Abstract
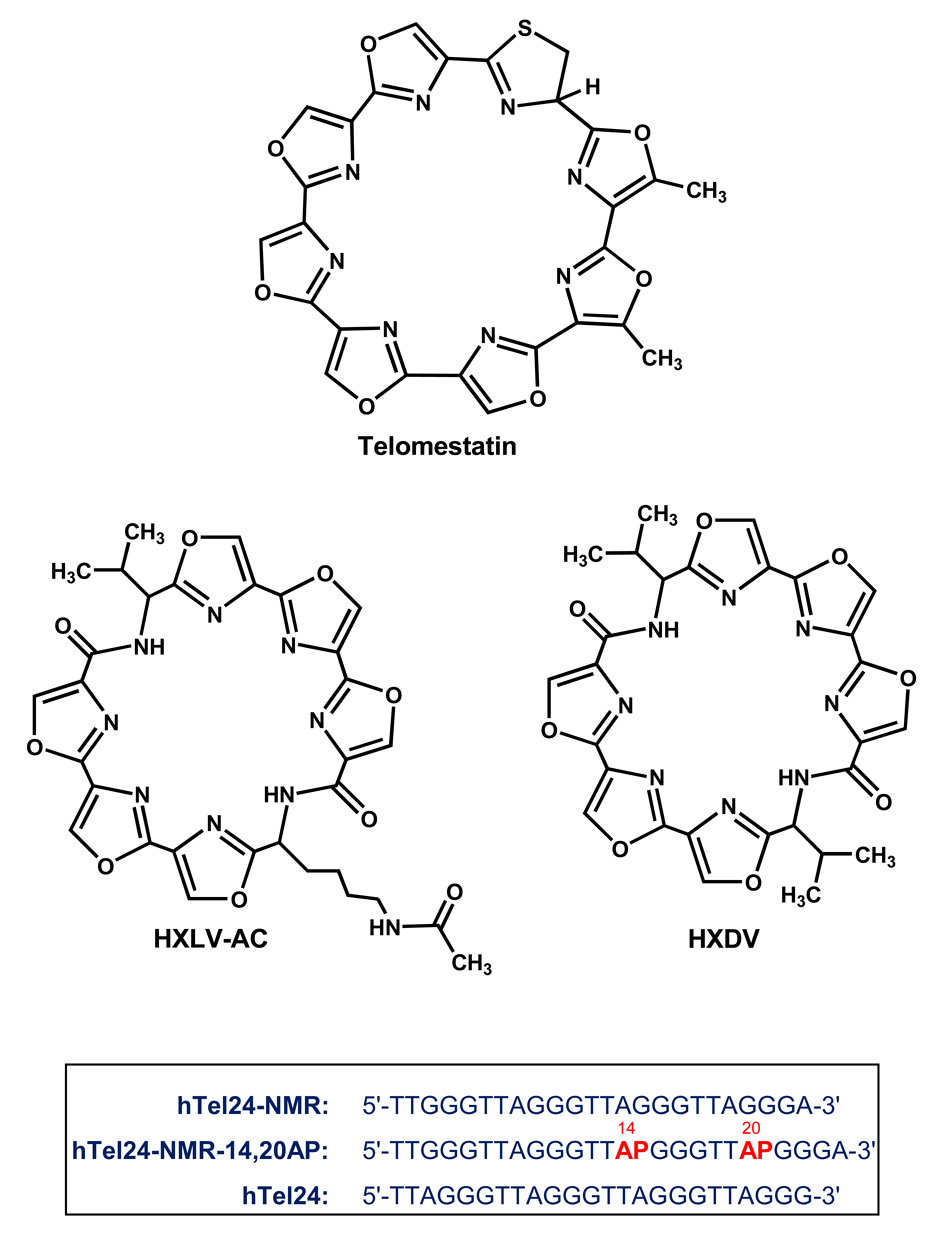
Oxazole-containing macrocycles, which include the natural product telomestatin, represent a promising class of anticancer agents that target G-quadruplex DNA. Two synthetic hexaoxazole-containing macrocyclic compounds (HXDV and HXLV-AC) have been characterized with regard to their cytotoxic activities versus human cancer cells, as well as the mode, thermodynamics, and specificity with which they bind to the intramolecular (3+1) G-quadruplex structural motif formed in the presence of K+ ions by human telomeric DNA. Both compounds exhibit cytotoxic activities versus human lymphoblast (RPMI 8402) and oral carcinoma (KB3-1) cells, with associated IC50 values ranging from 0.4 to 0.9 µM. The compounds bind solely to the quadruplex nucleic acid form, but not to the duplex or triplex form. Binding to the quadruplex is associated with a stoichiometry of two ligand molecules per DNA molecule, with one ligand molecule binding to each end of the host quadruplex via a nonintercalative “terminal capping” mode of interaction. For both compounds, quadruplex binding is primarily entropy driven, while also being associated with a negative change in heat capacity. These thermodynamic properties reflect contributions from favorable ligand-induced alterations in the loop configurational entropies of the quadruplex, but not from changes in net hydration. The stoichiometry and mode of binding revealed by our studies have profound implications with regard to the number of ligand molecules that can potentially bind the 3′-overhang region of human telomeric DNA.
1. Introduction
1.1 Telomere Maintenance, Telomerase, and the Shelterin Protein Complex
Telomeres are nucleoprotein structures located at the termini of eukaryotic chromosomes. These structures play a critical role in maintaining chromosomal integrity [1]. In this connection, the telomeres of human cells protect chromosomal ends from degradation and recombination events [1, 2]. Human telomeric DNA contains tandem repeats of the hexameric sequence 5′-T2AG3-3′ and ranges in size from 3 to 15 kb [3]. The 3′-ends of human telomeric DNA form single-stranded overhangs containing 16 to 35 repeats of the 5′-T2AG3-3′ sequence [4–8]. These single-stranded 3′-overhangs play an important role in determining telomeric structure and function [7, 8].
