Summary
Both the cadherin-catenin complex and Rho-family GTPases have been shown to regulate dendrite development. We show here a role for p120 catenin (p120ctn) in regulating spine and synapse formation in the developing mouse brain. p120catenin gene deletion in hippocampal pyramidal neurons in vivo resulted in reduced spine and synapse densities along dendrites. In addition, p120 catenin loss resulted in reduced cadherin levels and misregulation of Rho-family GTPases, with decreased Rac1 and increased RhoA activity. Analyses in vitro indicate that the reduced spine density reflects aberrant Rho-family GTPase signaling, whereas the effects on spine maturation appear to result from reduced cadherin levels and possibly aberrant Rho-family GTPase signaling. Thus, p120ctn acts as a signal coordinator between cadherins and Rho-family GTPases to regulate cyto-skeletal changes required during spine and synapse development.
Introduction
Dendritic spines are actin-rich protrusions that are the major postsynaptic sites of excitatory synaptic input. Changes in spine distribution and morphology have been observed in many neurological disorders (Fiala et al., 2002; Govek et al., 2005). During development, periods of high spine motility coincide with synapse formation and appear important for the establishment of connections with presynaptic partners (Dunaevsky et al., 1999; Fiala et al., 1998; Konur and Yuste, 2004; Stepanyants et al., 2004; Zhou et al., 2004; Ziv and Smith, 1996). Morphological plasticity of spines may contribute to changes in synaptic strength (Trachtenberg et al., 2002; Zhou et al., 2004). Recent studies indicate that the cadherin-catenin cell adhesion system and Rho-family GTPases are important regulators of spine and synapse number and morphology (Abe et al., 2004; Bozdagi et al., 2004; Govek et al., 2005; Togashi et al., 2002).
Cadherins and catenins have been localized to synapses where they become restricted to the region surrounding the active zone (Elste and Benson, 2006; Fannon and Colman, 1996; Uchida et al., 1996). Perturbations of cadherins affect axon targeting, synapse formation, and synaptic plasticity in vivo (Inoue and Sanes, 1997; Iwai et al., 2002; Prakash et al., 2005). In addition, cadherin inhibition disrupts spine maturation and post-synaptic protein localization (Okamura et al., 2004; Togashi et al., 2002). β-catenin promotes dendrite growth and branching in vitro (Yu and Malenka, 2003). Deletion of αN-catenin destabilizes spine heads (Abe et al., 2004). Deletion of β-catenin reduces the number of reserve pool vesicles at synapses in vivo and reduces synaptic vesicle localization in vitro (Bamji et al., 2003). Thus, the cadherin-catenin complex regulates both pre-synaptic and postsynaptic development.
Cadherins, through catenins, regulate several signaling pathways, including Rho-family GTPases (Hatzfeld, 2005; Reynolds and Roczniak-Ferguson, 2004). Rho-family GTPases regulate dendritic morphogenesis through regulation of the F-actin cytoskeleton (Govek et al., 2005; Nakayama et al., 2000; Tashiro et al., 2000; Tashiro and Yuste, 2004). Rac1 activation leads to the development and stabilization of dendritic spines, while Rac1 inhibition results in progressive spine loss. RhoA inactivation leads to increased spine density and spine neck length, while RhoA activation inhibits spine formation. Rho-family GTPase activation is controlled by gua-nine nucleotide exchange factors (GEFs), dissociation inhibitors (GDIs), and GTPase activating factors (GAPs). Mutations in several of these have been identified in patients with nonsyndromic mental retardation, cognitive disorders characterized by abnormal spine morphology (Govek et al., 2005).
The p120 catenin (p120ctn) family consists of four proteins: ARVCF, δ-catenin, p0071, and p120 catenin (Hatzfeld, 2005). These proteins have similar structures with N- and C-terminal sequences flanking a central domain consisting of 10 armadillo (arm) repeats. Each binds the membrane-proximal regions of cadherin cytoplasmic tails through the armadillo repeats. The p120ctn family is distinct structurally and functionally from β-catenin and its homolog plakoglobin, with the latter two proteins having central domains with 12 armadillo repeats that bind the C-terminal residues of cadherin cytoplasmic tails, a region distinct from that recognized by p12ctn family members. In contrast to the p120ctn family, β-catenin and plakoglobin provide a physical link through α-catenin to the F-actin cytoskeleton.
The p120ctn family regulates cadherin function and provides a link through which cadherin engagement controls cytoskeletal assembly and Rho-family GTPases. p120ctn family members promote the surface stability of cadherins (Chen et al., 2003; Davis et al., 2003). In their absence, cadherins are endocytosed and degraded. Kinesin-mediated trafficking of cadherins also promotes cell adhesion (Teng et al., 2005) and appears at least partially dependent upon p120ctn (Chen et al., 2003). p120ctn interacts with microtubules directly and through binding to kinesins (Chen et al., 2003; Franz and Ridley, 2004; Yanagisawa et al., 2004). In addition, the presence of a p120ctn family member appears essential to maintain the association of cadherins with β-catenin, and consequently the cytoskeleton, through recruitment of a protein tyrosine phosphatase that dephosphorylates β-catenin (Xu et al., 2004).
p120ctn controls cytoskeletal assembly at cadherin-mediated junctions through regulation of Rho-family GTPases, activating Rac1 and Cdc42, possibly through the exchange factor Vav2, and inactivating RhoA as a Rho GDI (Anastasiadis et al., 2000; Grosheva et al., 2001; Magie et al., 2002; Noren et al., 2000). The presence of p120ctn at cadherin-mediated junctions is required to recruit and activate Rac1 (Goodwin et al., 2003; Yap et al., 1998). In addition, at least some p120ctn family members recruit cortactin, a protein that promotes assembly of the F-actin cytoskeleton as well as spine morphogenesis (Hering and Sheng, 2003; Martinez et al., 2003).
Studies in Xenopus laevis have shown that p120ctn has important functions in early development that involve both regulation of Rho-family GTPases and control of transcription through interactions with the methyl CpG binding protein Kaiso (Park et al., 2005). Consistent with these observations, we have observed early embryonic lethality of mice homozygous for a mutant in the p120ctn gene with anatomical abnormalities consistent with those observed in Xenopus laevis (L.E . and L.R., unpublished data).
Both p120ctn and δ-catenin are widely expressed in neurons where both are localized to spines and synapses (Chauvet et al., 2003; Ho et al., 2000; Husi et al., 2000; Tanaka et al., 2000). In the present work, we characterize effects of p120ctn deletion on dendritic spine development through use of a floxed allele of p120ctn that bypasses the early embryonic lethality that we have observed in conventional p120ctn mutants (L.E. and L.R., unpublished data). Loss of p120ctn function in vivo results in a dramatic decrease in spine and synapse density and induces morphological changes in spine neck length and head width. Mechanistic analyses in vitro indicate that p120ctn controls spine density through regulation of Rho, independent of its association with cadherins, while it appears to regulate spine head width through interactions with cadherins directly.
Results
Targeting Strategy for Conditional Inactivation of the p120ctn Gene
To investigate the role of p120ctn in nervous system development, a conditional “floxed” allele of p120ctn (p120ctnflox) was generated by homologous recombination in ES cells. loxP sites were introduced into the p120ctn locus flanking exon 7 (see Figure S1 in the Supplemental Data). exon 7 encodes 27 amino acids of armadillo repeat 1, all of arm repeat 2, and 42 amino acids of arm repeat 3. In previous work, the presence of each of the first five arm repeats (1–5) has been shown to be essential for binding of p120ctn to the classical cadherins (Anastasiadis et al., 2000). Deletion of exon 7 is predicted to introduce a +2 frameshift in the translational reading frame. The presence of the frameshift plus an effective RNA surveillance mechanism that degrades mRNAs containing premature stop codons will result in rapid degradation of the entire mRNA (Singh and Lykke-Andersen, 2003). exon 7 is present in each of the four p120ctn isoforms generated by alternative splicing (Keirsebilck et al., 1998). Thus, Cre-mediated deletion is predicted to result in the elimination of p120ctn protein expression. This was confirmed by immunohistochemistry and SDS-PAGE analysis using Nand C-terminal-specific antibodies (Figures S1D–S1F and not shown). The presence of loxP sites in the p120ctnflox allele did not compromise protein expression or function. The homozygous p120ctnflox/flox mice were viable, fertile, and showed no obvious phenotype.
Generation of Dorsal Forebrain-Specific p120ctn Knockout Mice
To delete p120ctn in the dorsal forebrain, p120ctnflox/flox mice were mated to mice expressing Cre recombinase under the control of the emx1 promoter, which is expressed in neuroepithelial precursors of neurons and glia in the dorsal forebrain beginning at embryonic day 9 (Gorski et al., 2002). In the mutant mice (p120ctnflox/flox;emx1IREScre), p120ctn protein expression was absent from regions of the dorsal forebrain that express emx1IREScre as assayed by Western blot or immunocytochemistry (Figures S1D–S1F). The low levels of p120ctn remaining in the recombined tissues are almost certainly due to expression of p120ctn in blood vessels, interneurons, and other cell types which are not derived from precursors expressing Emx1.
Loss of p120ctn from the Forebrain Leads to a Reduced Spine Density in Hippocampal Pyramidal Neurons
Mutant mice lacking p120ctn in the dorsal forebrain (p120ctnflox/flox;emx1IREScre) were viable, fertile, and showed no obvious phenotype. Nissl staining revealed that the overall morphology, including laminar organization, of the cortex and hippocampus appeared normal at 6 weeks of age (Figures 1A and 1B).
Figure 1.
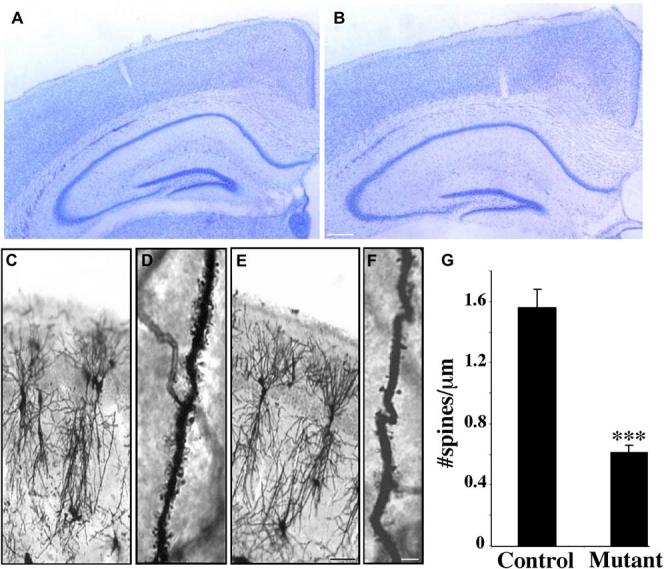
Effects of Deletion of p120ctn on Cortical and Hippocampal Development
(A and B) Nissl-stained coronal sections from (A) control (p120ctn flox/flox) and (B) mutant (p120ctnflox/flox;emx1IREScre) six-week-old male litter-mate mice (n = 3 pairs).
