl/5 mM 2-ME/50 μM EDTA (pH 6.5)] over 15 column volumes, and then maintaining an isocratic level of 100% Buffer E for 3 column volumes before changing back to Buffer D. Protein emergence was monitored by measuring A280 and selected fractions were tested for GPx4 activity by coupled assay using CuOOH or PCOOH as the peroxide substrate. Those fractions containing significant activity were pooled, concentrated, dialyzed extensively against 25 mM Tris-HCl/100 mM KSCN/5 mM 2-ME/50 μM EDTA/50 μM DFO (pH 7.7), and then stored at −20 °C in the presence of 30% (v/v) glycerol. Purity of the enzyme was assessed by SDS-PAGE, using a 4-12% polyacrylamide gel in MES buffer under reducing conditions and Coomassie Blue for staining. Densitometry was used to assess the degree of GPx4 purity in final preparations.
Immunoblot Analysis
The presence of GPx4 at various stages of purification was confirmed by Western blot analysis using a 4-12% polyacrylamide gel for electrophoresis and 0.45 μm polyvinylidene difluoride membrane for transblotting. Blots were blocked, treated with rabbit anti-human GPx4 antibody, then with peroxidase-conjugated anti-rabbit IgG, and finally analyzed using enhanced chemiluminescence. Additional details were as reported previously [23].
Determination of GPx4 Activity by Coupled Enzymatic Assay
Throughout the purification, GPx4 activity was monitored spectrophotometrically by coupled enzyme assay. The typical assay mixture (1.0 mL) contained 1 mM EDTA, 0.1% Triton X-100, 3 mM GSH, 0.2 mM NADPH, 0.14 units of glutathione reductase, 0.1 mM hydroperoxide (either CuOOH or PCOOH), and GPx4-containing sample (added last) in 0.1 M sodium phosphate buffer (pH 7.3) at 37 °C. The decay kinetics of NADPH absorbance at 340 nm were determined. Quantitation was based on NADPH's extinction coefficient of 6220 M−1cm−1 at 340 nm. A blank containing everything except GPx4 sample was monitored alongside. Any absorbance decay observed for the blank was subtracted from that of the complete reaction mixture to correct for non-specific activity. One unit of GPx4 activity is expressed as 1 μmol of CuOOH or PCOOH oxidized per min at 37 °C.
Determination of GPX4 Activity Based on 7α-OOH Reduction
A TLC-based approach similar to that previously described for assessing GPx4 activity in cell extracts was used [24]. The typical reaction mixture (0.4 or 0.5 mL) contained 3 mM GSH, 1 mM EDTA, 0.1 mM DFO, 0.1% Triton X-100, 25 μM 7α-OOH, and 25-100 nM GPx4 (added last) in PBS (25 mM sodium phosphate, 125 mM NaCl, pH 7.4). At various times during incubation at 37 °C, 50 μL samples were removed and added to 0.2 mL of ice-cold PBS to quench the reaction. Lipids were extracted using 0.4 mL of ice-cold chloroform/methanol (2:1, v/v). After centrifugation, 0.2 mL of the organic phase was dried under nitrogen to a film, which was stored at −20 °C if not used immediately. Before analysis, the film was dissolved in 20 μL of hexane/2-propanol (93:7, v/v). Normal phase high-performance thin-layer chromatography (HPTLC) with TMPD spray detection [20,24] was used for tracking GPx4-catalyzed reduction of 7α-OOH. Silica gel HPTLC plates (10 × 20 cm; 0.2 mm layer thickness) were from EM Science (Gibbstown, NJ). Each extract sample was applied to the plate in a hairline nitrogen stream, using a Linomat IV applicator (Camag Scientific, Wilmington, NC). The mobile phase was benzene/ethyl acetate (1:1, v/v), which allows 7α-OOH and 7α-OH to be separated from one another [24]. Immediately after chromatography, the plate was dried under a stream of argon and sprayed with a fine mist of freshly prepared 1% (w/v) TMPD in methanol/water/acetic acid (50:50:1, v/v/v). To minimize exposure to oxygen and keep background levels low, the TLC plate was covered tightly with a clean piece of pane glass immediately after spraying. The plate was photographed and 7α-OOH band intensities were measured densitometrically, using LabWorks version 4.6 from UVP BioImaging Systems. After assessing TMPD reactivity, the plate was sprayed lightly with 18 N sulfuric acid and heated for 10 min at 110 °C to develop the 7α-OH band, which was also measured densitometrically. Alternatively, both analytes could be determined simultaneously after sequential TMPD and sulfuric acid spraying, since TMPD reduces 7α-OOH to 7α-OH on the TLC plate [20]. Additional details were as described previously [20,24]. One unit of GPx4 activity based on this assay is taken as 1 μmol of 7α-OOH reduced (or 7α-OH formed) per min.