Without an active mechanism for the maintenance of telomeric structures, each round of human somatic cell division decreases telomeric length and ultimately the capacity for cellular replication [9–11]. The principle mechanism by which telomeres are maintained is through the actions of an enzyme termed telomerase [2, 12–14]. In human cells, telomerase is a ribonucleoprotein complex with reverse transcriptase activity that functions by adding multiple copies of the 5′-T2AG3-3′ sequence to the ends of telomeric 3′-overhangs [12–15]. Aside from its role in maintaining the integrity of the telomeric cap [12], telomerase also appears to be involved in the regulation of DNA damage response pathways [16].
In addition to telomerase, the telomeric binding proteins that form the shelterin complex are also involved in maintaining the structure of the telomere [2, 17, 18]. The shelterin proteins include POT1, TRF1, TRF2, TIN2, TPP1, and Rap1. TRF1 and TRF2 bind the duplex DNA domain of the telomere, while POT1 preferentially binds the single-stranded 3′-overhang. TIN2 binds TRF1 and TRF2, TPP1 binds TIN2 and POT1, and Rap1 binds TRF2. Thus, the shelterin complex appears to form a bridge between the duplex DNA region of the telomere and the single-stranded 3′-overhang [2]. It is thought to protect the 3′-end of the telomeric DNA from degradation while also modulating the accessibility of the 3′-overhang to telomerase [2]. References [2, 13, 14, 18] provide excellent reviews of the shelterin complex and telomerase, as well as their roles in telomere maintenance.
1.2 Telomerase and the Shelterin Proteins As Anticancer Drug Targets
Inhibition of telomerase activity is associated with telomeric shortening and subsequent activation of DNA damage responses that include cell cycle arrest, senescence, and apoptosis [19–26]. These antiproliferative effects make telomerase an appealing target for anticancer therapeutics [22, 23, 25–27], since the unlimited replication of cancer cells requires telomere maintenance [11]. This appeal is further bolstered by the observation that telomerase is overexpressed in 85 to 90% of human tumor cells, with most normal cells exhibiting little or no telomerase activity [22, 26–28]. Toward this end, telomerase-targeted therapy has been suggested for a number of cancers in which telomerase is overexpressed, including lung, malignant glioma, and leukemia [23–25, 27, 29].
Like telomerase inhibition, deprotection of telomeric DNA by inactivation of shelterin proteins can also activate DNA damage responses [30–32]. In certain human tumor cells (including melanoma, cervical carcinoma, fibrosarcoma, and ovarian carcinoma), telomere deprotection induced by overexpression of a dominant-negative form of TRF2 results in ATM-dependent apoptosis and senescence [30, 31]. Interestingly, inactivation of yeast Cdc13p (the yeast homolog of POT1) results in RAD9-dependent G2 arrest and MEC1-dependent apoptosis [33, 34]. Telomere deprotection may therefore activate a common ATM signaling pathway in both yeast and humans, since MEC1 is the functional yeast homolog of ATM.
1.3 Telomestatin Stabilizes Telomeric G-Quadruplex DNA, Inhibits Telomerase Activity, Causes Deprotection of Telomeric DNA, and Induces Cancer Cell Death
One of the more potent and selective inhibitors of telomerase to have been identified is telomestatin, a natural product isolated from Streptomyces anulatus 3533-SV4 [35, 36]. Telomestatin is a macrocyclic torand consisting of seven oxazole rings and one thiazoline ring (see structure in Fig. 1). It preferentially binds and stabilizes G-quadruplex relative to duplex DNA structures [36, 37]. The single-stranded 3′-overhangs of telomeric DNA can readily adopt thermodynamically stable G-quadruplex structures both in vitro [38–48] and in vivo [49, 50]. Telomestatin has been shown to bind and stabilize telomeric G-quadruplex DNA, which, in cancer cells, results in telomerase inhibition, telomeric shortening, anaphase bridge formation, and apoptosis [23, 25, 29, 35–37, 51–53]. Significantly, these effects are not observed in normal cells. The cytotoxic effects of telomestatin versus cancer cells have also been demonstrated in vivo using xenograft mouse models [25]. Recent studies have indicated that telomestatin induces the dissociation of TRF2 and POT1 from telomeres [52, 54, 55], suggesting that deprotection of telomeric DNA may also contribute to the cytotoxic activity of telomestatin versus cancer cells.
Fig. 1.
(Top) The chemical structures of telomestatin, HXDV, and HXLV-AC. (Bottom) Base sequences of the DNA oligomers discussed in this study. AP (in red) denotes the fluorescent base 2-aminopurine.
1.4 Development of Synthetic Oxazole-Containing Macrocyclic Compounds As G-Quadruplex Stabilizing Anticancer Agents
Compounds that selectively stabilize G-quadruplex DNA have the potential of inhibiting the activities of both telomerase and telomeric DNA binding proteins [56–61]. In addition to telomestatin, a diverse array of G-quadruplex-stabilizing compounds has been identified to date, including anthraquinones [62, 63], acridines [64–68], cationic porphyrins [69–78], bistriazoles [79], perylenes [80–82], ethidium derivatives [83, 84], fluorenones [85], pentacyclic acridinium salts [86–88], and fluoroquinophenoxazines [89, 90]. However, few of these compounds exhibit as high a degree of selectivity for G-quadruplex relative to duplex DNA as telomestatin [36]. This selectivity, coupled with the apoptotic activity exhibited by telomestatin in cancer cells, highlights oxazole-containing macrocyclic compounds as a promising new class of G-quadruplex stabilizing anticancer agents. Here, we describe studies of two synthetic macrocyclic hexaoxazoles that are toxic to human cancer cells. Aside from six oxazole moieties, one of these compounds (HXDV) also contains two valine residues, while the other (HXLV-AC) contains one valine residue and one lysine residue in which the 4-aminobutyl side chain has been N-acetylated (see structures of HXDV and HXLV-AC in Fig. 1). In addition to evaluating the antiproliferative properties of the two compounds versus two different human cancer cells lines, we also describe the mode, thermodynamics, and specificity with which the compounds bind to human telomeric G-quadruplex DNA.
2. Results and Discussion
2.1. Evaluating the Cytotoxic Activities of HXDV and HXLV-AC versus Human KB3-1 and RPMI 8402 Cancer Cells
We evaluated the cytotoxic activities of HXDV and HXLV-AV versus the human oral carcinoma cell line KB3-1, as well as the human lymphoblast cell line RPMI 8402. Both compounds exhibit similar cytotoxic potencies versus the two types of cancer cells, with IC50 values ranging from 0.4 to 0.9 µM (see Table 1). These cytotoxic activities compare favorably with those previously reported for telomestatin, which range from 0.8 to 5.0 µM [91].