(C–F) Golgi-stained pyramidal neurons from the CA1 hippocampus from control ([C] and [D]) and mutant ([E] and [F]) six-week-old male litter-mates (n = 3 pairs).
(G) Spine density quantification along dendrites from control and mutant mice (n = 3 pairs, ***p < 0.0001). Scale bars: 250 μm (B); 100 μm(E);20 μm(F).
To determine if absence of p120ctn resulted in abnormalities in neuronal fine structure, the hippocampi of control and mutant brains were visualized by Golgi staining. Compared to controls, the overall organization and development of the dendritic trees of individual pyramidal neurons in the CA1 hippocampus appeared relatively normal in the mutant (Figures 1C and 1E). Closer examination revealed a striking decrease in the density of spines along the dendrites of the mutant neurons (Figures 1D and 1F). Linear spine density in the mutant neurons was decreased to 39% of control (control: 1.56 ± 0.12 spines/μm; mutant: 0.61 ± 0.5 spines/μm) (Figure 1G). In addition, the mutant spines appeared smaller and shorter in length. Despite spine loss, the dendrites in the mutants retained a normal length and exhibited none of the classical signs of degeneration, such as beaded expansions or irregular diameter.
p120ctn Mutant Mice Have a Reduced Synapse Density in the Hippocampus
Dendritic spines are major sites of excitatory synaptic input. To determine if the reduced density of spines in the hippocampus of dorsal forebrain-specific p120ctn mutant mice was correlated with a similar reduction in synapse density, we used antibodies specific for synaptic vesicles and the postsynaptic density to measure synapse density. The absence of p120ctn resulted in reduced numbers of puncta that express the synaptic vesicle protein synaptophysin or the postsynaptic scaffold protein PSD95 in the stratum radiatum (SR) of the CA1 hippocampus (Figures 2A-2D). Both synaptophysin and PSD95 exhibited punctate patterns in both controls and mutants, but in the mutant there were 64% and 31% reductions in the densities of puncta expressing synaptophysin or PSD95, respectively (Figures 2E and 2F). Analysis of synapse density by electron microscopy confirmed this conclusion (Figures 2G and 2H). Quantification showed that the density of synapses was reduced by 42% in the mutant stratum radiatum (Figure 2I). Closer examination showed that there was a 48% reduction in the number of synapses formed on spines (Figure 2J), but not a significant difference in the number of synapses formed on the shafts of dendrites (Figure 2K). While the density of split or perforated synapses was low (Figure 2L), the absence of p120ctn resulted in an almost 3-fold increase in the number of these synapses. We quantified, but did not observe, significant differences in the average length of postsynaptic densities, average number of docked vesicles, and average total vesicle number (not shown).
Figure 2.
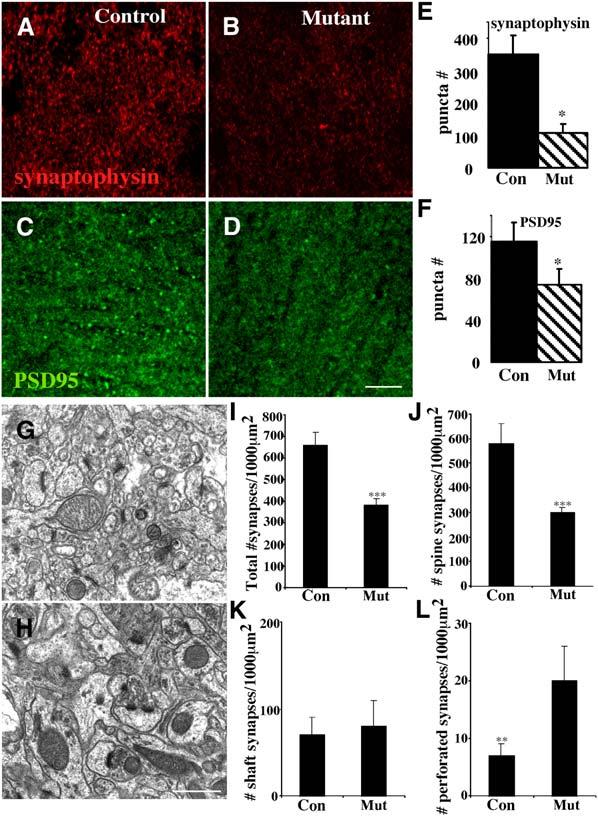
Reduced Synapse Density in the Stratum Radiatum of the p120ctn Mutant
(A–D) Imaged coronal sections of the stratum radiatum of the CA1 hippocampus from ([A] and [B]) control p120ctnflox/flox or ([C] and [D]) mutant p120ctnflox/flox;emx1IREScre six-week-old male littermates following labeling with anti-synaptophysin ([A] and [C]) or anti-PSD95 ([B] and [D]).
(E) Quantification of synaptophysin puncta: mean ± SEM (p < 0.07).
(F) Quantification of PSD95 puncta: mean ± SEM (p < 0.08). We measured 3520 μm2 per sample for (E) and (F). Electron micrographs of the CA1 stratum radiatum from (G) control p120ctnflox/flox or (H) mutant p120ctnflox/flox;emx1IREScre mice. Examples of spine-localized synapses, which contain the electron dense postsynaptic region (black arrowhead) juxtaposed to the presynaptic vesicle release site (active zone), are indicated. A perforated synapse is indicated in the mutant-derived section (white arrow, [H]).
(I–L) Quantification of EM analysis. (n = 3 pairs for all analyses).
Scale bars, 25 mm (D); 500 nm (H).
Significant Reduction in N-Cadherin Level in the p120ctn Mutant Mice
Recent studies have revealed that p120ctn and p120ctn family members, such as δ-catenin, play a significant role in determining surface and total cadherin levels; loss of p120ctn members results in a concomitant weakening of cell-cell adhesion by increasing cadherin turnover and lowering cadherin levels (Davis et al., 2003; Xiao et al., 2003). To determine if absence of p120ctn similarly reduced cadherin levels within the brain, we examined expression levels in the hippocampi of control and mutant mice. Compared to controls, indirect immunofluorescence revealed a 32% reduction in the density of N-cadherin puncta in the SR of the mutant hippocampus (Figure 3C). Quantification by Western blot showed the level of N-cadherin in the mutant was reduced significantly to 73% of control level. There were no significant reductions in the levels of β-catenin or α-catenin proteins (Figure 3B). Interestingly, reductions in N-cadherin and pan-cadherin levels have been observed in the brains of mice lacking δ-catenin (Israely et al., 2004). The presence of δ-catenin and possibly of other p120ctn family members appears sufficient to maintain significant levels of cadherins in the p120ctn mutant hippocampus.
Figure 3.
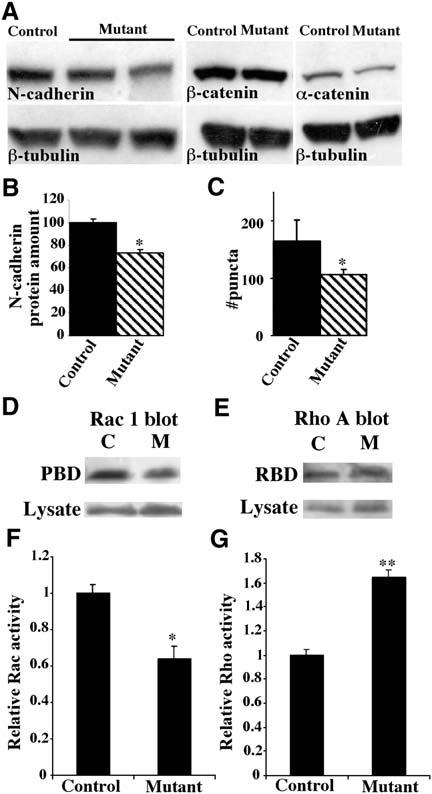
Reduced Cadherin and Altered Rac and Rho Activity in the p120ctn Mutant Hippocampus In Vivo
(A) Western blot analyses of cadherin and α- and β-catenin protein levels in hippocampal lysates of control or mutant six-week-old male littermates, quantitated in (B) (n = 3 pairs; p < 0.05).
(C) Quantification of N-cadherin puncta in stratum radiatum in control and mutant; mean ± SEM (n = 3 pairs; p < 0.1).
(D–G) Effects of p120ctn deletion on activities of hippocampal Rac and Rho. Active Rac1 and RhoA were precipitated from hippocampal extracts using GST fusions to the G protein binding domains of PAK and rhotekin, respectively. Following SDS-PAGE fractionation, extracts and precipitates were blotted with anti-Rac1 or anti-RhoA to determine quantities of Rac and Rho. Representative examples of precipitated active Rac (PBD), active Rho (RBD), and total Rac and Rho (lysate) are shown in (D) and (E). Active Rac and Rho were normalized to total quantities of each protein ([F] and [G]). Each experiment was repeated three times using independent extracts (*p < 0.03, **p < 0.005).
p120ctn Mutant Mice Have Reduced Rac1 and Increased RhoA Activity
Recent studies have shown that overexpression of p120ctn in cultured cells results in elevated activation of Rac1 and Cdc42 and reduced activation of RhoA (Anastasiadis et al., 2000; Franz and Ridley, 2004; Noren et al., 2000). To determine whether the absence of p120ctn affects the activities of Rho-family GTPases, we performed affinity precipitation assays to measure Rac1 and RhoA activity in hippocampal lysates (Ren et al., 1999). GST-fusion proteins containing the Rac1 binding domain of PAK (GST-PBD) and the RhoA binding domain of rhotekin (GST-RBD) were used to isolate active Rac1 and RhoA, respectively, with results normalized to the total level of each protein. Experiments are illustrated in Figures 3D and 3E, respectively. Loss of p120ctn in the hippocampus led to a 1.6-fold decrease in active Rac1 (Figure 3F) and a 1.65-fold increase in active RhoA (Figure 3G). The levels of endogenous Rac1 and RhoA were similar in control and mutant samples. As Rac and Rho regulate dendrite growth and spine morphogenesis, these results suggest that the spine deficit observed in the hippocampus of p120ctn mutant mice may be due, in part, to abnormal regulation of these G proteins.