Results and Discussion
Testis is known to be a rich source of GPx4 [18], which is reported to play an important role in sperm cell maturation and structural integrity [25]. During early stages of spermatogenesis, GPx4 may act as a vital antioxidant by modulating peroxide tone and detoxifying LOOHs, whereas in mature spermatozoa with relatively low GSH levels, it appears to play a structural role by inducing -Se-S- and -S-S- cross-linked protein aggregates and losing enzymatic activity in the process [25]. Recent data suggest that the nuclear isoform of GPx4 assists in the maintenance of sperm chromatin structural integrity by acting as a protein thiol peroxidase [26]. The relatively high expression of GPx4 in testis has made this the tissue of choice for isolating the enzyme in order to study its biochemical, structural, and kinetic properties [18]. This is emphasized by the fact that E. coli and other bacterial systems cannot express the recombinant enzyme unless the selenocysteine insertion sequence (SECIS) element is properly arranged, but even then, the yield is very low [16]. A mosquito expression system has also been described [27], but again, the output of active GPx4 protein was extremely low. By contrast, good yields of a recombinant mutant GPx4 containing an active site cysteine instead of selenocysteine have been obtained, although enzymatic activity was only ∼1% that of the natural enzyme [27]. For practical purposes, therefore, one must still rely on classical isolation procedures for obtaining reasonable amounts of this enzyme, as well as other selenoenzymes for that matter.
Stepwise column purification of testicular GPx4
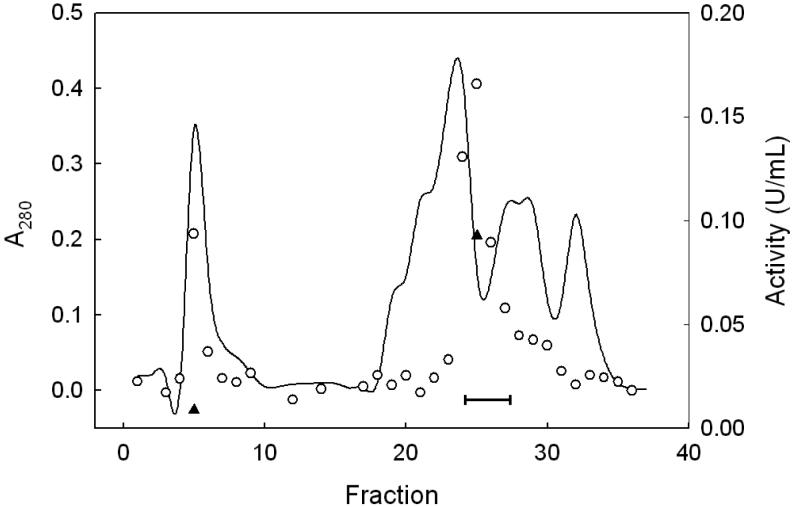
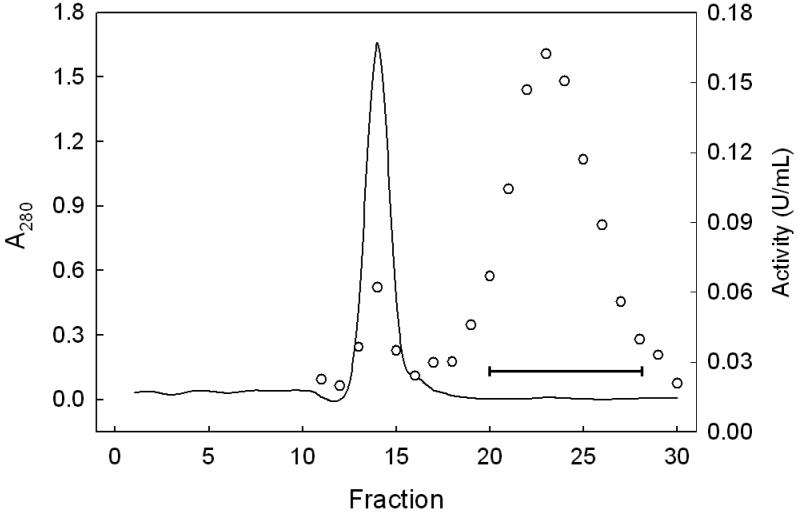
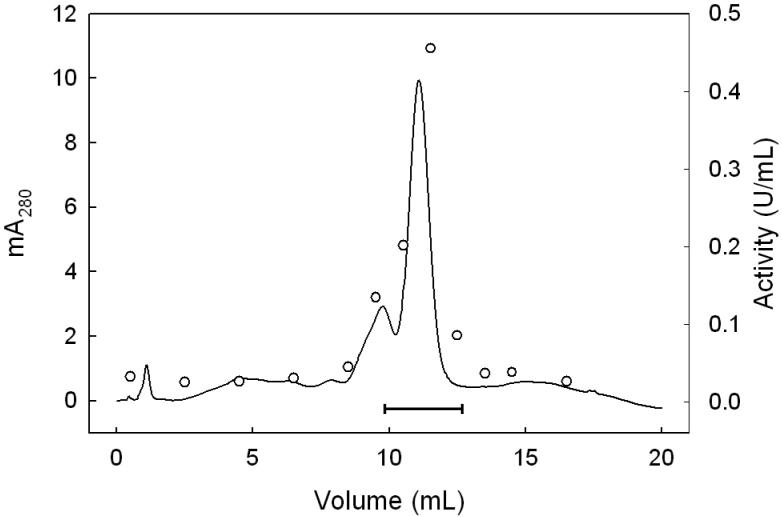
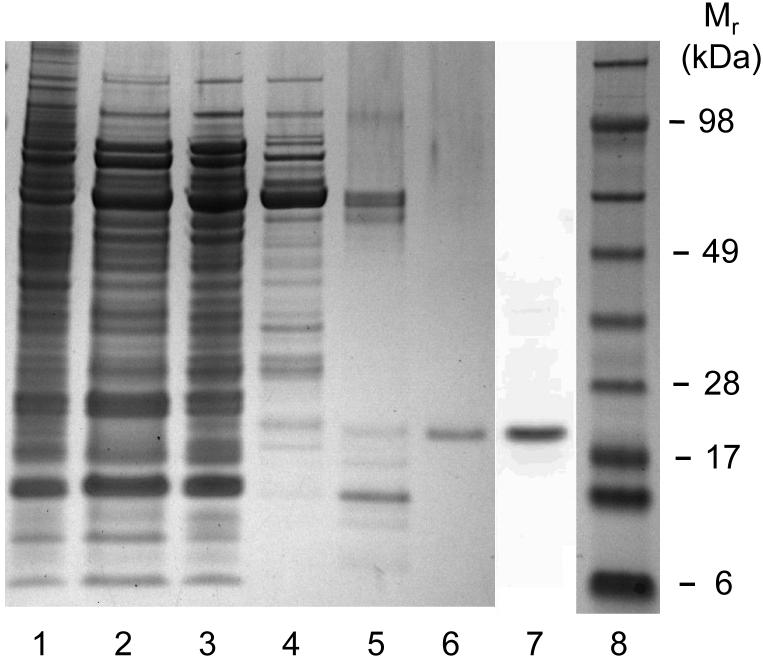
We describe a new purification procedure for rat testis GPx4 which omits the use of a BSP-GS-agarose column, as employed previously by Roveri et al. [18]. Not only is BSP-GS-agarose no longer commercially available and difficult to prepare de novo [19], but the rationale for its earlier use as a GPx4 affinity resin was not altogether clear [18]. Thus, it was shown that BSP alone can dose-dependently inactivate the enzyme, suggesting tight binding near the active site as a possible basis for affinity to BSP-GS-agarose; however, no direct evidence for this was given [18]. Moreover, the function of the glutathione moiety is not obvious, i.e. whether it participated in enzyme binding or merely served as a spacer for facilitating enzyme access to BSP groups. The approach we developed involves initial ammonium sulfate fractionation of a tissue homogenate, followed sequentially by DEAE-Sepharose, Sephadex G50 size exclusion, and HiTrap SP column chromatography, the latter being substituted for BSP-GS-agarose. As shown in Fig. 1, dialyzed protein obtained in the crude work-up at 70% (NH4)2SO4 saturation eluted from the weakly anionic DEAE-Sepharose column as four major protein (A280) peaks. The first was centered at fraction 5 (in the flow-through volume) using KME Buffer A for elution; the second was centered at fraction 23 after a linear gradient of 0-100% KME Buffer B was started at fraction 16. Fractions were checked for selenoperoxidase activity using a coupled enzymatic assay with CuOOH as the hydroperoxide substrate. A