Table 1.
Cytotoxicities of HXDV and HXLV-AC versus RPMI 8402 Human Lymphoblast and KB3-1 Human Oral Carcinoma Cellsa
| Compound | RPMI 8402 IC50 (µM) |
KB3-1 IC50 (µM) |
|---|---|---|
| HXDV | 0.4 ± 0.1 | 0.4 ± 0.1 |
| HXLV-AC | 0.8 ± 0.3 | 0.9 ± 0.2 |
Cytotoxicities were determined by standard MTT assay after four days of continuous compound exposure, and are reported as the compound concentrations at which cellular growth is 50% inhibited (IC50).
2.2 Defining the Nucleic Acid Binding Specificity of Oxazole-Containing Macrocycles
2.2.1 HXDV and HXLV-AC Binding Is Specific for Quadruplex Relative to Duplex or Triplex Nucleic Acid Forms
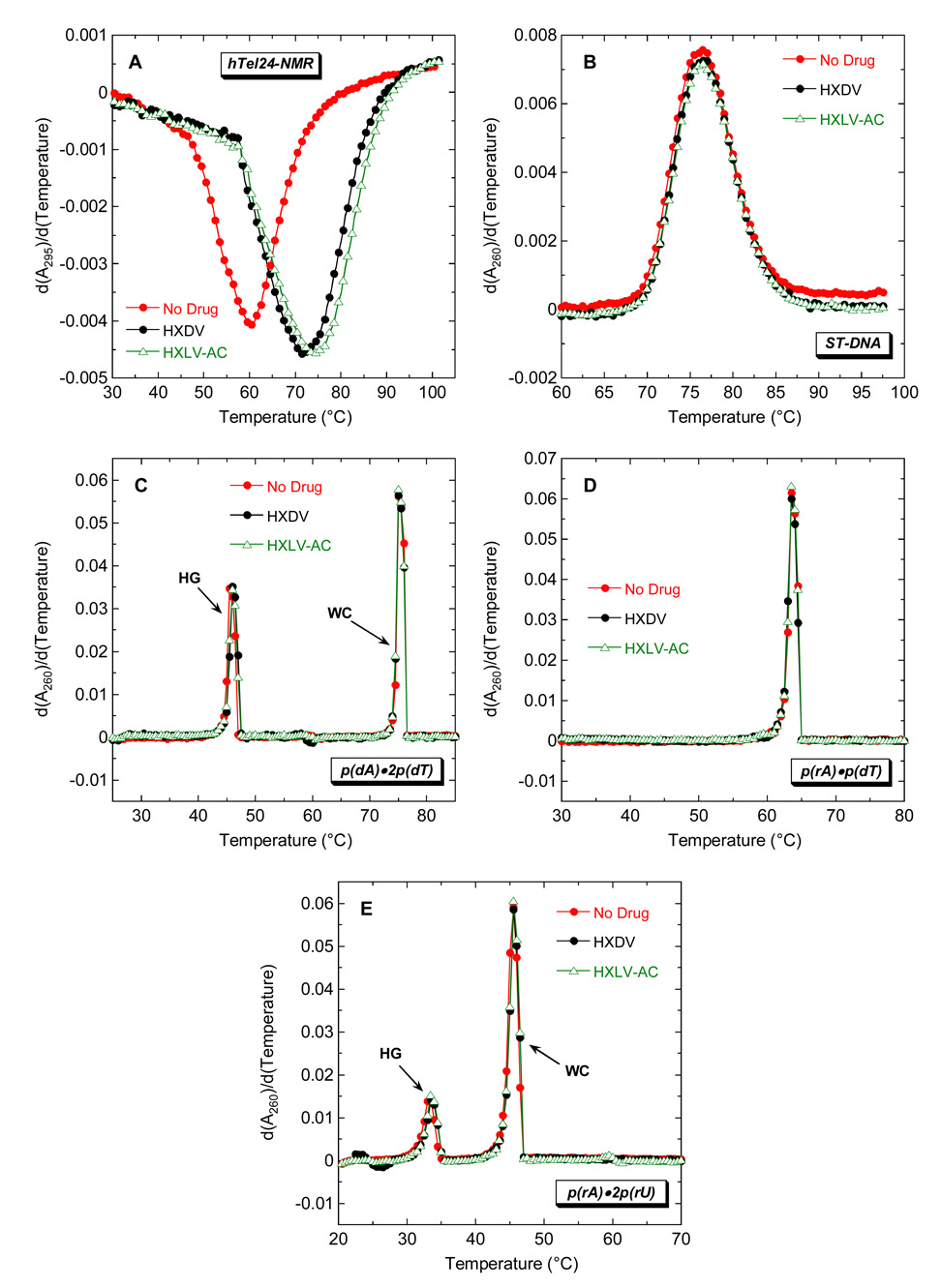
We evaluated the ability of HXDV and HXLV-AC to bind and thermally stabilize duplex, triplex, and quadruplex nucleic acids. Toward this end, we monitored the UV absorbances of the nucleic acids as a function of temperature in the absence and presence of either HXDV or HXLV-AC. The melting of duplex and triplex nucleic acids is generally associated with a hyperchromic shift at 260 nm [92, 93], while the melting of quadruplex nucleic acids is associated with a hypochromic shift at 295 nm [39, 94]. Thus, the temperature-dependent absorbances of duplexes and triplexes were monitored at 260 nm, with corresponding quadruplex absorbances being monitored at 295 nm. Salmon testes DNA (ST-DNA), p(rA)•p(rU), p(rA)•p(dT), p(dA)•2p(dT), and p(rA)•2p(rU) were used as representative models of duplex DNA, duplex RNA, hybrid duplex RNA•DNA, triplex DNA, and triplex RNA, respectively.