Impaired Dendrite Development in Cultured p120ctn−/− Hippocampal Pyramidal Neurons
To examine in more detail the role of p120ctn in dendrite morphogenesis, we examined dendrite development in vitro using neuronal cultures from control and mutant mice. At day 10 in vitro (DIV10), neurons were transfected with an EGFP expression plasmid to visualize cell morphologies. Tracings revealed that p120ctn loss resulted in simplification of dendritic branching patterns (Figure 4A). Sholl analysis showed that the mutant neurons had fewer dendritic branch crossings (Figure 4B). The mean number of dendritic terminal branch points was reduced to 40% of that in controls (Figure 4C). To determine if the deficit in dendrite formation reflected a requirement for p120ctn before or during culture in vitro, mutant neurons were cotransfected with a p120ctn expression plasmid. Expression of p120ctn in the mutant neurons resulted in comparatively normal dendrite development (Figure 4). Thus, deletion of p120ctn in neural precursors does not prevent later development of comparatively normal dendrites in neurons in which p120ctn expression is restored.
Figure 4.
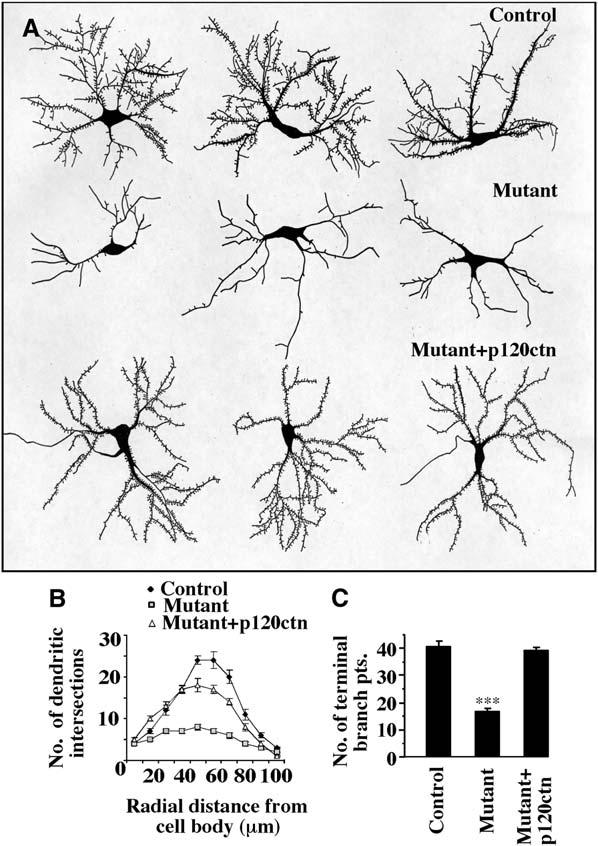
Reduced Dendritic Tree Complexity of p120ctn Mutant CA1 Hippocampal Pyramidal Neurons In Vitro
(A) Three representative camera lucida tracings are shown of cultured neurons of control p120ctnflox/flox, mutant p120ctnflox/flox;emx1IREScre, or mutant cotransfected with p120ctn. Neurons were transfected at DIV10 with an EGFP-encoding plasmid and analyzed at DIV14. The dendritic trees of the control and of the mutant plus p120ctn-cotransfected neurons are more complex than those of the mutant neurons.
(B) Sholl analysis of the neurons depicted in (A) [control (n = 7 cells); mutant (n = 9 cells); mutant plus p120ctn cDNA (n = 10 cells)].
(C) Quantification of the number of terminal dendritic branch points of the neurons depicted in (A). Dendritic branches with a length ≥ 7 μm were counted (control [n = 7 cells]; mutant [n = 9 cells]; mutant plus p120ctn cDNA [n = 9 cells]. ***p < 0.0001).
To determine if the in vivo deficit in spine formation was recapitulated in vitro, we analyzed cultured hippocampal neurons using EGFP after 14 days of culture (Figure 5). Absence of p120ctn resulted in a dramatic reduction in spine density (Figures 5A, 5A′, 5B, and 5B′). Quantification showed that spine density was significantly reduced to approximately 40% of that of controls (Figure 5I). Cotransfection of the mutant neurons at DIV10 with a p120ctn expression plasmid restored the spine density at DIV14 to a level similar to controls, indicating that p120ctn can promote spine formation over a comparatively short time span (Figures 5C, 5C′, and 5I). p120ctn expression in mutant neurons at DIV10 also enhanced dendrite development. As a result, the dendrites present at DIV14 were very similar to controls (Figures 5A and 5C).
Figure 5.
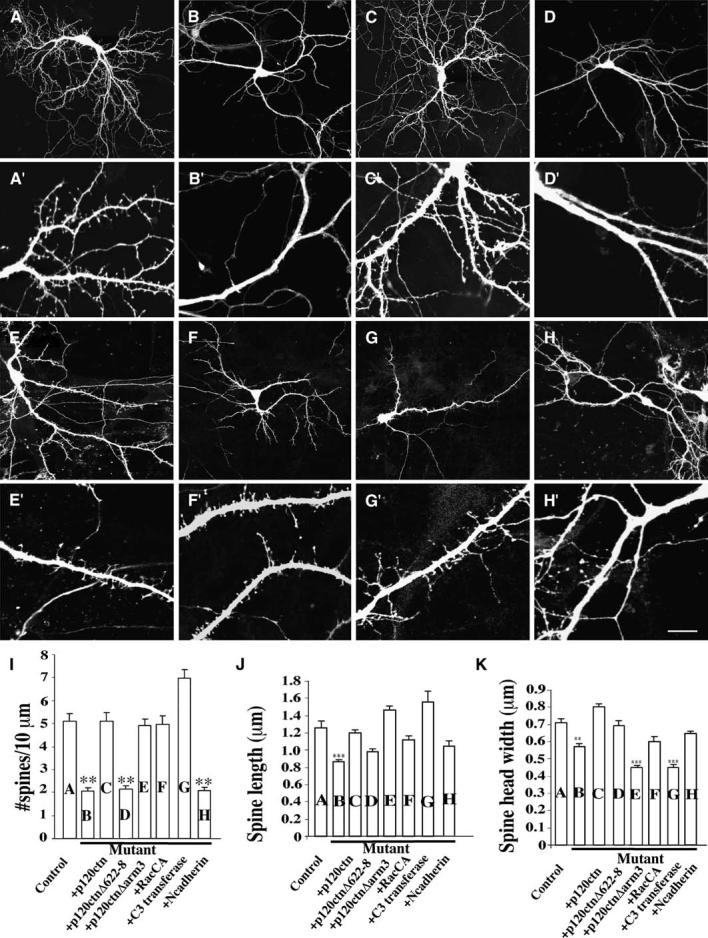
Reduced Spine Densities in p120ctn Mutant CA1 Hippocampal Pyramidal Neurons In Vitro
(A–H′) Representative examples of EGFP-transfected neurons cultured from control p120ctnflox/flox or mutant p120ctnflox/flox;emx1IREScre mice that were transfected at DIV10 and analyzed at DIV14. Lower (A–H) and higher magnification images (A′–H′) are shown. (A and A′) Control neuron (n = 17 cells; 940 protrusions). (B and B′) Mutant neuron (n = 22 cells; 930 protrusions). (C and C′) Mutant neuron cotransfected with p120ctn (n = 8 cells; 400 protrusions). (D and D′) Mutant neuron cotransfected with p120ctnΔ622–628 (n = 14 cells; 462 protrusions). (E and E′) Mutant neuron cotransfected with p120ctnΔarm3 (n = 8 cells; 506 protrusions). (F and F′) Mutant neuron cotransfected with constitutive-active Rac1 (n = 7 cells; 532 protrusions). (G and G′) Mutant neuron cotransfected with C3 transferase (n = 9 cells; 498 protrusions). (H and H′) Mutant neuron cotransfected with N-cadherin (n = 12 cell; 306 protrusions).
(I) Spine densities from each of the experimental conditions (**p < 0.0001).
(J and K) Mean spine length and head width. Four independent experiments were analyzed.
Scale bar, 40 μm ([A]–[K]); 10 μm ([A]′–[K]′). (**p < 0.001, ***p < 0.0001).
Analysis of the p120ctn-Regulated Signaling Pathways that Regulate Spine Morphogenesis
The p120ctn catenin family regulates many signaling pathways, including control of cadherin stability, Rho-family G protein activity, and signaling through protein kinases and phosphatases (Reynolds and Roczniak-Ferguson, 2004; Xu et al., 2004). Different p120ctn domains regulate these diverse signaling pathways. To determine which p120ctn domains control the spine density, two p120ctn deletion mutants, p120ctnΔarm3 and p120ctnΔ622–628, unable to bind cadherin and regulate Rho, respectively, were expressed in p120ctn mutant neurons between DIV10–14. p120ctnΔ622–628 prevents inhibition of Rho, but does not affect activation of Rac (P. Anastasiadis, personal communication). Analysis of spine density at DIV14 indicated that association of p120ctn with cadherins is not necessary to control spine density because p120ctnΔarm3 was as effective as wild-type (wt) p120ctn at restoring normal spine density (Figures 5C, 5C′, 5E, 5E′, and 5I). Many of these spines formed synaptic contacts as confirmed by colocalization of the spine heads with synaptophysin puncta (Figures S2D and S2D′). In contrast, expression of p120ctnΔ622–628 was completely ineffective at promoting spine formation in the mutant neurons, suggesting that regulation of spine density by p120ctn is through control of Rho activity (Figures 5D, 5D′, and 5I). To obtain further evidence that p120ctn controls spine density through Rho-family protein regulation, we examined the ability of a constitutively active Rac mutant, Rac1G12V, and a specific inhibitor of Rho, C3-transferase, to regulate spine density in mutant neurons. Expression of either protein dramatically increased the density of spines (Figures 5F, 5F′, 5G, and 5G′). Constitutively active Rac1 restored the normal density of spines, while RhoA suppression resulted in an abnormally high spine density (Figure 5I).