sharp peak of activity centered at fraction 25 was observed, along with a shoulder near fraction 29 (Fig. 1). It was reported previously [17] that DEAE-Sepharose chromatography cleanly separates GPx4 from the more abundant GPx1 isoform, the latter appearing in the weakly retained flow-through fractions. To confirm that the two enzymes had been separated, we spot-checked specific activity in various fractions, using PCOOH in coupled assay format. In doing this, we knew that while both GPx1 and GPx4 can catalyze the reduction of CuOOH, only GPx4 can act on PCOOH [10]. PCOOH reduction activity was found to be undetectable in fraction 5, whereas fraction 25 exhibited substantial activity (∼0.1 U/ml), consistent with GPx1 being present in the early peak fractions and GPx4 in the late peak fractions. Accordingly, fractions 24-27 were pooled, and material precipitated at 95% (NH4)2SO4 saturation was collected, dissolved in KME Buffer C, and subjected to Sephadex G-50 size-exclusion chromatography. As shown in Fig. 2, a major protein peak (A280) centered at fraction 14 was observed. This was well separated from a broad peak of CuOOH-based enzymatic activity, which maximized near fraction 23. There was relatively little A280 signal in this region, suggesting a large increase in specific enzymatic activity at this point. It is important to note that the presence of 0.1 M KSCN in KME Buffer C substantially reduced the amount of peroxidatic activity that emerged more rapidly with most of the protein (fractions 13-15, Fig. 2). Employed previously in GPx4 isolation [17], KSCN, appears to have disrupted the interaction of GPx4 with higher molecular weight proteins, presumably by acting as a chaotropic agent. Fractions 20-28 (Fig. 2) were pooled, dialyzed against KME Buffer D, concentrated to 0.5 mL using a 10 kDa cutoff spin-filter, and loaded onto a strongly cationic HiTrap SP column. After washing with 5 column volumes of KME Buffer D, elution was carried out with KME Buffer E, A280 being monitored. As shown in Fig. 3, a major protein peak centered at fraction 12 was observed. Measurement of CuOOH-based activity (Fig. 3) and PCOOH-based activity (not shown) also revealed a major peak at fraction 12, consistent with highly purified GPx4 protein. No significant activity against PCOOH was observed in fractions from the initial column wash (not shown), indicating that essentially all of the active GPx4 (see below) was in fractions 10-13. These fractions were pooled, dialyzed against 25 mM Tris-HCl/100 mM KSCN/5 mM 2-ME/50 μM EDTA/50 μM DFO (pH 7.7), and stored at −20 °C in this buffer supplemented with 30% (v/v) glycerol. DFO was included as an added precaution against enzyme inactivation by redox cycling of any trace iron [28]. SDS-PAGE electropherograms showing degree of GPx4 enrichment at the different stages of purification are shown in Fig. 4. As can be seen, pooled protein material recovered by gradient elution from the HiTrap SP column (Fig. 3) migrated as a single major band of Mr ∼20 kDa on SDS-PAGE with Coomassie Blue staining, consistent with it being GPx4 (Fig. 4, lane 6). This identity was confirmed by Western blot analysis using an anti-human GPx4 antibody (Fig. 4, lane 7). A densitometric trace across lane 6 indicated a purity level of at least 95% for this GPx4 preparation. It is important to note that achieving