We sought to use a human telomeric DNA sequence as our model for G-quadruplex DNA. Recent NMR studies by Patel and coworkers have shown that four-repeat human telomeric DNA sequences (including d(T2AG3)4, d[TAG3(T2AG3)3], and d[TAG3(T2AG3)3TT]) exhibit conformational heterogeneity in K+ solution, with the capability of forming two different intramolecular (3+1) G-quadruplexes in which three strands are oriented in one direction and the fourth strand is oriented in the opposite direction [45, 47, 48]. Although both quadruplexes contain the (3+1) core structure, they differ with regard to order of loop arrangement. These collective structural studies also revealed that the conformational preference of the telomeric DNA sequence could be fine-tuned by modulating the residues flanking the four G3 tracts. The d[TTG3(T2AG3)3A] variant of the human telomeric DNA sequence, which we hereafter term hTel24-NMR, was found to favor a single (3+1) G-quadruplex structure in K+ solution [45]. We therefore selected this sequence as our model for G-quadruplex DNA. It should be emphasized that all the UV melting studies were conducted in the presence of K+ ions.
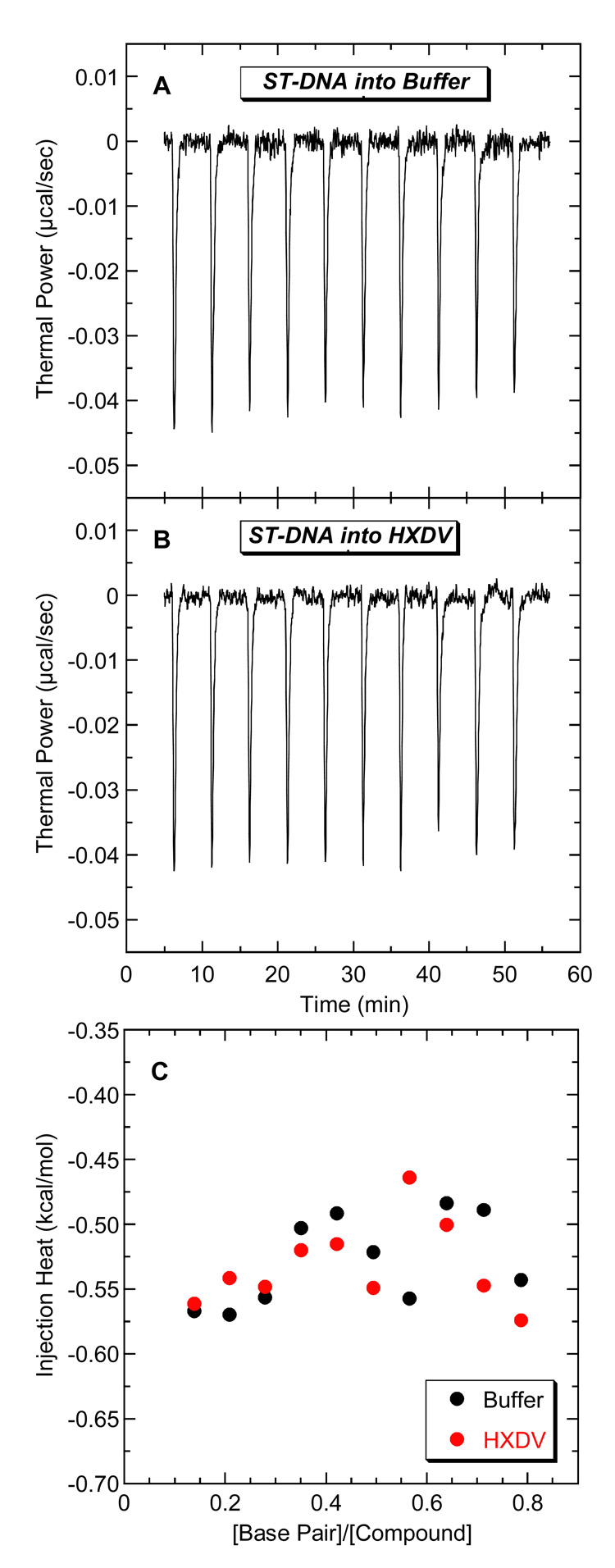
Fig. 2 shows representative UV melting profiles (depicted in their first-derivative forms) of hTel24-NMR (A), ST-DNA (B), p(dA)•2p(dT) (C), p(rA)•p(dT) (D), and p(rA)•2p(rU) (E) in the absence and presence of HXDV and HXLV-AC. Neither compound impacts the thermal transition of the ST-DNA and p(rA)•p(dT) duplexes, nor do they alter the Watson-Crick (WC) thermal transitions (which reflect the conversion of duplex and single strand to all single strand) of the p(dA)•2p(dT) and p(rA)•2p(rU) triplexes. These results are consistent with a little or no interaction between duplex DNA, duplex RNA, or hybrid duplex RNA•DNA and either compound. We used isothermal titration calorimetry (ITC) to further evaluate the ability of duplex DNA to serve as a binding substrate for HXDV and HXLV-AC. Fig. 3 shows the ITC profiles for the titration of ST-DNA into either buffer alone (A) or buffer containing HXDV (B). Note that the heats associated with sequential injections of ST-DNA into HXDV are essentially identical to the dilution heats associated with sequential injections of ST-DNA into buffer alone (Fig. 3C). We observed similar results for HXLV-AC (not shown). These collective observations are consistent with our UV melting results in confirming the absence of a measurable binding interaction between both compounds and duplex DNA.
Fig. 2.
First derivatives of the UV melting profiles of hTel24-NMR (A), ST-DNA (B), p(dA)•2p(dT) (C), p(rA)•p(dT) (D), and p(rA)•2p(rU) (E) in the absence (red) and presence of HXDV (black) or HXLV-AC (green). The melting profiles for the quadruplex formed by hTel24-NMR were acquired at 295 nm, while the melting profiles for the nucleic acid duplexes and triplexes were acquired at 260 nm. For the melting profiles of the triplexes, HG denotes the Hoogsteen transition of the triplex into duplex plus single strand, while WC denotes the Watson-Crick transition of the remaining duplex into single strands.
Fig. 3.
ITC profiles at 25 °C for the titration of ST-DNA (200 µM in base pair) into a solution of either buffer alone (A) or 20 µM HXDV (B). Each heat burst curve in panels A and B is the result of a 10 µL injection of ST-DNA. The solution conditions were 10 mM EPPS (pH 7.5) and sufficient KCl to bring the total K+ concentration to 50 mM. The data points in panel C reflect the integrated heats associated with the DNA injections into buffer (black) or HXDV (red).