To further distinguish whether the observed reduction in cadherin levels in the p120ctn mutant mice contributed to the spine density deficit, we determined whether cadherin overexpression restored spine numbers. Previous studies have shown that in cells with reduced cadherin as a consequence of p120ctn siRNA knockdown, cadherin levels can be restored by cadherin overexpression (Davis et al., 2003). We overexpressed full-length N-cadherin in p120ctn mutant neurons at DIV10 and analyzed spine density at DIV14. N-cadherin overexpression did not promote spine formation and restore spine numbers (Figures 5H, 5H′, and 5I).
While spine density was restored by Rac activation or Rho inhibition, spine morphology appeared abnormal in neurons lacking active Rho. This is consistent with previous observations indicating that misregulation of Rho-family G proteins perturbs spine morphology (Govek et al., 2005). Overall, our results suggest that modulation of Rho-family protein activity by p120ctn is sufficient to establish normal spine density.
Altered Morphology of Spines in p120ctn−/− Neurons In Vitro
Spine morphogenesis involves many proteins in addition to the Rho-family GTPases. Different proteins and signaling pathways have been implicated in initial steps of spine development, in which filopodial-like protrusions are formed, and later steps that control spine and synapse. In examinations of spine morphology, quantification of spine length showed an almost 66% reduction in length in mutant neurons (Figure 5J). When the mutant neurons were transfected with wild-type p120ctn, p120ctnΔarm3, N-cadherin, C3-transferase, or Rac1G12V, spine lengths were restored to normal (Figure 5J). Interestingly, expression of p120ctnΔ622–628 did not increase spine length significantly, similar to its failure to increase spine density.
Spine head width is a morphological feature used to assess spine maturation. The average head width was significantly reduced by approximately 20% in the mutant neurons (Figure 5K). This maturation deficit was completely rescued by expression of p120ctn. Interestingly, it was also rescued by p120ctnΔ622–628, indicating that p120ctn-mediated inhibition of Rho is not required. Consistent with this, suppression of Rho with C3-transferase did not rescue average head width. Thus, regulation by p120ctn of spine head width cannot be mediated solely through regulation of Rho. In contrast, p120ctnΔarm3, a mutant not able to bind to cadherins, was ineffective at restoring normal head width and its presence actually resulted in a further reduction in average head width below that observed in mutant neurons alone. Expression of N-cadherin or of active Rac did partially restore head width. Our data suggest that p120ctn must interact with cadherins and regulate Rac activity to control spine head shape, whereas modulation of Rho activity alone appears sufficient to regulate spine density and length.
p120ctn Deletion In Vitro also Results in Reduced Spine Density
To distinguish potential differences between prolonged versus acute loss of p120ctn on spine formation, we deleted p120ctn from cultured p120ctnflox/flox neurons at DIV10 through expression of Cre (Figure 6). At this time the neurons have already elaborated dendritic and axonal processes, but are just beginning to form spines. Cre recombinase-driven loss of p120ctn at DIV10 led to a 30% decrease in spine density, similar to what was observed in the p120ctnflox/flox;emx1IREScre cultured mutant neurons (Figure 6H). Spines were restored by expression of wild-type p120ctn, p120ctnΔarm3, Rac1-G12V, or C3-transferase, similar to what was seen in the rescue experiments performed in the p120ctnflox/flox; emx1IREScre cultured neurons. Similar to results using neurons from p120ctnflox/flox;emx1IREScre mice, expression of p120ctnΔ622–628, which is unable to regulate Rho activity, was not able to rescue spine density (Figures 6D, 6D′, and 6H). Thus, p120ctn must be present during the time of spine formation for normal spine development.
Figure 6.
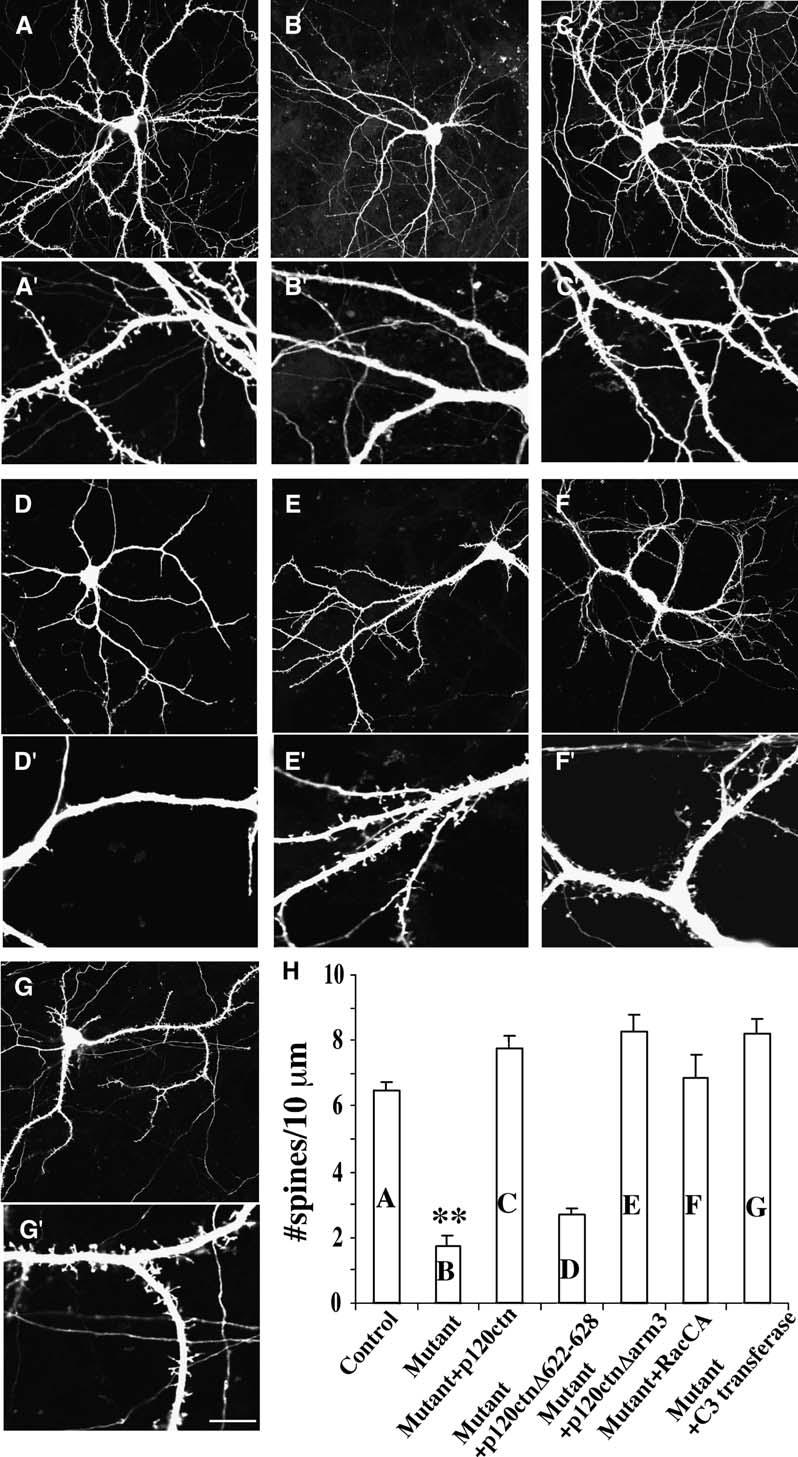
Reduced Spine Densities following Cre-Mediated Knockdown of p120ctn In Vitro
(A–G) Representative examples of EGFP-transfected hippocampal neurons cultured from p120ctn flox/flox mice transfected at DIV10 with indicated vectors and analyzed at DIV14. Low- (A–G) and high-magnification (A′–G′) images are shown. (A and A′) Control neuron (n = 9 cells). (B and B′) Neuron cotransfected with Cre (n = 9 cells). (C and C′) Neuron cotransfected with Cre and p120ctn (n = 7 cells). (D and D′) Neuron cotransfected with Cre and p120ctnΔ622–628 (n = 5 cells). (E and E′) Neuron cotransfected with Cre and p120ctnΔarm3 (n = 7 cells). (F and F′) Neuron cotransfected with Cre and rac1-V12 (RacCA) (n = 6 cells). (G and G′) Neuron cotransfected with Cre and C3 transferase (n = 7 cells).
(H) Mean spine density per 10 μm dendritic length (*p < 0.001; **p < 0.0001). Three independent experiments were analyzed.
Scale bar, 40 μm ([A]–[G]); 10 μm ([A′]–[G′]).
Interestingly, overexpression of p120ctn in cultured rat hippocampal neurons also resulted in altered spine morphology. By DIV14, control neurons have formed extensive dendritic trees from which protrude mature, stubby spines with broad heads (Figures 7A and 7A′). The average length of these spines was approximately 2 μm with virtually no spines having a length over 4 μm (Figure 7A″). In contrast, after p120ctn overexpression at DIV10, the spines assumed long filopodia-like morphologies with an average length substantially higher than 4 μm (Figures 7B, 7B′, and 7B″). In these neurons, spine lengths were highly variable, ranging from 2 to 10 μm. In contrast to controls, these spines did not have mature broad heads. This effect was prevented by coexpression of constitutively active RhoA (Figures 7C-7C″). Constitutively active Rho reduced spine density to approximately the same extent as observed in floxed murine p120ctn mutant neurons (Figures 7D-7D″). Rho suppression by C3-transferase increased spine length and suppressed spine maturation, but not as dramatically as observed following p120ctn overexpression (Figures 7E-7E″). The Rho phenotypes are similar to those in previous reports by others (Tashiro et al., 2000). The results indicate that p120ctn acts to control spine growth and maturation through Rho, but its effects are not completely explained by this pathway.
Figure 7.