this level required that size-exclusion chromatography be carried out before HiTrap SP and not the other way around, which left more protein contaminants. Table 1 shows PCOOH-based GPx4 enzymatic activity determined at the different stages of purification. Clearly, the largest increase in specific activity occurred in the G-50 size-exclusion step, it resulting in a 55-fold increase relative to the DEAE-Sepharose step, which gave only a 5-fold increase. The final HiTrap SP step produced an additional 1.2 fold increase, i.e. ∼420-fold overall, giving GPx4 with a specific activity of ∼22 U/mg protein. (In the coupled assay mixture used for this determination, 57 ng of enzyme was used.) The given activity remained constant for at least three months when the enzyme was stored under the conditions described (see above).
Figure 1.
DEAE-Sepharose column chromatography: protein concentration and peroxidase activity profiles. KME Buffer A was pumped through the column at 1.0 mL/min, with 10-mL fractions being collected. The linear elution gradient (0-100% KME Buffer B) was started at fraction 16 and continued to fraction 32, 100% Buffer B being used thereafter. Protein level was measured by absorbance at 280 nm (solid line). Enzymatic activity was determined by coupled spectrophotometric assay, using CuOOH (○) or PCOOH (▲) as the peroxide substrate. One unit (U) of activity corresponds to 1 μmol of CuOOH or PCOOH consumed per min. Bar indicates fractions that were pooled for further purification.
Figure 2.
Sephadex G-50 column chromatography: protein concentration and peroxidase activity profiles. The column bed volume was 100 mL and void volume ∼34 mL. Eluting KME Buffer C flowed through the column at 0.8 mL/min, with 3.2 mL fractions being collected. Protein level (A280) is represented by the solid line and CuOOH-based enzymatic activity by circles. Bar indicates fractions that were pooled for further purification
Figure 3.
Protein profile for HiTrap SP column chromatography. KME Buffer D was pumped through the column up to 3 mL of effluent (not shown), after which a linear gradient (0-100% KME Buffer E) was started, reaching 100% Buffer E at 15 mL. This was maintained for another 5 mL before switching back to 100% Buffer D. Fractions of 1.0 mL were collected throughout at a flow rate of 1.0 mL/min. Solid line represents protein concentration (A280); circles represent CuOOH-based enzymatic activity. Bar indicates fractions that were pooled for purity and activity determinations.
Figure 4.
SDS-PAGE-assessed total protein and immunodetectable protein at various stages of GPx4 purification. Proteins were dissolved in NuPAGE LDS sample buffer under reducing conditions and heated to 70 °C before loading onto a NuPAGE 4-12% acrylamide/bis-acrylamide gel. Protein bands (lanes 1-6) were visualized by Coomassie Blue staining or by immunoblotting (lane 7). Sample lanes with amounts of applied protein are as follows: (1) crude tissue lysate (20 μg); (2) supernatant from 50% saturated (NH4)2SO4 (20 μg); (3) dialyzed precipitate from 70% saturated (NH4)2SO4 (20 μg); (4) active fraction pool from DEAE-Sepharose column (5 μg); (5) active pool from Sephadex G-50 column (0.4 μg); (6) active pool from HiTrap SP column (0.2 μg); (7) anti-GPx4 immunoblot of active pool from HiTrap SP column; (8) protein molecular mass standards.
Table 1.