Not only does the presence of HXDV and HXLV-AC fail to impact the WC thermal transitions of the p(dA)•2p(dT) (Fig. 2C) and p(rA)•2p(rU) (Fig. 2E) triplexes, it also fails to alter the corresponding Hoogsteen (HG) thermal transitions (which reflect the conversion of the triplex to duplex and single strand) of the two triplexes. Thus, neither the DNA nor the RNA triplex form appears capable of serving as a binding substrate for HXDV or HXLV-AC.
Although HXDV and HXLV-AC do not alter the thermal stabilities of the nucleic acid duplexes and triplexes noted above, they do increase the thermal stability of hTel24-NMR (Fig. 2A). These ligand-induced thermal enhancements are indicative of ligand binding to the hTel24-NMR quadruplex. The transition temperature (Ttran) corresponding to the minimum of the hTel24-NMR melting profile increases from 60.0 ± 0.5 °C in the absence of ligand to 71.5 ± 0.5 °C in the presence of the HXDV and 74.0 ± 0.5 °C in the presence of HXLV-AC. Our ITC studies described below in section 2.5.1 reveal that the modestly (2.5 °C) greater extent of thermal enhancement (ΔTtran) induced in hTel24-NMR by HXLV-AC relative to HXDV corresponds to a modestly enhanced affinity for the host quadruplex. We have previously shown that HXDV binds and thermally stabilizes the tetramolecular quadruplex formed by r(UG4U) [95]. Taken together, our UV melting and ITC studies are consistent with HXDV and HXLV-AC binding solely to quadruplex and not to duplex or triplex nucleic acid forms. With the exception of telomestatin and the bistriazoles, few other G-quadruplex stabilizing compounds approach this degree of binding specificity [36, 37, 79, 96, 97].
2.2.2 The Quadruplex Binding Behavior of Oxazole-Containing Macrocycles Depends on the Number of Oxazoles
We sought to determine whether oxazole number can affect the quadruplex binding behavior of oxazole-containing macrocycles. To this end, we monitored the impact of a tetraoxazole derivative of HXDV (TXDV) on the thermal stability of hTel24-NMR. In contrast to HXDV, TXDV failed to alter hTel24-NMR thermal stability (not shown). We previously observed a similar result using the human telomeric sequence d(T2AG3)4 as the host quadruplex forming DNA [98]. Thus, the DNA quadruplex binding behavior of HXDV appears to be lost upon reduction of the oxazole number from six to four, an observation highlighting the importance of oxazole number on the G-quadruplex stabilizing properties of oxazole-containing macrocycles.
2.3 Oxazole-Containing Macrocycles Bind Human Telomeric (3+1) Quadruplex DNA via a “Terminal Capping” Rather Than an Intercalative Mode of Interaction
We have previously reported a fluorescence-based approach for defining the mode by which HXDV binds to human telomeric quadruplex DNA [95]. In this connection we constructed a series of variants of d(T2AG3)4 (which we hereafter term hTel24) in which the second, third, or fourth adenine residue from the 5′-end (corresponding to position 9, 15, or 21 in the sequence) was substituted with the fluorescent base analog 2-aminopurine (AP). NMR studies [42–46, 48] suggest that the predominant (3+1) G-quadruplex conformation adopted by hTel24 in K+ solution is similar to that adopted by hTel24-NMR. In support of this observation, both hTel24 and hTel24-NMR exhibit similar CD spectra in the presence of 50 mM K+ (see Fig. 4A). A significant feature of the predominant conformation adopted by hTel24 and hTel24-NMR is that the adenine residues in the edgewise loops (A15 and A21 in hTel24) are stacked on opposing terminal G-tetrads. Unlike these two adenine residues, the adenine residue in the double-chain reversal loop (A9 in hTel24) adopts an unstacked conformation. We have previously shown that HXDV binding to the AP-substituted hTel24 oligomers in K+ solution induced changes in quadruplex fluorescence that were consistent with the coupled destacking of the AP bases at positions 15 and 21, but not the base at position 9 [95]. We also used fluorescence resonance energy transfer (FRET)-based characterizations of end-labeled variants of hTel24 to rule out the possibility of an intercalative mode of interaction by demonstrating that HXDV binding was not associated with an increase in quadruplex length that would be expected from such a mode of interaction [95]. These collective results implied a at each end of the structure (schematically depicted in Fig. 5). It should be noted that in the presence of 50 mM K+, the CD spectra of hTel24 and hTel24-NMR in their HXDV- and HXLV-AC-bound states are similar (see Fig. 4B), suggesting that, like HXDV, HXLV-AC also binds human telomeric quadruplex DNA via a “terminal capping” mode of interaction. This type of interaction may prove general for oxazole-containing macrocyclic compounds, including telomestatin.
Fig. 4.
CD spectra of hTel24 and hTel24-NMR in the absence (A) and presence (B) of HXDV or HXLV-AC. The spectra were acquired at 25 °C under the solution conditions described in the legend to Fig. 3 Molar ellipticity [θ] is expressed in units of deg/M•cm, where M refers to moles of quadruplex per liter.
Fig. 5.
Schematic representation of the “terminal capping” model for the interactions between oxazole-containing macrocyclic compounds and human telomeric (3+1) quadruplex DNA. In the absence of ligand, the two adenine residues in the edgewise loops (depicted in magenta) are stacked on the terminal G-tetrads, while the adenine residue in the double-chain reversal loop (depicted in blue) does not engage in such stacking interactions. Upon the binding of a ligand molecule to each end of the quadruplex (i.e., “terminal capping” of the quadruplex), the adenine residues in the edgewise loops become destacked. Guanine bases are depicted in green, and thymine bases are depicted in orange. Ligand molecules are depicted in yellow.
