SUMMARY
The apolipoprotein E (APOE) ε4 allele is the best established genetic risk factor for late-onset Alzheimer’s disease (LOAD). We conducted genome-wide surveys of 502,627 single-nucleotide polymorphisms (SNPs) to characterize and confirm other LOAD susceptibility genes. In ε4 carriers from neuropathologically verified discovery, neuropathologically verified replication, and clinically characterized replication cohorts of 1411 cases and controls, LOAD was associated with six SNPs from the GRB-associated binding protein 2 (GAB2) gene and a common haplotype encompassing the entire GAB2 gene. SNP rs2373115 (p = 9 × 10−11) was associated with an odds ratio of 4.06 (confidence interval 2.81–14.69), which interacts with APOE ε4 to further modify risk. GAB2 was overexpressed in pathologically vulnerable neurons; the Gab2 protein was detected in neurons, tangle-bearing neurons, and dystrophic neuritis; and interference with GAB2 gene expression increased tau phosphorylation. Our findings suggest that GAB2 modifies LOAD risk in APOE ε4 carriers and influences Alzheimer’s neuropathology.
INTRODUCTION
Alzheimer’s disease (AD) afflicts about 10% of persons over 65 and almost half of those over 85 (Evans et al., 1989) and the number of afflicted persons continues to grow. To date, researchers have firmly established associations between four genes and AD risk. Whereas more than 150 mutations of the presenilin 1 (PS1), presenilin 2 (PS2), and amyloid precursor protein (APP) genes account for many early-onset AD cases with autosomal dominant inheritance, the apolipoprotein E (APOE) ε4 allele accounts for many cases of late-onset AD (LOAD), with dementia onset after age 60 (Papassotiropoulos et al., 2006; Corder et al., 1993, 1994; Farrer et al., 1997). The APOE gene has three common variants, ε2, ε3, and ε4. The ε2 allele is associated with the lowest LOAD risk, while each copy of the ε4 allele in a person’s APOE genotype is associated with a higher LOAD risk and a younger median age at dementia onset (Corder et al., 1994; Farrer et al., 1997). Although twin studies suggest that there are several susceptibility genes which, along with the APOE ε4 allele, contribute to up to 80% of LOAD cases (Gatz et al., 2006), discovery of other susceptibility genes has been elusive (Bertram et al., 2007).
To identify susceptibility genes for common and genetically complex disorders like LOAD, it has been proposed that it would help to conduct genome-wide surveys of at least 300,000 single-nucleotide polymorphisms (SNPs) in unrelated cases and controls, compare the most homogeneous samples, and consider interactions between major and minor genes (Papassotiropoulos et al., 2006; Kruglyak, 1999; Coon et al., 2007). We individually genotyped 502,627 SNPs to characterize and confirm LOAD susceptibility genes in three separate cohorts of LOAD cases and controls, including a discovery cohort of clinically and neuropathologically characterized brain donors, a replication cohort of similarly characterized brain donors, and a replication cohort of clinically characterized living subjects. The brain donor cohorts were selected to exclude clinically misdiagnosed cases and cognitively normal but neuropathologically affected elderly controls; the clinical cohort was selected to confirm genetic associations independent of any brain donor selection bias. Within each cohort, LOAD cases and controls were stratified into subgroups of APOE ε4 carriers and noncarriers, permitting us to investigate genes that modify LOAD risk in the ε4 carriers and genes that might otherwise be masked by disproportionately large ε4 effects.
RESULTS AND DISCUSSION
We recently demonstrated the feasibility of high-density genome-wide association studies in our neuropathologically characterized cases and controls, providing empirical support for the suggestion that the APOE locus is unparalleled in its contribution to LOAD risk (Coon et al., 2007). With the exception of an SNP only 14 kb pairs distal to and in linkage disequilibrium (LD) with the APOE ε4 variant on chromosome 19, no other SNP distinguished LOAD cases from controls after Bonferroni correction for multiple comparisons (Figure S1A at http://www.tgen.org/neurogenomics/data). For the previously noted reasons, we divided each cohort into two subgroups: allelic APOE ε4 carriers (Figure S1B) and APOE ε4 noncarriers (Figure S1C). We now report associations between a common gene and LOAD in APOE ε4 carriers in our three cohorts; we show that the implicated gene is associated with AD neuropathology in neuronal microarray and immunohistochemical studies; and we consider a possible mechanism by which GAB2 modifies AD risk in a small-interfering RNA (siRNA) study. Finally, we deposit all of the data into the public domain for use by the community (http://www.tgen.org/neurogenomics/data).
High-Density Genome-Wide Association Studies
Genome-wide genotyping was performed on each individual sample from a “neuropathological discovery cohort” of 736 brain donors, a “neuropathological replication cohort” of 311 brain donors, and an additional “clinical replication cohort” of 364 living subjects who were at least 65 years old at the time of their death or last clinical assessment and who were independently assessed for their APOE genotype. For the two neuropathological cohorts, brain tissue for DNA extraction, neuropathological diagnoses, and data were supplied by investigators from 20 of the National Institute on Aging (NIA)-sponsored Alzheimer’s Disease Centers (ADCs) (in accordance with agreements with the NIA, the ADCs, and the National Alzheimer’s Coordinating Center) and from the Netherlands Brain Bank. For the hypothesis-testing clinical replication cohort, DNA extracted from blood, clinical diagnoses, and data from subjects assessed in Rochester, MN were supplied by investigators from the Mayo Clinic.