Enzymatic activity at different stages of GPx4 purificationa
| Step | Total protein (mg) | Unitsb | Activity recovered (%) | Specific activity (U/mg) | Fold purification |
|---|---|---|---|---|---|
| Tissue lysate | 458 | 23.2 | 100 | 0.051 | 1.0 |
| 50% saturated (NH4)2SO4 supernatant | 133 | 5.73 | 22 | 0.043 | 0.8 |
| Dialyzed 70% saturated (NH4)2SO4 pellet | 54.0 | 3.43 | 15 | 0.064 | 1.3 |
| DEAE-Sepharose | 14.7 | 4.72 | 20 | 0.321 | 6.3 |
| Sephadex G-50 | 0.071 | 1.25 | 5 | 17.6 | 345 |
| HiTrap SP | 0.013 | 0.279 | 1 | 21.5 | 422 |
This preparation was from 10 g of rat testes
GPx4 activity determined by coupled enzymatic assay using PCOOH as peroxide substrate One unit represents one micromole of PCOOH reduced per min at 37 °C.
The GPx4 purification tracked in Table 1 is representative of four separate preparations in which final protein varied between 10 and 50 μg. Final enzyme yield (∼13 μg, 0.28 units) would be sufficient for more than 200 coupled enzymatic assays such as described in Table 1. For kinetic experiments, therefore, such levels would be quite adequate. We used a relatively modest amount of testis (10 g) for the purification described. If higher yields are needed, one could easily start with more tissue and scale-up the capacities of the ion exchange and size exclusion columns.
Roveri et al. [18] showed that rat testis GPx4 exists in at least two forms, a soluble cytosolic fraction and a CHAPS-liberated membrane-bound fraction, the bulk of which is in mitochondria. Units of enzymatic activity were found to be at least 3-times higher in mitochondria than in cytosol, but the two localized forms of the enzyme showed no significant differences in size, sequence, or substrate specificity. However, minor post-translational modifications and differences thereof between the two GPx4 forms were apparent [18]. Isolation of mitochondrial GPx4 required solubilization with an appropriate detergent such as CHAPS [18]. Since CHAPS was used in our procedure, the GPx4 that we isolated was probably a mixture of the mitochondrial and cytosolic forms.
Direct determination of GPx4 activity using 7α-OOH as a substrate
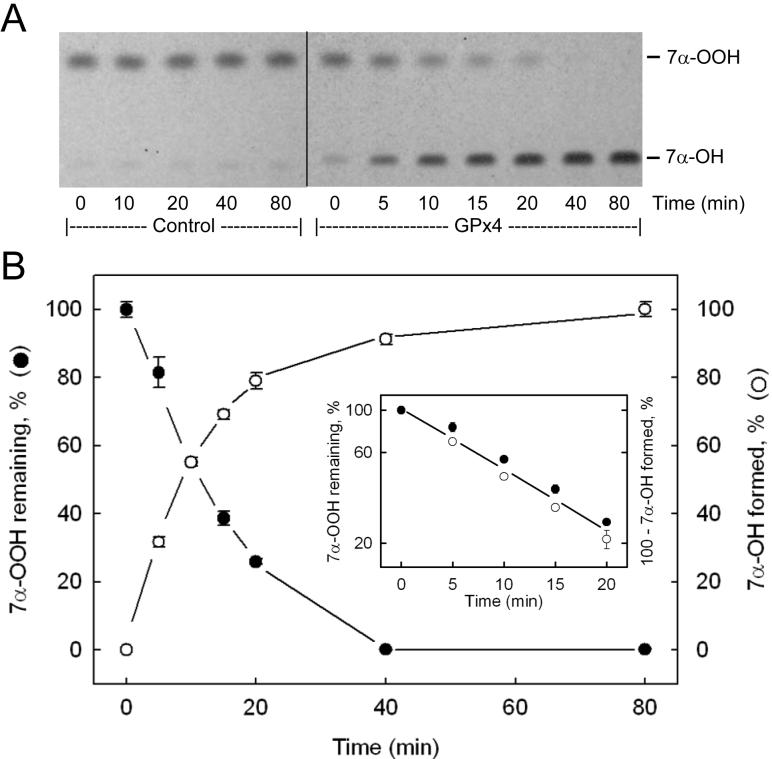
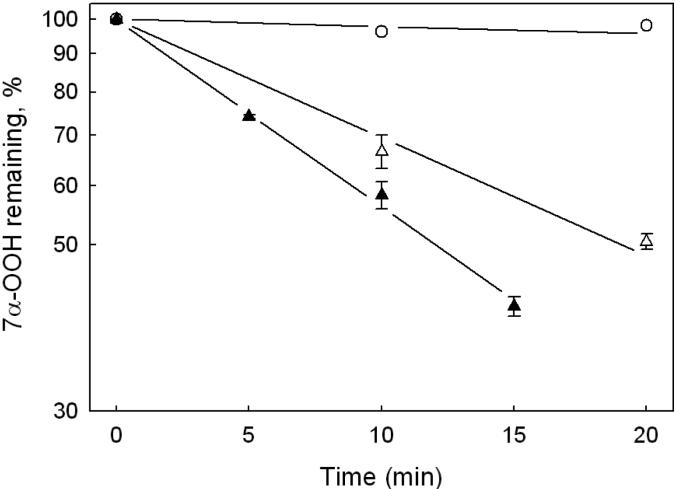
Activity of purified GPx4 was also assessed by a recently developed HPTLC assay, using TMPD as a redox-active spray indicator [20,24]. In addition to being more sensitive than the commonly used coupled enzymatic assay, the HPTLC approach detects peroxidase activity directly. Indirect measurement via coupled NADPH oxidation is potentially less accurate and open to more interferences, particularly when cell and tissue extracts are examined [24]. When reduction of a cholesterol-derived hydroperoxide such as 7α-OOH is monitored by the HPTLC method, it is possible to track on a single plate not only loss of the hydroperoxide, but also formation of the diol product, 7α-OH, since these species are well separated from one another [24]. Previous evidence indicated that cholesterol hydroperoxides, like phospholipid counterparts, are substrates for GPx4, but not GPx1 or other GPx isotypes [10]. Thus, another advantage of using 7α-OOH for determining GPx4 activity is that it appears to be a unique substrate for this enzyme, at least in the selenium-dependent class of peroxidases. Figure 5A shows an HPTLC chromatogram of lipid