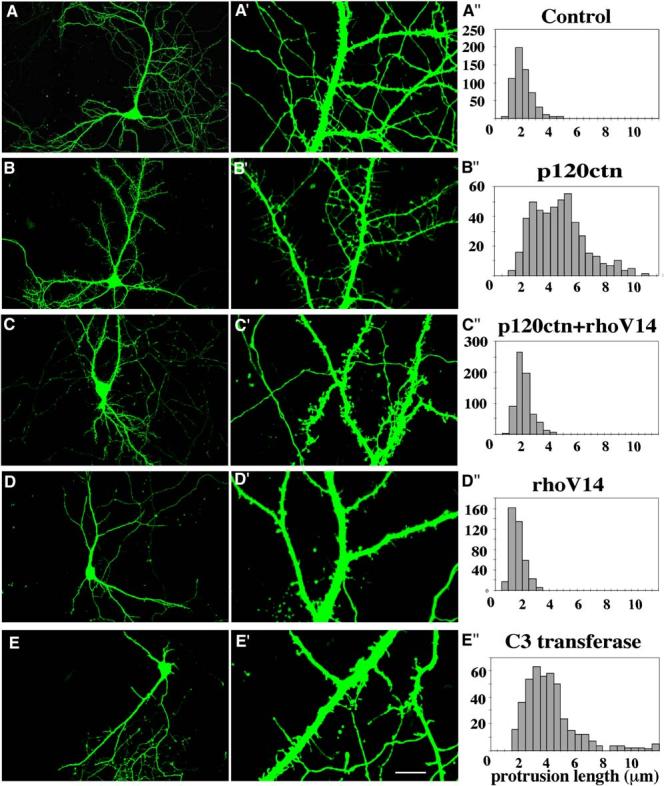
Altered Spine Morphologies in Rat Hippocampal Neurons Overexpressing Wild-Type or Mutant p120ctn
(A–J) Examples of DIV10 EGFP-transfected neurons analyzed at DIV14. Low- (A–E) and high- (A′–B′) magnification images are shown. (A and A′) Control neuron (n = 11 cells). (B and B′) Neuron cotransfected with p120ctn (n = 11 cells). (C and C′) Neuron cotransfected with p120ctn and RhoA-V14 (CA-Rho) (n = 9 cells). (D and D′) Neuron cotransfected with RhoA-V14 (CA-Rho) (n = 6 cells). (E and E′) Neuron cotransfected with C3 transferase (n = 7 cells). (A″–E″) Spine length distribution for each condition.
Scale bar, 40 μm ([A]–[E]); 10 μm ([A′]–[E′]).
p120ctn Deletion in Mature Neurons In Vitro Reduces Mature Spine Density
Recent studies have shown that loss of αN-catenin results in spine instability (Abe et al., 2004). Our culture results suggest that p120ctn contributes to spine maturation or stabilization in part through interaction with the cadherin complex. To determine if p120ctn is required for spine stabilization following formation, cultured p120ctnflox/flox hippocampal neurons were transfected with cre to delete p120ctn after 14 days of normal development, by which time mature spines are established. Loss of p120ctn led to the appearance of spines with features suggestive of increased morphological plasticity, such as an increase in protrusion length and the presence of spines with filopodia emerging from the spine head (Figures S3B and S3B′; see also Table S1 in the Supplemental Data). We also detected a decrease in the density of mature spines and an increase in the density of filopodial-like protrusions (Table S1). These results suggest that p120ctn is required to maintain or constrain spines in mature neurons and that loss of p120ctn may result in an increase in spine plasticity.
Reduced Synapse Density in Mutant Neurons In Vitro
To determine whether p120ctn regulates synaptic density in vitro, we quantified the number of potential synapses in control and p120ctn mutant neurons by staining with antibodies specific for synaptic proteins (PSD95, postsynaptic; synaptophysin, presynaptic; N-cadherin, presynaptic and postsynaptic). Analysis of PSD95 puncta along dendritic processes from single neurons was made possible by expression of GFP within a small proportion of the neurons. Punctate staining was detected along the GFP-labeled dendrites, with clusters associated with GFP-labeled spine heads (Figures 8F and 8G). In the neurons lacking p120ctn, PSD95 staining was still punctate, but the density of puncta was dramatically reduced to 35% of that observed in control neurons (Figures 8H-8J). A similar difference was observed between control and mutant neurons transfected with a PSD95-GFP reporter with the density of PSD95-GFP puncta in mutant dendrites approximately 40% of normal (not shown).
Figure 8.
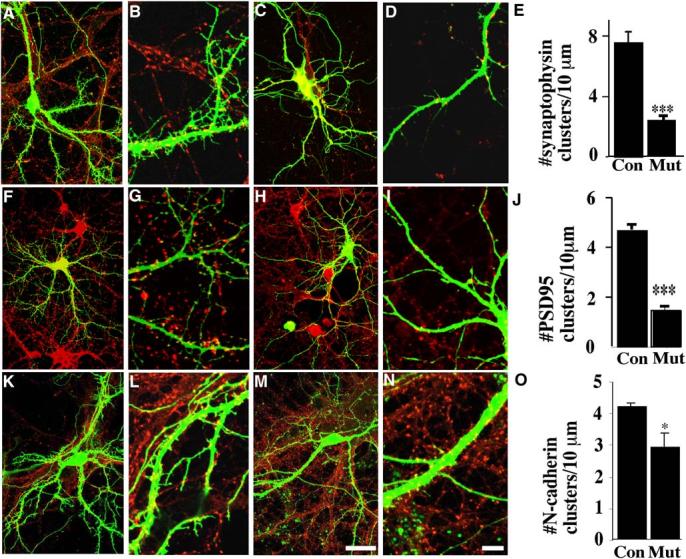
Reduced Density of Synaptic Clusters in p120ctn Mutant Neurons
(A–D) Examples of hippocampal neurons cultured from ([A] and [B]) control p120ctnflox/flox or ([C] and [D]) mutant p120ctnflox/flox;emx1IREScre mice transfected at DIV10 with EGFP and analyzed at DIV14 after labeling with anti-synaptophysin.
(E) Mean density of synaptophysin puncta (control, n = 6 cells; mutant, n = 7 cells) (***p < 0.0001). Examples of control ([F] and [G]) or mutant ([H] and [I]) neurons transfected at DIV10 with EGFP and analyzed at DIV14 after labeling with anti-PSD95.
(J) Mean density of PSD95 clusters (control and mutant, n = 8 cells) (***p < 0.0001). Examples of neurons cultured from control ([K] and [L]) or mutant ([M] and [N]) neurons transfected at DIV10 with EGFP and analyzed at DIV14 after labeling with anti-N-cadherin.
(O) Mean density per 10 μm length of N-cadherin puncta (control and mutant, n = 4 cells) (*p < 0.05). Approximately equal numbers of neurons from each of three independent experiments were used in these analyses. Scale bars, 40 μm for (A), (C), (F), (H), (K), and (M); 10 μm for (B), (D), (G), (I), (L), and (N).
We also quantified the densities of synaptic vesicle clusters in contact with GFP-labeled dendrites using anti-synaptophysin (Figures 8A-8D). Synaptophysin was distributed in a punctate pattern along the GFP-labeled dendrites in both control (p120ctnflox/flox) and mutant (p120ctnflox/flox;emx1IREScre) neurons, located adjacent to dendritic spine heads and less frequently along dendritic shafts. The density of synaptophysin puncta apposed to dendrites of mutant neurons was reduced by approximately 65% (Figure 8E).
Finally, we examined the pattern of cadherin expression in the mutant neurons by staining the cultures with anti-N-cadherin. It had been previously reported that p120ctn colocalizes with a subset of N-cadherin-positive synaptic puncta in cultured hippocampal neurons (Chauvet et al., 2003). N-cadherin protein was localized to spine heads and dendritic shafts in both control and mutant neurons with a 32% reduction in puncta density in mutant neurons (Figures 8K-8O).
Discussion
Our results indicate that the cadherin-associated protein, p120ctn, is required for normal spine and synapse development by hippocampal pyramidal neurons in vivo. The absence of p120ctn dramatically reduces the density of spines and synapses formed on their dendrites. Our results also demonstrate that p120ctn controls levels of active Rac and Rho as well as levels of N-cadherin. In its absence there are significant reductions in N-cadherin and active Rac1, together with an increase in active RhoA. To understand the mechanisms through which p120ctn regulates dendrite development, we have examined the development of hippocampal neurons lacking p120ctn in cell culture. Results indicate that absence of p20ctn diminishes spine density primarily through misregulation of Rho. p120ctn appears to act independently of its role in regulation of cadherin function. In addition, our results suggest that p120ctn functions in a cadherin-mediated pathway to promote spine maturation. p120ctn may promote spine maturation through control of cadherin-mediated adhesion. In addition, it may also regulate spine maturation through a signalling pathway initiated by cadherin ligation, such as Rac activation.
Our results are generally consistent with two recently published papers describing consequences of p120ctn ablation in the skin and submaxillary gland (Davis and Reynolds, 2006; Perez-Moreno et al., 2006). In each tissue, loss of p120ctn resulted in reduced surface expression of cadherins. Analyses of Rho activity indicated that there was elevated active Rho in mutant keratinocytes in vivo and in vitro (Perez-Moreno et al., 2006).
In our experiments, the absence of p120ctn resulted in global perturbations of Rac1 and RhoA activity in the forebrain with an elevation in active RhoA and reduction in active Rac1, consistent with prior studies on cultured cells where p120ctn overexpression was shown to stimulate Rac and inhibit Rho (Anastasiadis et al., 2000; Grosheva et al., 2001; Noren et al., 2000). Prior work has implicated Rac and Rho in dendrite growth and spine morphogenesis. In cultured hippocampal neurons, Rac1 inhibition results in loss of dendritic spines and synapses (Nakayama et al., 2000; Tashiro et al., 2000). Active RhoA leads to reduced spine density and length, and simplification of the dendritic tree (Nakayama et al., 2000; Tashiro et al., 2000; Tashiro and Yuste, 2004). Thus, our observations suggest that p120ctn deficiency reduces dendrite growth and spine density through regulation of these two Rho-family G proteins throughout the neuron and not solely at cadherin-dependent adhesion sites. In addition, spine density was restored in p120ctn-deficient neurons by activation of Rac or suppression of Rho. In attempted rescues of p120ctn deficiency, spine density was restored by a p120ctn mutant unable to interact with cadherins, but not by a mutant unable to inhibit Rho activity. This mutant promotes Rac activation, suggesting that misregulation of Rho alone is sufficient to cause the deficit in spine formation in p120ctn-deficient neurons. These experiments indicate that p120ctn deficiency inhibits spine formation through global activation of Rho. As Rac, Rho, and p120ctn mutants were overexpressed, our observations do not exclude the possibility that, when present at normal levels, p120ctn promotion of spine formation is facilitated through its interactions with cadherins.
Consistent with previous studies demonstrating roles for Rac and Rho in regulating dendrite development (Nakayama et al., 2000; Sin et al., 2002), we found that absence of p120ctn inhibits dendritic tree development by embryonic neurons in vitro. Except for the prominent deficit in spine formation, however, absence of p120ctn did not alter overall dendritic morphology of the same neurons at 6 weeks in vivo. There may be a transient delay of dendrite arbor development in vivo that is fully compensated before 6 weeks. Alternatively, many known regulators of dendritic growth could potentially compensate for absence of p120ctn in vivo. Intrinsic signals include other members of the p120ctn family, such as δ-catenin. A δ-catenin mutant has recently been characterized and also does not exhibit obvious deficits in brain structure or dendrite morphology in adult mice in vivo (Israely et al., 2004). In contrast to mice lacking p120ctn, however, mice lacking δ-catenin have apparently normal numbers of spines on CA1 neurons. Since δ-catenin also regulates Rac and Rho in vitro, it is somewhat surprising that this mutant does have an obvious deficit in spine formation.