The neuropathological discovery cohort included 446 LOAD cases (299 ε4 carriers and 147 ε4 noncarriers) and 290 controls (61 ε4 carriers and 229 ε4 noncarriers); the neuropathological replication cohort included 197 LOAD cases (113 ε4 carriers and 84 ε4 noncarriers) and 114 controls (27 ε4 carriers and 87 ε4 noncarriers); and the clinical replication cohort included 218 LOAD cases (115 ε4 carriers and 103 ε4 noncarriers) and 146 controls (29 ε4 carriers and 117 ε4 noncarriers). Brain donor cases satisfied clinical and neuropathological criteria for LOAD, and were age 73.5 ± 6.2 at death. Brain donor controls did not have significant cognitive impairment or significant neuropathological features of AD, and were age 75.8 ± 7.5 at death. Clinical cases satisfied criteria for probable AD, and were age 78.9 ± 7.8 at last clinical visit. Clinical controls did not have clinically significant cognitive impairment and were age 81.7 ± 6.6 at last clinical assessment.
We initially surveyed SNPs in the neuropathological discovery cohort to explore LOAD associations in the ε4 carrier and noncarrier subgroups. Within the discovery subgroup of APOE ε4 carriers, 10 of the 25 SNPs with the most significant LOAD-association significance levels (contingency test p = 9 × 10−5 to 1 × 10−7; uncorrected for multiple comparisons) were located in the GRB-associated binding protein 2 (GAB2) gene on chromosome 11q14.1 (Table 1). LOAD associations in six of these SNPs were then confirmed in both the neuropathological replication and clinical replication cohorts (Table 1). These ten SNPs were not significantly associated with LOAD in the APOE ε4 noncarrier group (contingency test p = 0.08 to 0.97) (Table S1A). Combining data from all 644 APOE ε4-carrying cases and controls, we found highly significant associations between LOAD and all ten GAB2 SNPs (contingency test p = 1.19 × 10−5 to 9.66 × 10−11), with five of the six consistently implicated alleles surviving the highly conservative Bonferroni correction for 312,316 independent comparisons (p = 1.55 × 10−7) (Table 1). When data from the ε4 carriers and noncarriers were analyzed together, as in our previous report (Coon et al., 2007), the ten GAB2 SNPs were still associated with LOAD (contingency test p = 0.013 to 2.7 × 10−6, Table S1B), but these associations did not survive Bonferroni correction.
Table 1.
GAB2 SNPs Implicated in the Neuropathological Discovery, Neuropathological Replication, and Clinical Replication Cohorts
| Neuropathology Discovery Cohort
|
Neuropathology Replication Cohort
|
Clinical Replication Cohort
|
Overall
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| dbsnp RS ID |
Position | Risk Allele |
Freq. Cases |
Freq. Controls |
p Valuea | OR (95% CI) |
Freq. Cases |
Freq. Controls |
p Valuea |
OR (95% CI) | Freq. Cases |
Freq. Controls |
p Valuea |
OR (95% CI) |
p Valuea |
OR (95% CI) |
| rs901104 | 77608147 | C | 0.89 | 0.72 | 4.56E-07 | 3.24 (2.01–5.20) |
0.87 | 0.73 | 4.52E-02 | 2.54 (0.90–7.17) |
0.89 | 0.80 | 1.47E-01 | 2.08 (0.76–5.88) |
1.99E-06 | 2.87 (1.84–4.50) |
| rs1385600 | 77613814 | T | 0.89 | 0.71 | 5.02E-07 | 3.18 (1.99–5.08) |
0.88 | 0.70 | 1.32E-02 | 3.00 (1.09–8.24) |
0.95 | 0.78 | 4.63E-02 | 5.56 (0.89–33.33) |
2.81E-09 | 3.65 (2.34–5.71) |
| rs1007837 | 77618724 | C | 0.89 | 0.73 | 4.78E-06 | 3.18 (1.99–5.08) |
0.89 | 0.73 | 3.20E-02 | 2.85 (1.00–8.11) |
0.88 | 0.80 | 2.04E-01 | 1.89 (0.70–5.00) |
3.97E-07 | 3.01 (1.94–4.68) |
| rs2510038 | 77643682 | C | 0.90 | 0.74 | 8.92E-05 | 2.44 (1.37–4.36) |
0.88 | 0.75 | 4.35E-02 | 2.53 (0.88–7.32) |
0.89 | 0.80 | 1.47E-01 | 2.08 (0.76–5.88) |
1.19E-05 | 2.72 (1.72–4.31) |
| rs4945261 | 77667908 | G | 0.89 | 0.72 | 5.66E-07 | 3.21 (1.99–5.15) |
0.88 | 0.73 | 4.20E-02 | 2.54 (0.90–7.17) |
0.95 | 0.78 | 4.63E-02 | 5.56 (0.89–33.33) |
3.06E-08 | 3.44 (2.18–5.43) |
| rs7101429 | 77670615 | A | 0.89 | 0.73 | 4.62E-05 | 2.13 (1.21–3.75) |
0.89 | 0.73 | 1.87E-02 | 2.84 (1.00–8.11) |
0.88 | 0.75 | 4.99E-02 | 2.50 (0.98–6.25) |
1.06E-06 | 2.96 (1.89–4.63) |
| rs10793294 | 77674051 | C | 0.82 | 0.66 | 2.98E-05 | 2.45 (1.60–3.77) |
0.80 | 0.59 | 1.34E-02 | 2.66 (1.06–6.57) |
0.88 | 0.61 | 2.17E-02 | 4.45 (1.18–16.95) |
1.59E-07 | 2.83 (1.90–4.21) |
| rs4291702 | 77678896 | C | 0.88 | 0.70 | 3.73E-06 | 3.19 (2.00–5.07) |
0.87 | 0.72 | 2.44E-02 | 2.70 (0.97–7.52) |
0.88 | 0.80 | 2.04E-01 | 1.89 (0.70–5.00) |
5.88E-07 | 2.96 (1.91–4.59) |
| rs7115850 | 77722719 | C | 0.86 | 0.67 | 1.60E-07 | 3.21 (2.04–5.05) |
0.86 | 0.72 | 3.91E-02 | 2.48 (0.90–6.81) |
0.98 | 0.72 | 3.45E-03 | 14.93 (1.61–140.85) |
2.80E-10 | 3.92 (2.51–6.11) |
| rs2373115 | 77768798 | G | 0.88 | 0.70 | 4.60E-07 | 3.21 (2.04–5.05) |
0.86 | 0.72 | 3.91E-02 | 2.48 (0.90–6.81) |
0.98 | 0.72 | 3.45E-03 | 14.93 (1.61–140.85) |
9.66E-11 | 4.06 (2.81–14.69) |
p values were computed using χ2 tests. For the overall comparison, the Cochran-Mantel-Haenszel χ2 test was used. All p values are uncorrected.