extracts recovered at various times during incubation of the highly purified (post-HiTrap SP) GPx4 represented in Table 1 with GSH and 7α-OOH. A control in which GPx4 was omitted from the incubation mixture is also represented. The plate in this case was sprayed first with TMPD, which converted the more rapidly migrating 7α-OOH to 7α-OH [24]. After subsequent spraying with 18 N H2SO4 and warming, two bright blue bands appeared, the upper representing residual 7α-OOH (now 7α-OH) and the lower, 7α-OH generated by GPx4 catalysis. Sample loads were restricted to the the linear dynamic range of the 7α-OH response. As shown in Fig. 5A, the GPx4-containing system exhibited a progressive loss of 7α-OOH and reciprocal buildup of 7α-OH over an 80-min incubation period, whereas the control showed no reaction. Figure 5B shows that 7α-OOH decay and 7α-OH accumulation was exponential in this system, a semilogarithmic plot of the data revealing apparent first-order kinetics with nearly identical rate constants for the two processes (Fig. 5B inset). From the rate constants and initial 7α-OOH concentration, we calculated initial rates, and from these, specific activities. The first-order plots in Fig. 6 show that the rate of 7α-OOH was proportional to GPx4 concentration, as expected. The average specific activity determined from these data (0.84 ± 0.21 U/mg protein) agrees closely with that obtained from the Fig. 5 data. We also used PCOOH for determining GPx4 activity by HPTLC, the value in this case being similar to that determined by coupled assay, i.e. ∼20 U/mg (Table 1). However, since the alcohol product, PCOH, is not resolved from PCOOH substrate on HPTLC [24], only decay of the latter could be tracked, in contrast to 7α-OOH (Fig. 5A). Previous studies showed that phospholipid hydroperoxides are much better substrates for GPx4 than cholesterol hydroperoxides [10,11]. The ∼25-fold greater final specific activity obtained with PCOOH (Table 1) versus 7α-OOH (Figs. 5 and 6) can be explained on this basis.
Figure 5.
HPTLC-assessed GPx4 activity based on 7α-OOH reduction. Each 0.41 mL reaction mixture contained 3 mM GSH, 1 mM EDTA, 0.1 mM DFO, 0.1% Triton X-100, 25 μM 7α-OOH, and GPx4 (2.1 μg/mL) in PBS at 37 °C. At the indicated time points, samples were extracted and recovered lipid fractions were analyzed by HPTLC, using a TMPD spray followed by H2SO4 for analyte detection. (A) HPTLC profile showing changes in 7α-OOH and 7α-OH levels as a function of incubation time. Profile for a control mixture containing everything except GPx4 is also shown. (B) Plot of time-dependent changes in 7α-OOH (●) and 7α-OH (○) level, as determined by densitometry. The inset shows first-order plots for 7α-OOH loss (●) and 7α-OH accumulation (○), the specific enzymatic activity determined from the former being 0.84 ± 0.01 U/mg, and from the latter, 0.80 ± 0.05 U/mg. Values are means ± deviations from duplicate determinations.
Figure 6.
Kinetics of GPx4-catalyzed 7α-OOH reduction. Each 0.5 mL reaction mixture contained 3 mM GSH, 1 mM EDTA, 0.1 mM DFO, 0.1% Triton X-100, 25 μM 7α-OOH, and GPx4 in PBS at 37 °C. GPx4 concentration was either 0.9 μg/mL (△) or 1.8 μg/mL (▲). First-order plots of 7α-OOH decay as a function of incubation time are shown. Data from a control lacking GPx4 are plotted alongside (○). Values are means from duplicate experiments.