2.4 Defining the Stoichiometry with which HXDV and HXLV-AC Bind Human Telomeric (3+1) GQuadruplex DNA
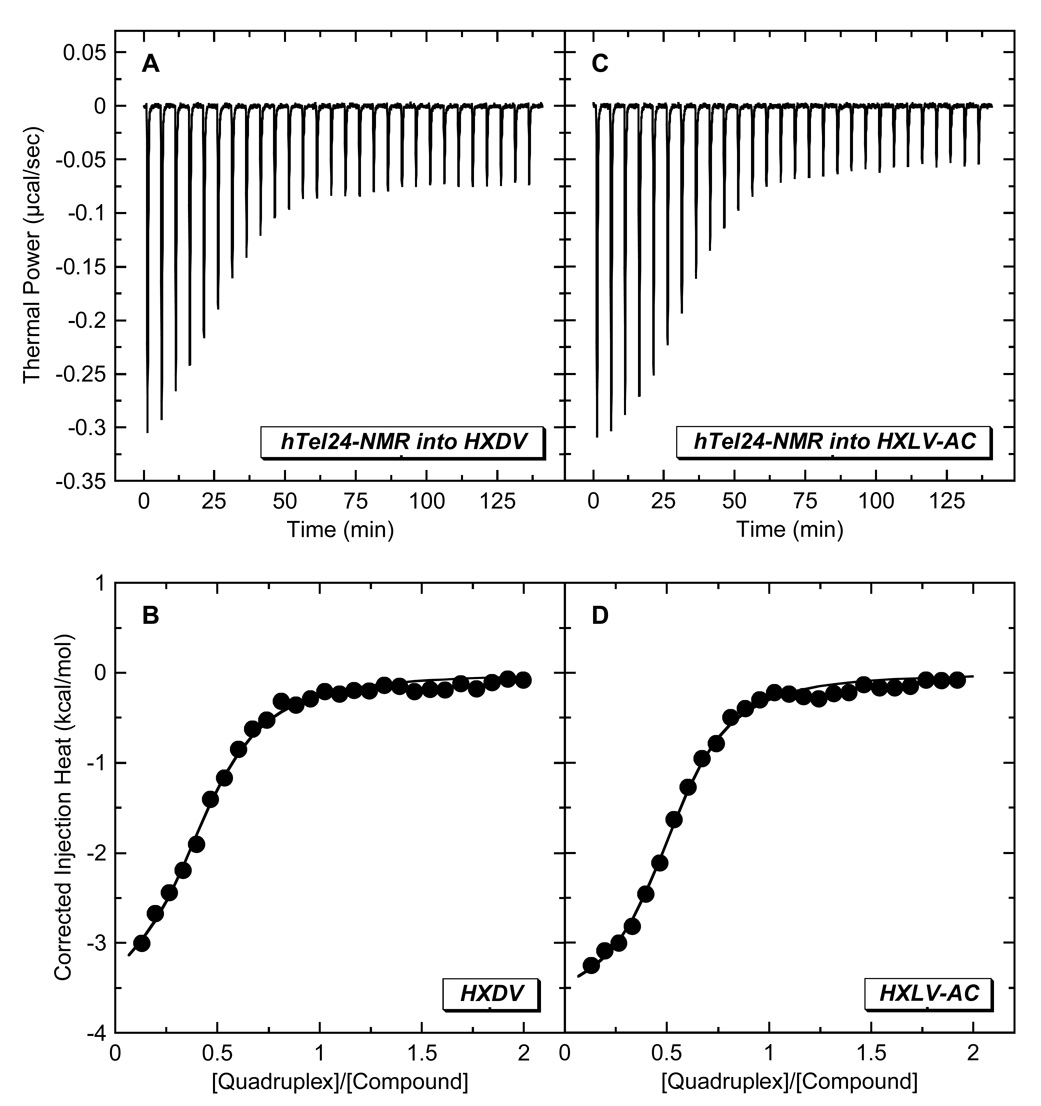
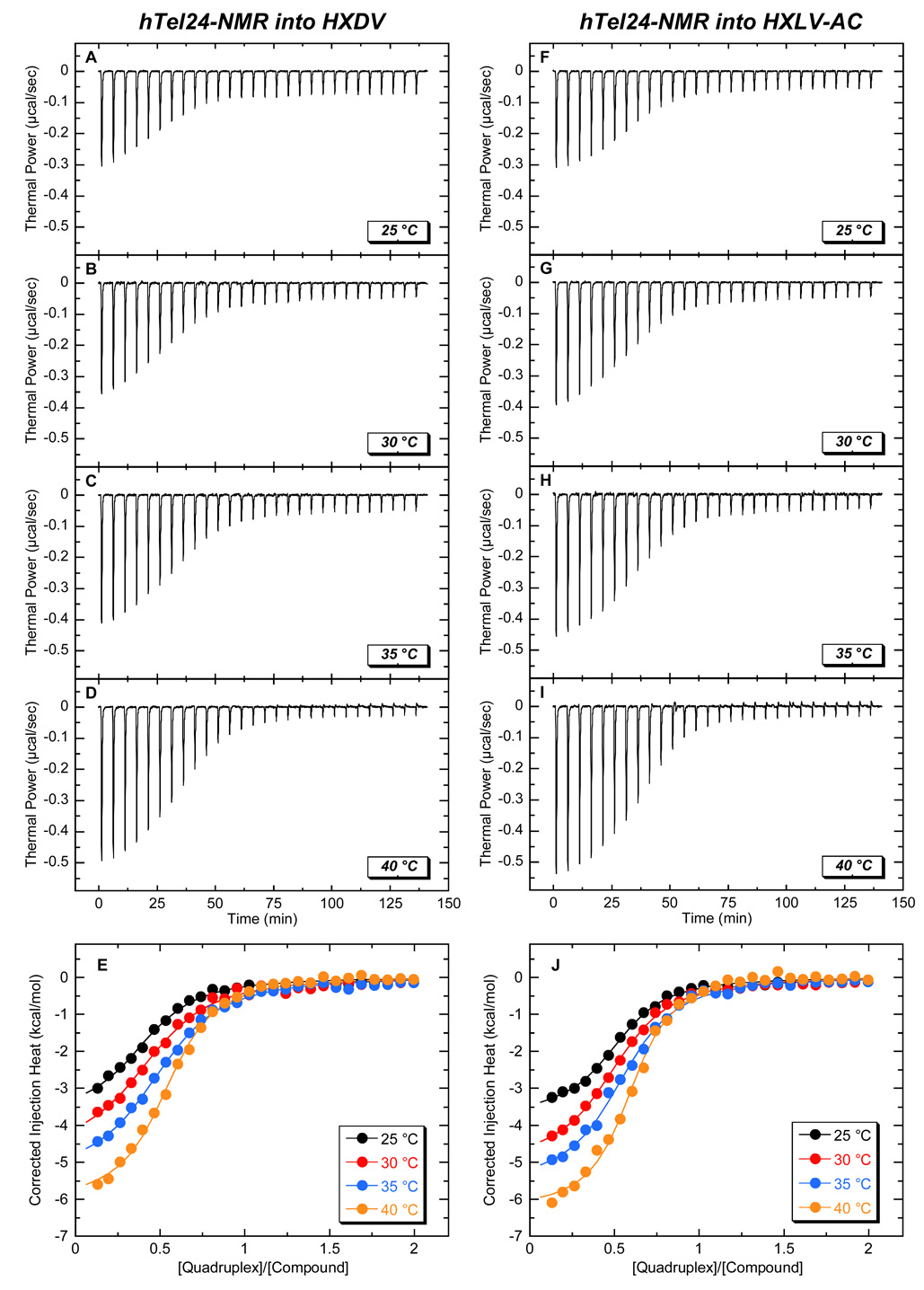
The “terminal capping” model for the ligand-quadruplex interaction invokes the binding of one ligand molecule to each end of the quadruplex. We sought to determine experimentally the stoichiometry with which HXDV and HXLV-AC bind the host quadruplex. To this end, we used ITC to characterize the binding of the two compounds to a variant of hTel24 (hTel24-NMR) whose high-resolution solution structure in the presence of K+ ions was determined by Patel and coworkers using NMR-based techniques [45]. At concentrations greater than 100 µM, HXDV and HXLV-AC form visible precipitates in aqueous solution. These limiting ligand solubilities precluded our being able to conduct ITC studies using the conventional approach of sequentially injecting the small molecule (ligand) into the macromolecule (quadruplex) solution. Instead, we conducted reverse ITC titrations of quadruplex into ligand, with representative profiles acquired at 25 °C in the presence of 50 mM K+ being shown in Fig. 6. These profiles were analyzed with a model for one set of binding sites to yield the binding parameters listed in Table 2. Significantly, both HXDV and HXLV-AC bind hTel24-NMR with a stoichiometry (N) of approximately two ligand molecules per quadruplex, a stoichiometry fully consistent with the “terminal capping” mode of interaction implied by the results described above in section 2.3. It is also of interest to note that both ends of the hTel24-NMR quadruplex appear to present the compounds with essentially identical binding sites, as implied by the ITC profiles being well described by a model for one set of binding sites.
Fig. 6.
ITC profiles at 25 °C for the titration of hTel24-NMR (200 µM in strand) into a solution of either 20 µM HXDV (A,B) or 20 µM HXLV-AC (C,D). Each heat burst curve in panels A and C is the result of a 10 µL injection of DNA, with the solution conditions being as described in the legend to Fig. 3. The data points in panels B and D reflect the corrected injection heats, which were derived by integration of the corresponding heat burst curves in panels A and C, followed by subtraction of the corresponding dilution heats derived from control titrations of DNA into buffer alone. The solid lines in panels B and D reflect calculated fits of the data with a model for one set of binding sites.