The PLINK analysis toolset (http://pngu.mgh.harvard.edu/~purcell/plink/index.shtml) was used for whole-genome analysis. Haploview v3.32 was used to determine the LD structure of the chromosome 11q14.1 region surrounding GAB2 in each of the three APOE ε4-stratified cohorts (Figure 1). Three haplotype blocks are present in this region: one block upstream of GAB2, roughly corresponding to the ALG8 locus; one 189 kb-pair block encompassing most of the GAB2 locus; and one downstream block corresponding to the NARS2 locus. These blocks were consistent with the LD structure of the HapMap CEPH populations. The GAB2 gene is completely encompassed by a single haplotype block extending from rs901104 to rs2373115 (SNPs 5–22 in Figure 1), which has three major haplotypes: an extremely common “GAB2 risk haplotype,” a common “GAB2 protective haplotype,” and a relatively uncommon GAB2 haplotype unrelated to LOAD risk in APOE ε4 carriers (Figure 1). In all three cohorts, the GAB2 CT-AAG-CAGATCAGACG haplotype was associated with higher LOAD risk, the GAB2 TC-GCA-TGAGGTGTCTT haplotype was associated with a lower LOAD risk, and the CT-AAG-CAGAGCA GCCG was unrelated to LOAD risk in the APOE ε4 carriers (Figure 1).
Figure 1. Linkage Disequilibrium Structure and Haplotype Significance Levels for the Region Encompassing GAB2.
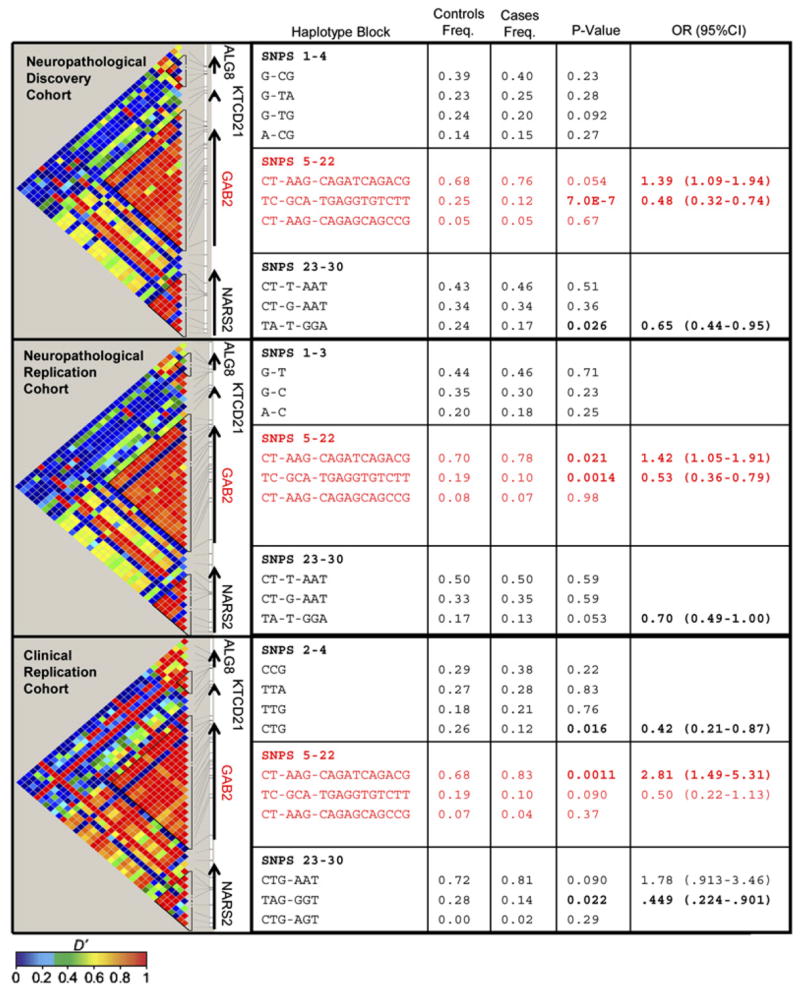
Plots follow the Haploview GOLD heatmap color scheme. SNP numbers correspond to the following dbSNP ID identification numbers: 1:rs579711, 2:rs977978, 3:rs637149, 4:rs977977, 5:rs901104, 6:rs1385600, 7:rs11237419, 8:rs1007837, 9:rs2450130, 10:rs2510054, 11:rs11237429, 12:rs2510038, 13:rs2511170, 14:rs4945261, 15:rs7101429, 16:rs10793294, 17:rs4291702, 18:rs11602622, 19:rs10899467, 20:rs2458640, 21:rs10793302, 22:rs2373115, 23:rs12280198, 24:rs12287010, 25:rs17136630, 26:rs4945276, 27:rs1996172, 28:rs7395344, 29:rs11237522, and 30:rs7950813. In all three cohorts, there was a GAB2 risk haplotype, a protective haplotype, and a haplotype unrelated to LOAD risk in APOE ε4 carriers.
Data from the 1411 subjects (including 644 APOE ε4 carriers and 767 noncarriers) in all three cohorts were combined to characterize odds ratios (ORs) and 95% confidence intervals (CIs) for rs2373115, the most significant SNP in our screen (Table 2). In ε4 carriers, LOAD cases had a risk genotype frequency of 0.88 and controls had a frequency of 0.71. In comparison with the other ε4 carriers, those with rs2373115 genotype GG had a significantly higher risk of LOAD (OR 2.36, 95% CI 1.55–3.58) than those with genotype GT and TT. In contrast, APOE ε4 noncarriers with rs2373115 genotype GG did not have a higher LOAD risk than the other ε4 noncarriers (OR 1.01, 95% CI 0.74–1.38).