Summary
We describe a new procedure for purifying natural GPx4 to apparent homogeneity. The strategy used is relatively straightforward and does not require an affinity resin such as BSP-GS-agarose, which is no longer commercially available. Even if available, this resin could be potentially problematic because BSP is an avid GPx4 inhibitor [18] and any leaching of it during an affinity run would be detrimental to high activity yield. There is a sustained interest in GPx4 because of its many different biological effects, ranging from protection against damaging peroxidative stress and regulation of cyclooxygenase/lipoxygenase activities [2,3,6,7] to an integral structural role in spermatogenesis [25,26]. The isolation procedure we have developed should prove valuable in basic studies aimed at better characterizing GPx4's structural, kinetic, and regulatory properties.
Acknowledgements
This work was supported by USPHS Grant CA72630 from the National Cancer Institute. The assistance of Vlad Levchenko and Gabriella Papale in the early stages of this work is greatly appreciated. We also thank Antonella Roveri in Fulvio Ursini's laboratory for helpful suggestions regarding GPx4 isolation.
Abbreviations used
- CHAPS
3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate
- DFO
desferrioxamine
- GPx1
glutathione peroxidase isotype-1
- GPx4
glutathione peroxidase isotype-4
- HPTLC-TPD
high-performance thin layer chromatography with N,N,N′,N′-tetramethyl-p-phenylene-diamine detection
- LOOH(s)
lipid hydroperoxide(s)
- 2-ME
2-mercaptoethanol
- PCOOH
phosphatidylcholine hydroperoxide
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine
- SDS-PAGE
sodium dodecyl sulfate-polyacrylamide gel electrophoresis
- TMPD
N,N,N′,N′-tetramethyl-p-phenylene-diamine
- 7α-OOH
3β-hydroxycholest-5-ene-7α-hydroperoxide
- 7α-OH
cholest-5-ene-3β, 7α-diol
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sies H. Biochemistry of oxidative stress. Angew. Chem. Int. Ed. Engl. 1986;25:1058–1071. [Google Scholar]
- 2.Girotti AW. Lipid hydroperoxide generation, turnover, and effector action in biological systems. J. Lipid Res. 1998;39:1529–1542. [PubMed] [Google Scholar]
- 3.Vanda Papp L, Lu J, Holmgren A, Khanna KK. From selenium to selenoproteins: synthesis, identity, and their role in human health. Antioxid. Redox. Signal. 2007;9:775–806. doi: 10.1089/ars.2007.1528. [DOI] [PubMed] [Google Scholar]
- 4.Flohe L, Budde H, Hofmann B. Peroxiredoxins in antioxidant defense and redox regulation. BioFactors. 2003;19:3–10. doi: 10.1002/biof.5520190102. [DOI] [PubMed] [Google Scholar]
- 5.Ursini F, Bindoli A. The role of selenium peroxidases in the protection against oxidative damage of membranes. Chem. Phys. Lipids. 1987;44:255–276. doi: 10.1016/0009-3084(87)90053-3. [DOI] [PubMed] [Google Scholar]
- 6.Brigelius-Flohe R. Tissue-specific function of individual glutathione peroxidases. Free Radic. Biol. Med. 1999;27:951–965. doi: 10.1016/s0891-5849(99)00173-2. [DOI] [PubMed] [Google Scholar]
- 7.Kuhn H, Borchert A. Regulation of enzymatic lipid peroxidation: the interplay of peroxidizing and peroxide reducing enzymes. Free Radic. Biol. Med. 2002;33:154–172. doi: 10.1016/s0891-5849(02)00855-9. [DOI] [PubMed] [Google Scholar]
- 8.Ursini F, Maiorino M, Sevanian A. Membrane hydroperoxides. In: Sies H, editor. Oxidative stress: oxidants and antioxidants. Academic Press; New York: 1991. pp. 319–336. [Google Scholar]
- 9.Van Kuijk FJGM, Sevanian A, Handelman GJ, Dratz EA. A new role for phospholipase A2: protection of membranes from lipid peroxidation damage. Trends Biochem. Sci. 1987;12:31–34. [Google Scholar]
- 10.Thomas JP, Mairoino M, Ursini F, Girotti AW. Protective action of phospholipid hydroperoxide glutathione peroxidase against membrane-damaging lipid peroxidation. J. Biol. Chem. 1990;265:454–461. [PubMed] [Google Scholar]
- 11.Thomas JP, Geiger PG, Maiorino M, Ursini F, Girotti AW. Enzymatic reduction of phospholipid and cholesterol hydroperoxides in artificial bilayers and lipoproteins. Biochim. Biophys. Acta. 1990;1045:252–260. doi: 10.1016/0005-2760(90)90128-k. [DOI] [PubMed] [Google Scholar]
- 12.Yant LJ, Ran Q, Rao L, Van Remmen H, Shibatani T, Belter JG, Motta L, Richardson A, Prolla TA. Free Radic. Biol. Med. 2003;34:496–502. doi: 10.1016/s0891-5849(02)01360-6. [DOI] [PubMed] [Google Scholar]