Table 2.
ITC-Derived Parameters for the Binding of HXDV and HXLV-AC to hTel24-NMR at 25 °C in the Presence of 50 mM K+a
| Compound | K (M−1) | ΔH (kcal/mol) | N |
|---|---|---|---|
| HXDV | (3.0 ± 0.4) × 105 | −1.7 ± 0.1 | 2.2 ± 0.1 |
| HXLV-AC | (5.5 ± 0.6) × 105 | −2.1 ± 0.1 | 1.9 ± 0.1 |
Buffer conditions were 10 mM EPPS (pH 7.5) and sufficient KCl to bring the total K+ concentration to 50 mM. The listed values of K, ΔH, and N were derived from fits of the ITC profiles shown in Fig. 6 with a model for one set of binding sites. The indicated uncertainties reflect the standard deviations of the experimental data points from the fitted curves.
2.5 Defining the Thermodynamic Forces That Drive the Binding of HXDV and HXLV-AC to Human Telomeric (3+1) G-Quadruplex DNA
2.5.1 At 25 °C, HXLV-AC Binds hTel24-NMR with a 1.8-Fold Greater Affinity Than HXDV, with this Differential Affinity Being Enthalpic in Origin
Further inspection of the data in Table 2 reveals that HXDV and HXLV-AC bind hTel24-NMR with association constants (K) of (3.0 ± 0.4) × 105 and (5.5 ± 0.6) × 105 M−1, respectively. Thus, at 25 °C, HXLV-AC binds hTel24-NMR with an approximately 1.8-fold greater affinity than HXDV. Although modest, this difference in affinity exceeds the experimental uncertainty and correlates with the 2.5 °C greater ΔTtran associated with the hTel24-NMR binding of HXLV-AC relative to HXDV (see section 2.2.1). These results imply that substitution of one valine functionality in HXDV with an N-acetylated lysinyl moiety affords a modest enhancement in affinity for human telomeric quadruplex DNA. A comparison of the enthalpy changes (ΔH) associated with the binding of HXLV-AC and HXDV to hTel24-NMR at 25 °C (see Table 2) indicates that the binding of HXLV-AC is 0.4 kcal/mol more negative (favorable) than the binding of HXDV. As will be discussed below in section 2.5.3, the entropic contributions (-TΔS) to the binding of both compounds are similar. Viewed as a whole, these results indicate that the enhanced affinity of HXVL-AC for the host quadruplex relative to HXDV is enthalpic in origin. This observation may reflect enhanced van der Waals and/or hydrogen bonding contacts afforded by the N-acetylated lysinyl moiety of HXVL-AC, since such contacts are typically manifested enthalpically.