Table 2.
GAB2 LOAD Odds Ratios in APOE ε4 Carriers and Noncarriers
| APOE ε4 group | APOE ε4 OR (95% CI) | rs2373115 genotype | Controls (n) | Cases (n) | % LOAD | rs2373115 OR (95% CI)a |
|---|---|---|---|---|---|---|
| APOE ε4− | GG | 314 | 232 | 42.4% | 1.12 (0.82–1.53) | |
| GT/TT | 121 | 100 | 45.2% | |||
| APOE ε4+ | GG | 63 | 406 | 86.6% | 2.88 (1.90–4.36) | |
| GT/TT | 54 | 121 | 69.1% | |||
| All samples | 6.07 (4.63–7.95) | GG | 377 | 638 | 62.9% | 1.34 (1.06–1.70) |
| GT/TT | 175 | 221 | 55.8% |
All rs2373115 ORs are calculated using GG versus GT/TT.
Whereas we confirmed a younger age at dementia onset in the APOE ε4 carriers than in noncarriers (age 75.5 ± 7.2 versus 77.8 ± 7.9, p = 2.4 × 10−4, two-tailed unpaired t test; unequal variance is statistical test used for all tests), there was no significant effect of rs2373115 genotype on age at dementia onset in either the ε4 carriers (t test, p = 0.32) or noncarriers (t test, p = 0.84).
Neuronal Microarray Studies
To provide converging evidence that GAB2 is biologically relevant to AD neuropathology, expression profiling using the Affymetrix Human Genome U133 Plus 2.0 array was used to characterize and compare GAB2 expression in laser-capture microdissected non-tangle-bearing neurons of cases and controls in six brain regions differentially affected by AD. LOAD cases had significantly greater neuronal GAB2 expression in the posterior cingulate cortex (9 cases, 13 controls, 4.50-fold change, t test, p = 0.00039) and hippocampus (9 cases, 13 controls, 2.94-fold change, t test, p = 0.00085), and no significant expression differences in the entorhinal cortex (10 cases, 13 controls, 1.20-fold change, t test, p = 0.46), middle temporal gyrus (13 cases, 12 controls, 1.44-fold change, t test, p = 0.14), superior frontal gyrus (22 cases, 11 controls, 1.25-fold change, t test, p = 0.47), or primary visual cortex (17 cases, 12 controls, 1.53-fold change, t test, p = 0.14).
The hippocampus is known to be especially vulnerable to AD-related neurofibrillary tangles (Braak and Braak, 1991), neuronal loss, and brain atrophy (Bobinski et al., 2000). It is preferentially involved in AD-related memory impairment (Jack et al., 1999) and is associated with the highest cerebral Gab2 expression in the rodent brain (Lein et al., 2007). The posterior cingulate cortex is known to be preferentially vulnerable to AD-related hypometabolic abnormalities and fibrillar amyloid deposition, and is also involved in AD-related memory impairment (Reiman et al., 1996, 2005; Johnson et al., 2006; Buckner et al., 2005; Mintun et al., 2006). While the entorhinal cortex and temporal and prefrontal regions are also affected by AD neuropathology, the visual cortex is relatively spared. Using a repeated measures analysis of variance to analyze neuronal GAB2 gene expression data from the same eight LOAD cases and ten controls, there was a significant group-by-region interaction (p = 0.011), with LOADrelated increases in neuronal GAB2 gene expression that were greater in the posterior cingulate cortex and hippocampus than in the visual cortex.
Tau Phosphorylation siRNA Study
In addition to its other properties, GAB2 is the principal activator of the phosphatidylinositol 3-kinase (PI3K) signaling pathway (Pratt et al., 2001). PI3K activates Akt, which in turn promotes glycogen synthase kinase-3 (Gsk3) phosphorylation/inactivation. This mechanism suppresses Gsk3-dependent phosphorylation of tau at AD-related hyperphorylated tau residues, the principal component of neurofibrillary tangles, and prevents apoptosis of confluent cells (Baki et al., 2004; Kang et al., 2005). Based on these findings, we hypothesized that Gab2 might function to protect cells from neuronal tangle formation and cell death and that a loss-of-function GAB2 haplotype would diminish such protection. We thus postulated that interference with GAB2 expression using siRNA treatment would increase tau phosphorylation at the serine-262 residue known to be hyperphosphorylated in AD. As shown in Figure 2, GAB2 siRNA treatment was associated with a 1.70-fold increase in serine-262 phosphorylated tau. This increase was not attributable to a concomitant increase in total tau levels. Additional siRNA and protein validation studies are now being performed to determine the extent to which GAB2 affects tau phosphorylation at additional relevant epitopes.
Figure 2. siRNA Knockdown of GAB2 Increases Tau Phosphorylation.