A recent study also has indicated a role in spine-like protrusion formation by Drosophila sensory neurons for the single Drosophila p120ctn/δ-catenin homolog (Li et al., 2005). Although these processes do not make synaptic connections, they are actin-rich and morphologically similar to immature mammalian neuron spines. Interactions with Rac, Rho, and cadherin-mediated signaling in regulation of spine-like protrusion formation were not explored in this paper.
Our data also suggest that p120ctn deficiency affects spine maturation through a cadherin signaling pathway that promotes spine head expansion. In experiments using p120ctn-deficient neurons, a p120ctn deletion mutant able to associate with cadherins, but not regulate Rho, restored spine head width, while a mutant unable to associate with cadherins was ineffective even though it regulates Rho function normally (Anastasiadis et al., 2000). Consistent with this, N-cadherin overexpression partially rescued spine head width, although it did not suppress the deficiency in spine density. The cadherin-catenin complex has been previously shown to regulate the formation, shape, and dynamics of spines and synapses. Inhibition of cadherin function through direct cadherin inhibition or deletion of αN-catenin increases spine length and decreases spine head width in hippocampal neurons; αN-catenin deletion also destabilizes spines and their synaptic contacts (Abe et al., 2004; Togashi et al., 2002). Cadherin inhibition also inhibits formation of PSD95 clusters. In contrast, deletion of β-catenin or αN-catenin does not disrupt PSD95 clustering (Bamji et al., 2003; Togashi et al., 2002).
p120ctn may influence spine and synapse morphogenesis, in part, through its promotion of cadherin stability. Consistent with observations in other cells (Davis et al., 2003; Xiao et al., 2003), our data demonstrate that p120ctn loss results in reduced cadherin levels, probably because of the accelerated endocytosis and degradation of cadherins observed in these other cells. Lower cadherin levels may reduce adhesive interactions at synapses that influence spine head size and synapse formation (Murase et al., 2002).
In addition, p120ctn enhances cadherin function through stabilization of cadherin interactions with the cytoskeleton (Xu et al., 2004). In neuroepithelial cells, p120ctn promotes cadherin-dependent cell adhesion and neurite outgrowth by recruitment of the tyrosine kinase Fer. Fer- and p120ctn-dependent recruitment of the protein tyrosine phosphatase PTP1B enhances the interaction of cadherins with β-catenin through dephosphorylation of a phosphotyrosine residue on β-catenin. In hippocampal neurons, activity-dependent dephosphorylation of this phosphotyrosine residue increases the affinity of β-catenin for cadherins and promotes recruitment of β-catenin to spines, increasing synapse size and spontaneous synaptic transmission (Murase et al., 2002). The pathway through which p120ctn controls PTP1B activity may also control cadherin function in neurons. This can be explored with cadherin-α-catenin fusions that bypass the requirement for β-catenin to link cadherins to the cytoskeleton (Pacquelet and Rorth, 2005).
Studies have also shown that cadherins are distributed diffusely throughout the plasma membrane of dendritic filopodia, but become concentrated at axon contacts (Togashi et al., 2002). This clustering may depend upon interactions with the cytoskeleton and be promoted by p120ctn. Work has shown that association of p120ctn with the cadherin juxtamembrane domain recruits Rac1 to cadherin contacts, leading to adhesion strengthening through cadherin clustering and adhesion contact zone extension (Gavard et al., 2004; Yap et al., 1998).
Finally, p120ctn interacts with kinesins and microtubules and facilitates directed cadherin transport to adhesion sites (Chen et al., 2003; Franz and Ridley, 2004; Yanagisawa et al., 2004). In neuroepithelia, the kinesin KIF3 is crucial for N-cadherin transport and organization of adhesive contacts (Teng et al., 2005). The role of kinesins in cadherin transport in neurons has not been explored.
p120ctn may also influence spine maturation as an effector of cadherin-initiated signaling. First, cadherin ligation has been shown to promote Rac recruitment of Rac to adhesion sites through p120ctn-dependent mechanisms (Yap et al., 1998). Active Rac is required to establish normal head width (Tashiro and Yuste, 2004), so cadherin-coupled p120ctn may promote spine head enlargement through Rac-dependent regulation of the Arp2/3 complex. δ-catenin, a p120ctn homolog, recruits cortactin, an F-actin cross-linking protein that also promotes F-actin polymerization through activation of the Arp2/3 complex (Martinez et al., 2003). Recently, p120ctn has also been shown to interact with cortactin (A. Bershadsky and B. Geiger, personal communication). Cortactin promotes spine formation and exhibits rapid redistribution between the dendritic shaft and spine in response to stimuli that control spine formation and shape (Gray et al., 2005; Hering and Sheng, 2003). Cortactin also controls maturation of spines through interactions with the postsynaptic density protein Shank (Hering and Sheng, 2003). Shank forms a complex with the scaffold Git1, the protein kinase PAK, and the GEF β-Pix, each of which promote spine maturation (Park et al., 2003; Zhang et al., 2005). Consequently, p120ctn may control initiation and maturation of spines in part through interactions with cortactin.
It will be interesting to determine whether signaling pathways that impact spine formation and morphology, such as electrical activity and glutamate and Eph receptor activation, act through regulation of p120ctn function. Both extracellular and intracellular proteins modulate p120ctn's phosphorylation state (Cozzolino et al., 2003; Esser et al., 1998; Shibamoto et al., 1995; Xia et al., 2003). Membrane-localized, cadherin-coupled p120ctn is phosphorylated on tyrosine and serine/threonine residues, whereas unbound p120ctn has a lower level of tyrosine phosphorylation (Reynolds and Roczniak-Ferguson, 2004). Phosphorylation may determine whether p120ctn has adhesion promoting versus adhesion inhibiting effects (Aono et al., 1999; Kinch et al., 1995; Ozawa and Ohkubo, 2001). Tyrosine phosphorylation of p120ctn correlates with increased cadherin binding affinity and serine/threonine phosphorylation correlates with negative regulation of cadherin adhesion. Membrane localization also clearly correlates with the ability of p120ctn to inhibit Rho activity (Grosheva et al., 2001). Unfortunately, with few exceptions (Ozawa and Ohkubo, 2001), these studies are mostly correlative.
Finally, it will be interesting to characterize the roles of p120ctn family members in synaptic plasticity. The CA1 synapses from a mouse lacking δ-catenin exhibited several abnormal properties, including deficits in short- and long-term plasticity (paired-pulse facilitation and long-term potentiation) (Israely et al., 2004). p120ctn is associated with NMDA receptors (Husi et al., 2000), so it is strategically placed to influence synapse function. As potential targets, EphR expression and localization are regulated by cadherins (Orsulic and Kemler, 2000; Zantek et al., 1999). Several studies have noted synaptic plasticity deficits in ephrin and EphR receptor mutants (Klein, 2004).
In conclusion, work presented here demonstrates an important role for the cadherin-associated protein p120ctn in promoting dendritic spine development. Our work indicates that p120ctn promotes spine development through multiple signaling pathways, including regulation of Rho-family GTPases. It will be important to characterize these signaling pathways in more detail and to examine their roles in brain function.
Experimental Procedures
Generation and Genotyping of the Floxed Allele of p120ctn
This allele was generated by standard techniques. Details are described in Supplemental Experimental Procedures.
Western Blotting, Immunocytochemistry, and Nissl Staining
Standard methods for blots, immunocytochemistry, and Nissl staining are described in detail in the Supplemental Experimental Procedures. Blots were probed with anti-p120ctn (Transduction labs), Ncadherin (Zymed), α-catenin (Zymed), or β-catenin (Zymed) mAbs. Anti-β-tubulin mAb (Sigma) was used for normalization. For immunofluoresence, sections were incubated with rabbit anti-synaptophysin (Zymed), mouse anti-PSD95 (D. Bredt), or rabbit anti-N-cadherin (D. Coleman).
Golgi Staining
Six-week-old male littermate brains were processed in parallel (n = 3) and stained using modified Golgi-Cox impregnation of neurons following the manufacturer's protocol (FD NeuroTechnologies). For spine density quantification the number of spines were counted along equivalent length dendritic segments proximal to the cell body for pyramidal neurons from the CA1 region of the hippocampi of p120ctnflox/flox and p120ctnflox/flox;emx1IREScre mice.
Electron Microscopy
Mice were perfused with 0.9% NaCl, followed by 2.5% glutaraldehyde and 1% paraformaldehyde in 0.1 sodium cacodylate buffer (pH 7.4). Following overnight fixation, 100 μm vibratome sections were cut, dehydrated, and embedded in Epon-Araldite. Semithin sections were stained with toluidine blue to identify the CA1 region of the hippocampus. After trimming, ultrathin sections were cut, stained with uranyl acetate and lead citrate, and photographed at the San Francisco VA Hospital EM facility. Quantification was carried out as described (Bamji et al., 2003).
Quantification of Fluorescent Puncta in Brain Sections
Anti-synaptophysin, anti-PSD95, or anti-N-cadherin puncta were examined with a Zeiss confocal microscope (100 ×, Zeiss Plan-Neofluor). The CA1 stratum radiatum layer was imaged for p120ctnflox/flox and p120ctnflox/flox;emx1IREScre mice (n = 3 pairs). All images were taken by Kalman averaging (4×) with the same laser settings. Binary images were obtained by using the background threshold of the p120ctnflox/flox sections for the corresponding sections from p120ctnflox/flox;emx1IREScre littermates. For particle analysis, the threshold was set at 5 pixels, the estimated minimal size of the labeled puncta, and particle numbers in areas of equivalent size (3172 μm2) were measured using NIH Image 1.62.
Hippocampal Cultures and Transfection
Hippocampal cultures were prepared as described (Bamji et al., 2003) and are detailed in the Supplemental Data. Neurons from individual hippocampi were cultured separately with retrospective genotyping. Comparisons were made using cultures from a single litter. Four independent litters were analyzed. Spines were defined as headed dendritic protrusions up to 2 μm and filopodia as headless protrusions > 2 μm.