- 13.Ran Q, Van Remmen H, Gu M, Qi W, Roberts LJ, II, Prolla T, Richardson A. Embryonic fibroblasts from Gpx4+/− mice: a novel model for studying the role of membrane peroxidation in biological processes. Free Radic. Biol. Med. 2003;35:1101–1109. doi: 10.1016/s0891-5849(03)00466-0. [DOI] [PubMed] [Google Scholar]
- 14.Ran Q, Liang H, Gu M, Qi W, Walter CA, Roberts LJ, II, Herman B, Richardson A, Van Remmen H. Transgenic mice overexpressing glutathione peroxidase 4 are protected against oxidative stress-induced apoptosis. J. Biol. Chem. 2004;279:55137–55146. doi: 10.1074/jbc.M410387200. [DOI] [PubMed] [Google Scholar]
- 15.Ran Q, Gu M, Van Remmen H, Strong R, Roberts JL, Richardson A. Glutathione peroxidase 4 protects cortical neurons from oxidative injury and amyloid toxicity. J. Neurosci. Res. 2006;84:202–208. doi: 10.1002/jnr.20868. [DOI] [PubMed] [Google Scholar]
- 16.Su D, Li Y, Gladyshev WN. Selenocysteine insertion directed by the 3′-UTR SECIS element in Escherichia coli. Nucleic Acids Res. 2005;33:2486–2492. doi: 10.1093/nar/gki547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ursini F, Maiorino M, Valente M, Ferri L, Gregolin C. Purification from pig liver of a protein which protects liposomes and biomembranes from peroxidative degradation and exhibits glutathione peroxidase activity on phosphatidylcholine hydroperoxides. Biochim. Biophys. Acta. 1982;710:197–212. doi: 10.1016/0005-2760(82)90150-3. [DOI] [PubMed] [Google Scholar]
- 18.Roveri A, Maiorino M, Nisii C, Ursini F. Purification and characterization of phospholipid hydroperoxide glutathione peroxidase from rat testis mitochondrial membranes. Biochim. Biophys. Acta. 1994;1208:211–221. doi: 10.1016/0167-4838(94)90106-6. [DOI] [PubMed] [Google Scholar]
- 19.Whelan G, Hoch J, Combes B. A direct assessment of the importance of conjugation for biliary transport of sulfobromophthalein sodium. J. Lab. Clin. Med. 1970;75:542–557. [PubMed] [Google Scholar]
- 20.Kriska T, Girotti AW. Separation and quantitation of peroxidized phospholipids using high-performance thin-layer chromatography with tetramethyl-p-phenylenediamine detection. Anal. Biochem. 2004;327:97–106. doi: 10.1016/j.ab.2003.12.021. [DOI] [PubMed] [Google Scholar]
- 21.Kriska T, Marathe GK, Schmidt JC, McIntyre TM, Girotti AW. Phospholipase action of platelet-activating factor acetylhydrolase, but not paraoxonase-1, on long fatty acyl chain phospholipid hydroperoxides. J. Biol. Chem. 2007;282:100–108. doi: 10.1074/jbc.M608135200. [DOI] [PubMed] [Google Scholar]
- 22.Girotti AW, Korytowski W. Cholesterol as a singlet oxygen detector in biological systems. Methods Enzymol. 2000;319:85–100. doi: 10.1016/s0076-6879(00)19011-1. [DOI] [PubMed] [Google Scholar]
- 23.Hurst R, Korytowski W, Kriska T, Esworthy RS, Chu F-F, Girotti AW. Hyperresistance to cholesterol hydroperoxide-induced peroxidative injury and apoptotic death in a tumor cell line that overexpresses glutathione peroxidase isotype-4. Free Radic. Biol. Med. 2001;31:1051–1065. doi: 10.1016/s0891-5849(01)00685-2. [DOI] [PubMed] [Google Scholar]
- 24.Kriska T, Girotti AW. Thin layer chromatographic method for determining the enzymatic activity of peroxidases catalyzing the two-electron reduction of lipid hydroperoxides. J. Chromatogr. B. 2005;827:58–64. doi: 10.1016/j.jchromb.2005.03.045. [DOI] [PubMed] [Google Scholar]
- 25.Ursini F, Heim S, Kiess M, Maiorino M, Roveri A, Wissing J, Flohe L. Dual function of the selenoprotein PHGPx during sperm maturation. Science. 1999;285:1393–1396. doi: 10.1126/science.285.5432.1393. [DOI] [PubMed] [Google Scholar]
- 26.Conrad M, Moreno SG, Sinowatz F, Ursini F, Kolle S, Roveri A, Brielmeier M, Wurtz W, Maiorino M, Bornkamm GW. The nuclear form of phospholipid hydroperoxide glutathione peroxidase is a protein thiol peroxidase contributing to sperm chromatin stability. Mol. Cell. Biol. 2005;25:7637–7644. doi: 10.1128/MCB.25.17.7637-7644.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schnurr K, Borchert A, Gerth C, Anton M, Kuhn H. Bacterial and nonbacterial expression of wild-type and mutant human phospholipid hydroperoxide glutathione peroxidase and purification of the mutant enzyme in the milligram scale. Protein Express. Purif. 2000;19:403–410. doi: 10.1006/prep.2000.1262. [DOI] [PubMed] [Google Scholar]