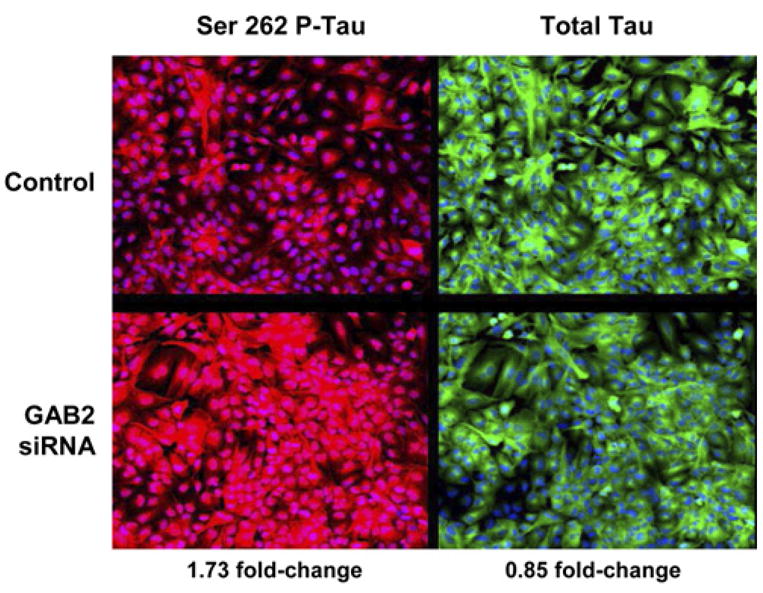
In comparison with vehicle treatment (red, upper left), GAB2 siRNA treatment resulted in a 1.70-fold increase in tau phosphorylation at the serene-262 residue (red, lower left), which is phosphorylated in LOAD neurofibrillary tangle-bearing neurons. This fold-change was not attributable to an increase in total tau (green, upper and lower right).
Immunohistochemical Validation
Gab2 immunohistochemistry was assessed in hippocampus and posterior cingulate cortex in LOAD cases. In hippocampus, Gab2 immunoreactivity was observed in structures with the morphology of dystrophic neurites or neuropil threads, neurons, and corpora amylacea. The putative neurons were almost entirely dystrophic in appearance (Figure 3A) or had cytoplasmic inclusions resembling neurofibrillary tangles (Figure 3B). Dystrophic neurons and neurites (Figure 3C) and neurofibrillary tangle-bearing cells (Figure 3D) were also revealed by the Gab2 antibody in posterior cingulate. Here, however, many relatively normal neurons were observed as well, with long stretches of immunoreactive apical dendrites ascending through the cortical layers (Figures 3C and 3D).
Figure 3. Gab2 Colocalizes with Dystrophic Neurons in the AD Brain.
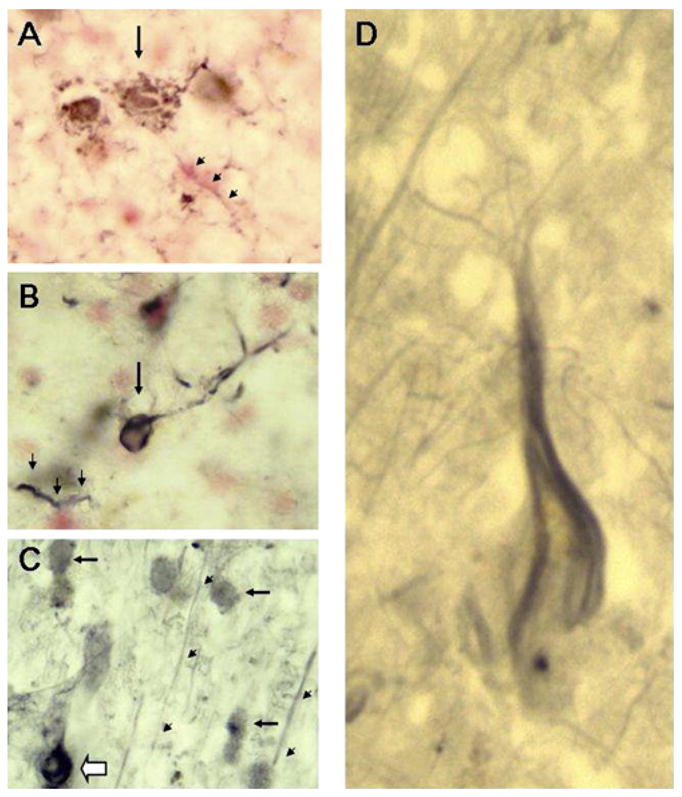
(A) LOAD hippocampus (neutral red counterstain) (40× objective). The arrow indicates a highly dystrophic cell with the size and morphology of a cortical pyramidal neuron. Arrowheads point to one of many structures in the sections that resemble dystrophic neurites or neuropil threads. (B) LOAD hippocampus (neutral red counterstain) (40×). The arrow denotes a putative neurofibrillary tangle-containing neuron. Arrowheads again indicate a dystrophic neurite. (C) LOAD posterior cingulate gyrus (40×). Filled arrows point to normal-appearing putative neurons. The open arrow points to a cell with the features of a neurofibrillary tangle-bearing neuron. Immunoreactive structures clearly resembling pyramidal cell apical dendrites were also observed ascending through the cortical layers (arrowheads). (D) LOAD posterior cingulate cortex (100× objective). Shown is a Gab2 immunoreactive cell with the flame-shaped cytoplasmic inclusion typical of the neurofibrillary tangle.
Discussion
In order to characterize and confirm associations between the GAB2 gene and LOAD risk in APOE ε4 carriers, our studies capitalized on the genome-wide survey of more than 300,000 SNPs, two clinically characterized and neuropathologically verified cohorts of AD cases and controls, a third cohort of clinically well characterized subjects, and stratification of the samples with respect to carriers and noncarriers of a major LOAD susceptibility gene, APOE. Six SNPs that are part of a common haplotype block encompassing the entire GAB2 gene were implicated in three independent cohorts. Maximal significance of the association was at SNP rs2373115 (p = 9 × 10−11) with an odds ratio of 4.06 (CI 2.81–14.69). An odds ratio of 24.64 (CI 7.44–116.79) for overall genetic risk is achieved when both ε4 and the GAB2 rs2373115 risk alleles are present. Data from a microarray study of laser-capture microdissected neurons in LOAD cases and controls, immunohistochemistry, and an siRNA study provided converging evidence for the relevance of GAB2 to the neuropathology of LOAD, raising testable hypotheses about the mechanisms by which GAB2 could modify LOAD risk in ε4 carriers and provide targets at which to aim new treatments.