RhoA and Rac1 Activity Assays
The RhoA and Rac1 assays were performed as described (Noren et al., 2000; Ren et al., 1999). Additional details are provided in the Supplemental Data.
Supplementary Material
Acknowledgments
We thank S. Bamji, H. Beggs, T. Elul, and B. Rico for discussions; Z. Huang, J. Arikkath, J. Linton, S. Lee, D. Marciano, and T. Elul for comments on the manuscript; A. Reynolds, P. Anastasiadis, and M. Davis for discussions and reagents; M. Symons, H. Bourne, D. Bredt, and D. Coleman for reagents; and S. Huling for assistance with electron microscopy. This work was supported by the Howard Hughes Medical Institute. L.F.R. is an H.H.M.I. investigator.
Footnotes
Supplemental Data
The Supplemental Data for this article can be found online at http://www.neuron.org/cgi/content/full/51/1/43/DC1/.
References
- Abe K, Chisaka O, Van Roy F, Takeichi M. Stability of dendritic spines and synaptic contacts is controlled by alpha N-catenin. Nat. Neurosci. 2004;7:357–363. doi: 10.1038/nn1212. [DOI] [PubMed] [Google Scholar]
- Anastasiadis PZ, Moon SY, Thoreson MA, Mariner DJ, Crawford HC, Zheng Y, Reynolds AB. Inhibition of RhoA by p120 catenin. Nat. Cell Biol. 2000;2:637–644. doi: 10.1038/35023588. [DOI] [PubMed] [Google Scholar]
- Aono S, Nakagawa S, Reynolds AB, Takeichi M. p120(ctn) acts as an inhibitory regulator of cadherin function in colon carcinoma cells. J. Cell Biol. 1999;145:551–562. doi: 10.1083/jcb.145.3.551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamji SX, Shimazu K, Kimes N, Huelsken J, Birchmeier W, Lu B, Reichardt LF. Role of beta-catenin in synaptic vesicle localization and presynaptic assembly. Neuron. 2003;40:719–731. doi: 10.1016/s0896-6273(03)00718-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdagi O, Valcin M, Poskanzer K, Tanaka H, Benson DL. Temporally distinct demands for classic cadherins in synapse formation and maturation. Mol. Cell. Neurosci. 2004;27:509–521. doi: 10.1016/j.mcn.2004.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvet N, Prieto M, Fabre C, Noren NK, Privat A. Distribution of p120 catenin during rat brain development: potential role in regulation of cadherin-mediated adhesion and actin cytoskeleton organization. Mol. Cell. Neurosci. 2003;22:467–486. doi: 10.1016/s1044-7431(03)00030-7. [DOI] [PubMed] [Google Scholar]
- Chen X, Kojima S, Borisy GG, Green KJ. p120 catenin associates with kinesin and facilitates the transport of cadherin-catenin complexes to intercellular junctions. J. Cell Biol. 2003;163:547–557. doi: 10.1083/jcb.200305137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozzolino M, Stagni V, Spinardi L, Campioni N, Fiorentini C, Salvati E, Alema S, Salvatore AM. p120 Catenin is required for growth factor-dependent cell motility and scattering in epithelial cells. Mol. Biol. Cell. 2003;14:1964–1977. doi: 10.1091/mbc.E02-08-0469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis MA, Reynolds AB. Blocked acinar development, E-cadherin reduction, and intraepithelial neoplasia upon ablation of p120-catenin in the mouse salivary gland. Dev. Cell. 2006;10:21–31. doi: 10.1016/j.devcel.2005.12.004. [DOI] [PubMed] [Google Scholar]
- Davis MA, Ireton RC, Reynolds AB. A core function for p120-catenin in cadherin turnover. J. Cell Biol. 2003;163:525–534. doi: 10.1083/jcb.200307111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunaevsky A, Tashiro A, Majewska A, Mason C, Yuste R. Developmental regulation of spine motility in the mammalian central nervous system. Proc. Natl. Acad. Sci. USA. 1999;96:13438–13443. doi: 10.1073/pnas.96.23.13438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elste AM, Benson DL. Structural basis for developmentally regulated changes in cadherin function at synapses. J. Comp. Neurol. 2006;495:324–335. doi: 10.1002/cne.20876. [DOI] [PubMed] [Google Scholar]
- Esser S, Lampugnani MG, Corada M, Dejana E, Risau W. Vascular endothelial growth factor induces VE-cadherin tyrosine phosphorylation in endothelial cells. J. Cell Sci. 1998;111:1853–1865. doi: 10.1242/jcs.111.13.1853. [DOI] [PubMed] [Google Scholar]
- Fannon AM, Colman DR. A model for central synaptic junctional complex formation based on the differential adhesive specificities of the cadherins. Neuron. 1996;17:423–434. doi: 10.1016/s0896-6273(00)80175-0. [DOI] [PubMed] [Google Scholar]
- Fiala JC, Feinberg M, Popov V, Harris KM. Synaptogenesis via dendritic filopodia in developing hippocampal area CA1. J. Neurosci. 1998;18:8900–8911. doi: 10.1523/JNEUROSCI.18-21-08900.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiala JC, Spacek J, Harris KM. Dendritic spine pathology: cause or consequence of neurological disorders? Brain Res. Brain Res. Rev. 2002;39:29–54. doi: 10.1016/s0165-0173(02)00158-3. [DOI] [PubMed] [Google Scholar]
- Franz CM, Ridley AJ. p120 catenin associates with microtubules: inverse relationship between microtubule binding and Rho GTPase regulation. J. Biol. Chem. 2004;279:6588–6594. doi: 10.1074/jbc.M312812200. [DOI] [PubMed] [Google Scholar]
- Gavard J, Lambert M, Grosheva I, Marthiens V, Irinopoulou T, Riou JF, Bershadsky A, Mege RM. Lamellipodium extension and cadherin adhesion: two cell responses to cadherin activation relying on distinct signalling pathways. J. Cell Sci. 2004;117:257–270. doi: 10.1242/jcs.00857. [DOI] [PubMed] [Google Scholar]
- Goodwin M, Kovacs EM, Thoreson MA, Reynolds AB, Yap AS. Minimal mutation of the cytoplasmic tail inhibits the ability of E-cadherin to activate Rac but not phosphatidylinositol 3-kinase: direct evidence of a role for cadherin-activated Rac signaling in adhesion and contact formation. J. Biol. Chem. 2003;278:20533–20539. doi: 10.1074/jbc.M213171200. [DOI] [PubMed] [Google Scholar]
- Gorski JA, Talley T, Qiu M, Puelles L, Rubenstein JL, Jones KR. Cortical excitatory neurons and glia, but not GABAergic neurons, are produced in the Emx1-expressing lineage. J. Neurosci. 2002;22:6309–6314. doi: 10.1523/JNEUROSCI.22-15-06309.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govek EE, Newey SE, Van Aelst L. The role of the Rho GTPases in neuronal development. Genes Dev. 2005;19:1–49. doi: 10.1101/gad.1256405. [DOI] [PubMed] [Google Scholar]
- Gray NW, Kruchten AE, Chen J, McNiven MA. A dynamin-3 spliced variant modulates the actin/cortactin-dependent morphogenesis of dendritic spines. J. Cell Sci. 2005;118:1279–1290. doi: 10.1242/jcs.01711. [DOI] [PubMed] [Google Scholar]
- Grosheva I, Shtutman M, Elbaum M, Bershadsky AD. p120 catenin affects cell motility via modulation of activity of Rho-family GTPases: a link between cell-cell contact formation and regulation of cell locomotion. J. Cell Sci. 2001;114:695–707. doi: 10.1242/jcs.114.4.695. [DOI] [PubMed] [Google Scholar]
- Hatzfeld M. The p120 family of cell adhesion molecules. Eur. J. Cell Biol. 2005;84:205–214. doi: 10.1016/j.ejcb.2004.12.016. [DOI] [PubMed] [Google Scholar]
- Hering H, Sheng M. Activity-dependent redistribution and essential role of cortactin in dendritic spine morphogenesis. J. Neurosci. 2003;23:11759–11769. doi: 10.1523/JNEUROSCI.23-37-11759.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C, Zhou J, Medina M, Goto T, Jacobson M, Bhide PG, Kosik KS. delta-catenin is a nervous system-specific adherens junction protein which undergoes dynamic relocalization during development. J. Comp. Neurol. 2000;420:261–276. [PubMed] [Google Scholar]
- Husi H, Ward MA, Choudhary JS, Blackstock WP, Grant SG. Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat. Neurosci. 2000;3:661–669. doi: 10.1038/76615. [DOI] [PubMed] [Google Scholar]
- Inoue A, Sanes JR. Lamina-specific connectivity in the brain: regulation by N-cadherin, neurotrophins, and glycoconjugates. Science. 1997;276:1428–1431. doi: 10.1126/science.276.5317.1428. [DOI] [PubMed] [Google Scholar]
- Israely I, Costa RM, Xie CW, Silva AJ, Kosik KS, Liu X. Deletion of the neuron-specific protein delta-catenin leads to severe cognitive and synaptic dysfunction. Curr. Biol. 2004;14:1657–1663. doi: 10.1016/j.cub.2004.08.065. [DOI] [PubMed] [Google Scholar]
- Iwai Y, Hirota Y, Ozaki K, Okano H, Takeichi M, Uemura T. DN-cadherin is required for spatial arrangement of nerve terminals and ultrastructural organization of synapses. Mol. Cell. Neurosci. 2002;19:375–388. doi: 10.1006/mcne.2001.1081. [DOI] [PubMed] [Google Scholar]
- Keirsebilck A, Bonne S, Staes K, van Hengel J, Nollet F, Reynolds A, van Roy F. Molecular cloning of the human p120ctn catenin gene (CTNND1): expression of multiple alternatively spliced isoforms. Genomics. 1998;50:129–146. doi: 10.1006/geno.1998.5325. [DOI] [PubMed] [Google Scholar]
- Kinch MS, Clark GJ, Der CJ, Burridge K. Tyrosine phosphorylation regulates the adhesions of ras-transformed breast epithelia. J. Cell Biol. 1995;130:461–471. doi: 10.1083/jcb.130.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein R. Eph/ephrin signaling in morphogenesis, neural development and plasticity. Curr. Opin. Cell Biol. 2004;16:580–589. doi: 10.1016/j.ceb.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Konur S, Yuste R. Developmental regulation of spine and filopodial motility in primary visual cortex: reduced effects of activity and sensory deprivation. J. Neurobiol. 2004;59:236–246. doi: 10.1002/neu.10306. [DOI] [PubMed] [Google Scholar]