Although only one genotyping platform was used in all three cohorts, our findings are unlikely to be attributable to any platform-related bias in genotyping calls because the observed association was not limited to a single SNP but was related to a large haplotype block in agreement with the LD structure of the HapMap CEPH population. Furthermore, all six of the implicated SNPs had high-quality SNiPer-HD scores (greater than 0.45), indicating that the data for each SNP clustered well (see Figure S2 for cluster diagrams).
Individual genotype data for all samples across >300,000 high-quality SNiPer-HD calls (see Experimental Procedures for QC metrics) is made available to the community as a .ped file at http://www.tgen.org/neurogenomics/data. This genome-wide scan data will enable replication of putative common LOAD risk alleles, and also enable further discovery of both independent and combinatorial genetic associations.
The GAB2 haplotype block spans 189 kb and includes at least 614 known SNPs. Four of the six hundred and fourteen known SNPs in this locus are nonsynonymous coding SNPs, which are generally considered to be the best candidates for functional variation. However, all four of these SNPs are reported to have minor allele frequencies of 0.0% in the CEPH population (The International HapMap Consortium, 2005), and therefore are not candidates for the common functional variant on the GAB2 risk haplotype.
GAB2 is a scaffolding protein involved in multiple signaling pathways, which could affect AD-related tau, amyloid, metabolic, or other aspects of AD pathology and cell survival in different ways (Koncz et al., 2002; Gu et al., 2001; Zompi, Gu and Colucci, 2004), and it has been found to be coexpressed with other putative AD-related genes (Li et al., 2004a). Discovery of this LOAD susceptibility gene, if further replicated, provides new opportunities to investigate LOAD pathogenesis, predisposition, treatment, and prevention. Genome-wide studies using even higher density platforms and compound genetic analyses in sufficiently large samples of well-characterized cases and controls promise to play increasingly important roles in the scientific understanding, evaluation, treatment, and prevention of AD and other common and genetically complex disorders. In the interim, public access to the raw genotyping data from our series will provide valuable information to assess the contribution of other putative risk loci to this devastating disease.
EXPERIMENTAL PROCEDURES
High-Density Genome-Wide Association Study
The 500K GeneChip (Affymetrix, Santa Clara, CA) was used to survey 502,267 SNPs in each subject as recently described (Coon et al., 2007). Genotypes were extracted using both SNiPer-HD (Hua et al., 2007) and BRLMM (Affymetrix) software. 312,316 SNPs were analyzed after excluding those that were monomorphic, did not cluster into three distinct Gaussian distributions, clustered poorly, had Hardy Weinberg equilibrium p values less than 0.01, had minor allele frequencies less than 2%, or exhibited less than 98% concordance between the SNiPer-HD and BRLMM calls. The software program STRUCTURE (Pritchard et al., 2000) was employed to test for underlying genetic stratification using 5000 randomly selected SNPs and including at least 100 SNPs per chromosome. The initial analysis yielded empirical evidence of three populations. Since 14 subjects belonged to a population far removed from the rest of the study population, they were eliminated from further analyses. STRUCTURE then was used to demonstrate a comparable admixture of the two populations in the cases and controls. After stratifying the LOAD cases and controls for presence or absence of the APOE ε4 allele, allelic χ2 statistics were computed for each SNP. APOE genotypes were obtained in each subject by either pyrosequencing (Ahmadian et al., 2000) or restriction fragment length polymorphism (RFLP) analysis (Lai et al., 1998).
LD mapping was performed by importing genotypes into the Haplo-View program v3.32. Pairwise LD values (as measured by D′) reflect the likelihood that two genetic markers are inherited together.
Neuronal Microarray Studies
Brain samples (mean post-mortem interval of 2.5 hr) from six brain regions that are either histopathologically or metabolically relevant to LOAD and aging were collected at the Sun Health Research Institute. Expression profiling was performed as described previously (Liang et al., 2007). Direct case-to-control comparisons were performed to analyze expression differences in each region.
Immunohistochemical Validation
Gab2 immunoreactivity in LOAD hippocampus and posterior cingulate cortex was examined using an affinity-purified goat polyclonal antibody directed against a C-terminal epitope of Gab2 (Santa Cruz Biotechnology, Santa Cruz, CA). Blocks were obtained from rapid autopsy LOAD cases (<3 hr postmortem) (n = 5). Hippocampus sections were derived from blocks that were fixed for 24 hr in 4% paraformaldehyde and sectioned at 40 μm on a freezing microtome. Posterior cingulate sections were derived from snap-frozen blocks that were sectioned at 6 μm on a cryostat. Immunohistochemical protocols were as previously described (Li et al., 2004b). Immunoreactivity was visualized with nickel-intensified diaminobenzidine.
Tau Phosphorylation siRNA Study
Neuroglioma cells overexpressing wild-type tau protein were grown in 96-well plates and transfected with siRNA directed at GAB2 mRNA. Following 4 days of transfection, cells were fixed, permeablized, and immunostained with antibodies against total tau protein and tau protein phosphorylated on serine-262. A FITC- and Cy5-conjugated secondary antibody cocktail was then applied. After incubation and washing, images were captured and quantitated using the InCell imager 3000 (General Electric). The fold increase in serine-262 phosphorylated tau levels was calculated against control samples that had been transfected with a scrambled siRNA sequence.
Supplementary Material
Acknowledgments
These studies were supported by Kronos Life Sciences Laboratories, the National Institute on Aging (Arizona Alzheimer’s Disease Center P30 AG19610, RO1 AG023193, Mayo Clinic Alzheimer’s Disease Center P50 AG16574, and Intramural Research Program), the National Alzheimer’s Coordinating Center (U01 AG016976), and the state of Arizona. We thank our research volunteers and their families for their generous participation; Drs. Creighton Phelps, Marcelle Morrison-Bogorad, Marilyn Miller, and Walter Kukull for their assistance in the acquisition of tissue samples and data; and directors, pathologists, and technologists from the following ADCs and brain banks: Lucia Sue (Sun Health Research Institute and Arizona Alzheimer’s Disease Center); Ruth Seemann and Dan Brady (National Institute on Aging); Juan C. Troncoso and Olga Pletnikova (John Hopkins, P50 AG05146); Harry Vinters and Justine Pomakian (University of California, Los Angeles, P50 AG16570); Christine Hulette (The Kathleen Price Bryan Brain Bank, Duke University Medical Center, P50 AG05128, RO1 NS39764, RO1 MH60451, and GlaxoSmithKline); Dikran Horoupian and Ahmad Salehi (Stanford University, P30 AG17824); Jean Paul Vonsattel (New York Brain Bank, Taub Institute, Columbia University, P50 AG08702); E. Tessa Hedley-Whyte and Karlotta Fitch (Massachusetts General Hospital, P50 AG05134); Roger Albin, Lisa Bain, and Eszter Gombosi (University of Michigan, P50 AG08671); William Markesbery, Sonya Anderson (University of Kentucky, P50 AG05144); Dennis W. Dickson and Natalie Thomas (Mayo Clinic, Jacksonville, P50 AG16574 and P50 AG25711); Carol A. Miller, Jenny Tang, and Dimitri Diaz (University of Southern California, P50 AG05142); Dan McKeel, John C. Morris, Eugene Johnson, Jr., Virginia Buckles, and Deborah Carter (Washington University, St. Louis, P50 AG 05681); Thomas Montine and Aimee Schantz (University of Washington, P50 AG05136); John Q. Trojanowski, Virginia M. Lee, Vivianna Van Deerlin, and Terry Schuck (University of Pennsylvania); Ann C. McKee and Carol Kubilus (Boston University, P30 AG13846); Bruce H. Wainer and Marla Gearing (Emory University, AG025688); Charles L. White III, Roger Rosenberg, Marilyn Howell, and Joan Reisch (University of Texas, Southwestern Medical School, P30-AG12300); William Ellis and Mary Ann Jarvis (University of California, Davis, P30 AG AG01542); David A. Bennett, Julie A. Schneider, Karen Skish, and Wayne T. Longman (Rush University Medical Center, P30 AG10161); and Deborah C. Mash, Margaret J. Basile, and Mitsuko Tanaka University of Miami/NPF Brain Endowment Bank). Additional support was provided by the Johnnie B. Byrd Sr. Alzheimer’s Disease and Research Institute, the Swiss National Science Foundation (PP00B-68859), the Verum foundation, the Bisgrove charitable donation, the NIH Neuroscience Blueprint (U24NS051872), the ENDGAME Consortium (UO1HL084744), and the National Institute on Aging (K01AG024079).
References
- Ahmadian A, Gharizadeh B, Gustafsson AC, Sterky F, Nyren P, Uhlen M, Lundeberg J. Single-nucleotide polymorphism analysis by pyrosequencing. Anal Biochem. 2000;280:103–110. doi: 10.1006/abio.2000.4493. [DOI] [PubMed] [Google Scholar]
- Baki L, Shioi J, Wen P, Shao Z, Schwarzman A, Gama-Sosa M, Neve R, Robakis NK. PS1 activates PI3K thus inhibiting GSK-3 activity and tau overphosphorylation: effects of FAD mutations. EMBO J. 2004;23:2586–2596. doi: 10.1038/sj.emboj.7600251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39:17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- Bobinski M, de Leon MJ, Wegiel J, Desanti S, Convit A, Saint Louis LA, Rusinek H, Wisniewski HM. The histological validation of post mortem magnetic resonance imaging-determined hippocampal volume in Alzheimer’s disease. Neuroscience. 2000;95:721–725. doi: 10.1016/s0306-4522(99)00476-5. [DOI] [PubMed] [Google Scholar]
- Braak H, Braak E. Neuropathological staging of Alzheimer’s-related changes. Acta Neuropathol (Berl) 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- Buckner R, Snyder AZ, Shannon BJ, LaRossa G, Sachs R, Fotenos AF, Sheline YI, Klunk WE, Mathis CA, Morris JC, Mintun MA. Molecular, structural and functional characterization of Alzheimer’s disease: evidence for a relationship between default activity, amyloid and memory. J Neurosci. 2005;25:7709–7717. doi: 10.1523/JNEUROSCI.2177-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coon KD, Myers AJ, Craig DW, Webster JA, Pearson JV, Hu-Lince D, Zismann VL, Beach T, Leung D, Bryden L, et al. A high density whole-genome association study reveals that APOE is the major susceptibility gene for sporadic late-onset Alzheimer’s disease. J Clin Psychiatry. 2007;68:613–618. doi: 10.4088/jcp.v68n0419. [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Strittmatter WJ, Schmechel DE, Gaskell PC, Small GW, Roses AD, Haines JL, Pericak-Vance MA. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. doi: 10.1126/science.8346443. [DOI] [PubMed] [Google Scholar]
- Corder EH, Saunders AM, Risch NJ, Strittmatter WJ, Schemchel DE, Gaskell PC, Jr, Rimmler JB, Locke PA, Conneally PM, Schmader KE, et al. Protective effect of apolipoprotein E type 2 allele for late onset Alzheimer disease. Nat Genet. 1994;7:180–184. doi: 10.1038/ng0694-180. [DOI] [PubMed] [Google Scholar]