- Li W, Li Y, Gao FB. Abelson, enabled, and p120 catenin exert distinct effects on dendritic morphogenesis in Drosophila. Dev. Dyn. 2005;234:512–522. doi: 10.1002/dvdy.20496. [DOI] [PubMed] [Google Scholar]
- Magie CR, Pinto-Santini D, Parkhurst SM. Rho1 interacts with p120ctn and alpha-catenin, and regulates cadherin-based adherens junction components in Drosophila. Development. 2002;129:3771–3782. doi: 10.1242/dev.129.16.3771. [DOI] [PubMed] [Google Scholar]
- Martinez MC, Ochiishi T, Majewski M, Kosik KS. Dual regulation of neuronal morphogenesis by a delta-catenin-cortactin complex and Rho. J. Cell Biol. 2003;162:99–111. doi: 10.1083/jcb.200211025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase S, Mosser E, Schuman EM. Depolarization drives beta-Catenin into neuronal spines promoting changes in synaptic structure and function. Neuron. 2002;35:91–105. doi: 10.1016/s0896-6273(02)00764-x. [DOI] [PubMed] [Google Scholar]
- Nakayama AY, Harms MB, Luo L. Small GTPases Rac and Rho in the maintenance of dendritic spines and branches in hippocampal pyramidal neurons. J. Neurosci. 2000;20:5329–5338. doi: 10.1523/JNEUROSCI.20-14-05329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noren NK, Liu BP, Burridge K, Kreft B. p120 catenin regulates the actin cytoskeleton via Rho family GTPases. J. Cell Biol. 2000;150:567–580. doi: 10.1083/jcb.150.3.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamura K, Tanaka H, Yagita Y, Saeki Y, Taguchi A, Hiraoka Y, Zeng LH, Colman DR, Miki N. Cadherin activity is required for activity-induced spine remodeling. J. Cell Biol. 2004;167:961–972. doi: 10.1083/jcb.200406030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orsulic S, Kemler R. Expression of Eph receptors and ephrins is differentially regulated by E-cadherin. J. Cell Sci. 2000;113:1793–1802. doi: 10.1242/jcs.113.10.1793. [DOI] [PubMed] [Google Scholar]
- Ozawa M, Ohkubo T. Tyrosine phosphorylation of p120(ctn) in v-Src transfected L cells depends on its association with E-cadherin and reduces adhesion activity. J. Cell Sci. 2001;114:503–512. doi: 10.1242/jcs.114.3.503. [DOI] [PubMed] [Google Scholar]
- Pacquelet A, Rorth P. Regulatory mechanisms required for DE-cadherin function in cell migration and other types of adhesion. J. Cell Biol. 2005;170:803–812. doi: 10.1083/jcb.200506131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park E, Na M, Choi J, Kim S, Lee JR, Yoon J, Park D, Sheng M, Kim E. The Shank family of postsynaptic density proteins interacts with and promotes synaptic accumulation of the beta PIX guanine nucleotide exchange factor for Rac1 and Cdc42. J. Biol. Chem. 2003;278:19220–19229. doi: 10.1074/jbc.M301052200. [DOI] [PubMed] [Google Scholar]
- Park JI, Kim SW, Lyons JP, Ji H, Nguyen TT, Cho K, Barton MC, Deroo T, Vleminckx K, Moon RT, McCrea PD. Kaiso/p120-catenin and TCF/beta-catenin complexes coordinately regulate canonical Wnt gene targets. Dev. Cell. 2005;8:843–854. doi: 10.1016/j.devcel.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Perez-Moreno M, Davis MA, Wong E, Pasolli HA, Reynolds AB, Fuchs E. p120-Catenin Mediates Inflammatory Responses in the Skin. Cell. 2006;124:631–644. doi: 10.1016/j.cell.2005.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash S, Caldwell JC, Eberl DF, Clandinin TR. Drosophila N-cadherin mediates an attractive interaction between photoreceptor axons and their targets. Nat. Neurosci. 2005;8:443–450. doi: 10.1038/nn1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren XD, Kiosses WB, Schwartz MA. Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. EMBO J. 1999;18:578–585. doi: 10.1093/emboj/18.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds AB, Roczniak-Ferguson A. Emerging roles for p120-catenin in cell adhesion and cancer. Oncogene. 2004;23:7947–7956. doi: 10.1038/sj.onc.1208161. [DOI] [PubMed] [Google Scholar]
- Shibamoto S, Hayakawa M, Takeuchi K, Hori T, Miyazawa K, Kitamura N, Johnson KR, Wheelock MJ, Matsuyoshi N, Takeichi M, et al. Association of p120, a tyrosine kinase substrate, with E-cadherin/catenin complexes. J. Cell Biol. 1995;128:949–957. doi: 10.1083/jcb.128.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sin WC, Haas K, Ruthazer ES, Cline HT. Dendrite growth increased by visual activity requires NMDA receptor and Rho GTPases. Nature. 2002;419:475–480. doi: 10.1038/nature00987. [DOI] [PubMed] [Google Scholar]
- Singh G, Lykke-Andersen J. New insights into the formation of active nonsense-mediated decay complexes. Trends Biochem. Sci. 2003;28:464–466. doi: 10.1016/S0968-0004(03)00176-2. [DOI] [PubMed] [Google Scholar]
- Stepanyants A, Tamas G, Chklovskii DB. Class-specific features of neuronal wiring. Neuron. 2004;43:251–259. doi: 10.1016/j.neuron.2004.06.013. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Shan W, Phillips GR, Arndt K, Bozdagi O, Shapiro L, Huntley GW, Benson DL, Colman DR. Molecular modification of N-cadherin in response to synaptic activity. Neuron. 2000;25:93–107. doi: 10.1016/s0896-6273(00)80874-0. [DOI] [PubMed] [Google Scholar]
- Tashiro A, Yuste R. Regulation of dendritic spine motility and stability by Rac1 and Rho kinase: evidence for two forms of spine motility. Mol. Cell. Neurosci. 2004;26:429–440. doi: 10.1016/j.mcn.2004.04.001. [DOI] [PubMed] [Google Scholar]
- Tashiro A, Minden A, Yuste R. Regulation of dendritic spine morphology by the rho family of small GTPases: antagonistic roles of Rac and Rho. Cereb. Cortex. 2000;10:927–938. doi: 10.1093/cercor/10.10.927. [DOI] [PubMed] [Google Scholar]
- Teng J, Rai T, Tanaka Y, Takei Y, Nakata T, Hirasawa M, Kulkarni AB, Hirokawa N. The KIF3 motor transports N-cadherin and organizes the developing neuroepithelium. Nat. Cell Biol. 2005;7:474–482. doi: 10.1038/ncb1249. [DOI] [PubMed] [Google Scholar]
- Togashi H, Abe K, Mizoguchi A, Takaoka K, Chisaka O, Takeichi M. Cadherin regulates dendritic spine morphogenesis. Neuron. 2002;35:77–89. doi: 10.1016/s0896-6273(02)00748-1. [DOI] [PubMed] [Google Scholar]
- Trachtenberg JT, Chen BE, Knott GW, Feng G, Sanes JR, Welker E, Svoboda K. Long-term in vivo imaging of experience-dependent synaptic plasticity in adult cortex. Nature. 2002;420:788–794. doi: 10.1038/nature01273. [DOI] [PubMed] [Google Scholar]
- Uchida N, Honjo Y, Johnson KR, Wheelock MJ, Takeichi M. The catenin/cadherin adhesion system is localized in synaptic junctions bordering transmitter release zones. J. Cell Biol. 1996;135:767–779. doi: 10.1083/jcb.135.3.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia X, Mariner DJ, Reynolds AB. Adhesion-associated and PKC-modulated changes in serine/threonine phosphorylation of p120-catenin. Biochemistry. 2003;42:9195–9204. doi: 10.1021/bi034597h. [DOI] [PubMed] [Google Scholar]
- Xiao K, Allison DF, Buckley KM, Kottke MD, Vincent PA, Faundez V, Kowalczyk AP. Cellular levels of p120 catenin function as a set point for cadherin expression levels in microvascular endothelial cells. J. Cell Biol. 2003;163:535–545. doi: 10.1083/jcb.200306001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Craig AW, Greer P, Miller M, Anastasiadis PZ, Lilien J, Balsamo J. Continuous association of cadherin with beta-catenin requires the non-receptor tyrosine-kinase Fer. J. Cell Sci. 2004;117:3207–3219. doi: 10.1242/jcs.01174. [DOI] [PubMed] [Google Scholar]
- Yanagisawa M, Kaverina IN, Wang A, Fujita Y, Reynolds AB, Anastasiadis PZ. A novel interaction between kinesin and p120 modulates p120 localization and function. J. Biol. Chem. 2004;279:9512–9521. doi: 10.1074/jbc.M310895200. [DOI] [PubMed] [Google Scholar]
- Yap AS, Niessen CM, Gumbiner BM. The juxtamembrane region of the cadherin cytoplasmic tail supports lateral clustering, adhesive strengthening, and interaction with p120ctn. J. Cell Biol. 1998;141:779–789. doi: 10.1083/jcb.141.3.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Malenka RC. Beta-catenin is critical for dendritic morphogenesis. Nat. Neurosci. 2003;6:1169–1177. doi: 10.1038/nn1132. [DOI] [PubMed] [Google Scholar]
- Zantek ND, Azimi M, Fedor-Chaiken M, Wang B, Brackenbury R, Kinch MS. E-cadherin regulates the function of the EphA2 receptor tyrosine kinase. Cell Growth Differ. 1999;10:629–638. [PubMed] [Google Scholar]
- Zhang H, Webb DJ, Asmussen H, Niu S, Horwitz AF. A GIT1/PIX/Rac/PAK signaling module regulates spine morphogenesis and synapse formation through MLC. J. Neurosci. 2005;25:3379–3388. doi: 10.1523/JNEUROSCI.3553-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Homma KJ, Poo M.-m. Shrinkage of dendritic spines associated with long-term depression of hippocampal synapses. Neuron. 2004;44:749–757. doi: 10.1016/j.neuron.2004.11.011. [DOI] [PubMed] [Google Scholar]
- Ziv NE, Smith SJ. Evidence for a role of dendritic filopodia in synaptogenesis and spine formation. Neuron. 1996;17:91–102. doi: 10.1016/s0896-6273(00)80283-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