- Evans DA, Funkenstein HH, Albert MS, Scherr PA, Cook NR, Chown MJ, Hebert LE, Hennekens CH, Taylor JO. Prevalence of Alzheimer’s disease in a community population of older persons: higher than previously reported. JAMA. 1989;262:2551–2556. [PubMed] [Google Scholar]
- Farrer LA, Cupples LA, Haines JL, Hyman B, Kukull WA, Mayeux R, Myers RH, Pericak-Vance MA, Risch N, van Duijn CM. Effects of age, sex and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A metaanalysis. APOE and Alzheimer Disease Meta Analysis Consortium. JAMA. 1997;278:1349–1356. [PubMed] [Google Scholar]
- Gatz M, Reynolds CA, Fratiglioni L, Johansson B, Mortimer JA, Berg S, Fiske A, Pedersen NL. Role of genes and environments for explaining Alzheimer disease. Arch Gen Psychiatry. 2006;63:168–174. doi: 10.1001/archpsyc.63.2.168. [DOI] [PubMed] [Google Scholar]
- Gu H, Saito K, Klaman LD, Shen J, Fleming T, Wang Y, Pratt JC, Lin G, Lim B, Kinet JP, Neel BG. Essential role for Gab2 in the allergic response. Nature. 2001;412:186–190. doi: 10.1038/35084076. [DOI] [PubMed] [Google Scholar]
- Hua J, Craig DW, Brun M, Webster J, Zismann V, Tembe W, Joshipura K, Huentelman MJ, Dougherty ER, Stephan DA. SNiPer-HD: Improved genotype calling accuracy by an expectation-maximization algorithm for high-density SNP arrays. Bioinformatics. 2007;23:57–63. doi: 10.1093/bioinformatics/btl536. [DOI] [PubMed] [Google Scholar]
- Jack CR, Jr, Petersen RC, Xu YC, O’Brien PC, Smith GE, Ivnik RJ, Boeve BF, Waring SC, Tangalos EG, Kokmen E. Prediction of AD with MRI-based hippocampal volume in mild cognitive impairment. Neurology. 1999;52:1397–1403. doi: 10.1212/wnl.52.7.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SC, Schmitz TW, Moritz CH, Meyerand ME, Rowley HA, Alexander AL, Hansen KW, Gleason CE, Carlsson CM, Ries ML, et al. Activation of brain regions vulnerable to Alzheimer’s disease: The effect of mild cognitive impairment. Neurobiol Aging. 2006;27:1604–1612. doi: 10.1016/j.neurobiolaging.2005.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang DE, Yoon IS, Repetto E, Busse T, Yermian N, Ie L, Koo EH. Presenilins mediate phosphatidylinositol 3-kinase/AKT and ERK activation via select signaling receptors. Selectivity of PS2 in platelet-derived growth factor signaling. J Biol Chem. 2005;208:31537–31547. doi: 10.1074/jbc.M500833200. [DOI] [PubMed] [Google Scholar]
- Koncz G, Bodor C, Kovesdi D, Gati R, Sarmay G. BCR mediated signal transduction in immature and mature B cells. Immunol Lett. 2002;82:41–49. doi: 10.1016/s0165-2478(02)00017-2. [DOI] [PubMed] [Google Scholar]
- Kruglyak L. Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat Genet. 1999;22:139–144. doi: 10.1038/9642. [DOI] [PubMed] [Google Scholar]
- Lai E, Riley J, Purvis I, Roses A. A 4-Mb high-density single nucleotide polymorphism-based map around human APOE. Genomics. 1998;54:31–38. doi: 10.1006/geno.1998.5581. [DOI] [PubMed] [Google Scholar]
- Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:160–161. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- Li K, Liu C, Sun W, Yuan S, Yu T. A system for enhancing genome-wide coexpression dynamic study. Proc Natl Acad Sci USA. 2004a;101:15561–15566. doi: 10.1073/pnas.0402962101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Strohmeyer R, Liang Z, Lue LF, Rogers J. CCAAT/Enhancer binding protein δ (C/EBP-δ) expression and elevation in Alzheimer’s disease. Neurobiol Aging. 2004b;25:991–999. doi: 10.1016/j.neurobiolaging.2003.10.016. [DOI] [PubMed] [Google Scholar]
- Liang WS, Dunckley T, Beach TG, Grover A, Mastroeni D, Walker DG, Caselli RJ, Kukull WA, McKeel D, Morris JC, et al. Gene expression profiles in anatomically and functionally distinct regions of the normal aged human brain. Physiol Genomics. 2007;28:311–322. doi: 10.1152/physiolgenomics.00208.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mintun MA, Larossa GN, Sheline YI, Dence CS, Lee SY, Mach RH, Klunk WE, Mathis CA, DeKosky ST, Morris JC. [11C]PIB in a nondemented population: potential antecedent marker of Alzheimer disease. Neurology. 2006;67:446–452. doi: 10.1212/01.wnl.0000228230.26044.a4. [DOI] [PubMed] [Google Scholar]
- Papassotiropoulos A, Fountoulakis M, Dunckley T, Stephan DA, Reiman EM. Genetics, transcriptomics and proteomics of Alzheimer’s disease. J Clin Psychiatry. 2006;67:652–670. doi: 10.4088/jcp.v67n0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt JC, Igras VE, Maeda H, Baksh S, Gelfand EW, Burakoff SJ, Neel BG, Gu H. Cutting edge: gab2 mediates an inhibitory phosphatidylinositol 3′-kinase pathway in T cell antigen receptor signaling. J Immunol. 2001;165:4158–4163. doi: 10.4049/jimmunol.165.8.4158. [DOI] [PubMed] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiman EM, Caselli RJ, Yun LS, Chen K, Bandy D, Minoshima S, Thibodeau SN, Osborne D. Preclinical evidence for Alzheimer’s disease in persons homozygous for the epsilon 4 allele for apolipoprotein E. N Engl J Med. 1996;334:752–758. doi: 10.1056/NEJM199603213341202. [DOI] [PubMed] [Google Scholar]
- Reiman EM, Chen K, Alexander GE, Caselli RJ, Bandy D, Osborne D, Saunders AM, Hardy J. Correlations between apolipoprotein E ε4 gene dose and brain-imaging measurements of regional hypometabolism. Proc Natl Acad Sci USA. 2005;102:8299–8302. doi: 10.1073/pnas.0500579102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zompi S, Gu H, Colucci F. The absence of Grb2-associated binder 2 (Gab2) does not disrupt NK cell development and functions. J Leukoc Biol. 2004;76:896–903. doi: 10.1189/jlb.0304179. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