2.5.2 HXLV-AC and HXDV Binding to hTel24-NMR Is Associated with a Negative Heat Capacity Change
We used the reverse ITC approach described above to monitor the binding of HXLV-AC and HXDV to hTel24-NMR at temperatures ranging from 25 to 40 °C. The resulting temperature-dependent ITC binding profiles are shown in Fig. 7. These binding profiles were analyzed with a model for one set of binding sites to yield the binding parameters listed in Table 3. Inspection of these data reveals two significant features: (i) The affinity constants for the binding of both compounds remain essentially unchanged over the temperature range of 25 to 35 °C, and then increase approximately two-fold at 40 °C. (ii) The enthalpy changes associated with the binding of both compounds become more negative (exothermic) with increasing temperature.
Fig. 7.
ITC profiles for the titration of hTel24-NMR (200 µM in strand) into a solution of 20 µM HXDV (A-E) or HXLV-AC (F-J) at temperatures ranging from 25 to 40 °C. Analysis and presentation of the raw (A-D, F-I) and dilution-corrected (E, J) data are as described in the legend to Fig. 6, with the solution conditions being as described in the legend to Fig. 3.
Table 3.
Temperature Dependence of the ITC-Derived Parameters for the Binding of HXDV and HXLV-AC to hTel24-NMR in the presence of 50 mM K+a
| Compound | Temperature (°C) | K (M−1) | ΔH (kcal/mol) | N |
|---|---|---|---|---|
| HXDV | 25 | (3.0 ± 0.4) × 105 | −1.7 ± 0.1 | 2.2 ± 0.1 |
| HXDV | 30 | (2.8 ± 0.4) × 105 | −2.4 ± 0.1 | 2.0 ± 0.1 |
| HXDV | 35 | (3.5 ± 0.3) × 105 | −3.1 ± 0.1 | 1.9 ± 0.1 |
| HXDV | 40 | (6.9 ± 0.5) × 105 | −3.8 ± 0.1 | 1.8 ± 0.1 |
| HXLV-AC | 25 | (5.5 ± 0.6) × 105 | −2.1 ± 0.1 | 1.9 ± 0.1 |
| HXLV-AC | 30 | (4.2 ± 0.5) × 105 | −2.8 ± 0.1 | 1.9 ± 0.1 |
| HXLV-AC | 35 | (5.4 ± 0.5) × 105 | −3.4 ± 0.1 | 1.8 ± 0.1 |
| HXLV-AC | 40 | (1.1 ± 0.1) × 106 | −4.0 ± 0.1 | 1.7 ± 0.1 |
Buffer conditions were as described in the footnote to Table 2. The listed values of K, ΔH, and N were derived from fits of ITC profiles (as shown in Fig. 7 for HXDV) with a model one set of binding sites. The indicated uncertainties reflect the standard deviations of the experimental data points from the fitted curves.
The heat capacity change (ΔCp) for a binding interaction can be determined from the temperature dependence of the observed binding enthalpy using the standard relationship:
| (1) |
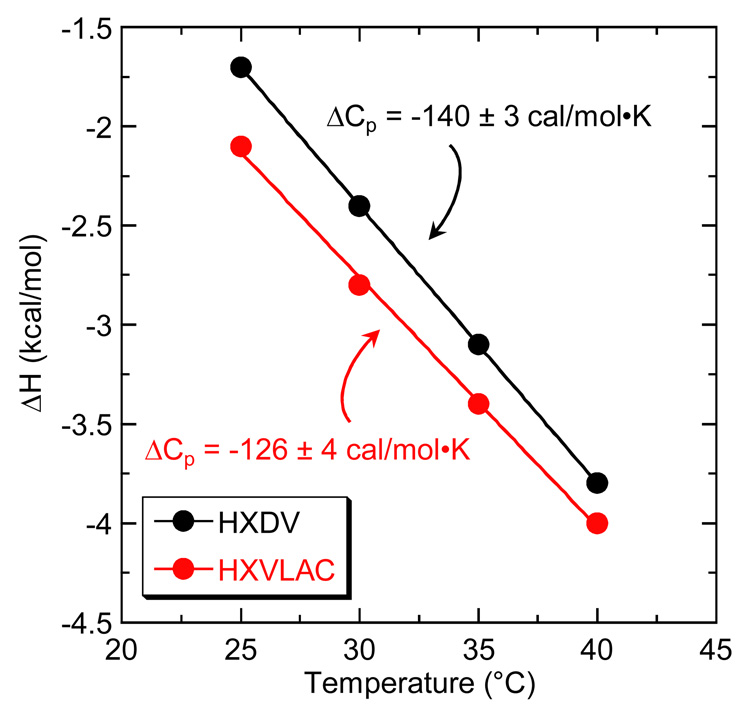
Fig. 8 graphically depicts the ΔH data listed in Table 3 in the form of ΔH versus temperature plots. The data points in these plots were fit by linear regression, with the slopes of the resulting lines yielding ΔCp estimates of −126 ± 4 and −140 ± 3 cal/mol•K for the binding of HXLV-AC and HXDV, respectively. Thus, the interactions of HXLV-AC and HXDV with hTel24-NMR are associated with similar negative heat capacity changes, the magnitudes of which fall within the range (−100 to −550 cal/mol•K) frequently observed for both ligand-nucleic acid and ligand-protein interactions [99–104].
Fig. 8.
Plots of ΔH versus temperature for the binding of HXDV (black) and HXLV-AC (red) to hTel24-NMR. The experimental data points were fit by linear regression (solid lines), with the ΔCp values derived from the slopes of the fitted lines being indicated.
One of the more common molecular interpretations for ligand-macromolecule binding reactions associated with observed negative ΔCp values is a hydrophobic effect that stems from the release of constrained water molecules accompanying the burial of nonpolar surfaces [105–110]. As discussed below in section 2.6.1, HXDV binding to hTel24-NMR does not appear to be coupled to a net change in hydration. Thus, binding-induced release of constrained water molecules is unlikely to account for the observed negative ΔCp values for the binding of HXDV and HXLV-AC to hTel24-NMR. Another factor that can contribute to observed negative ΔCp values for binding reactions is the coupling linkage of temperature-dependent equilibria (e.g., conformational changes and protonation reactions) [104, 111–115]. In this connection, Kozlov and Lohman have demonstrated that the binding of E. coli SSB protein to single-stranded oligo(dA) is coupled to the destacking of adenine bases, with this coupled destacking equilibrium dominating the observed negative ΔCp for the binding reaction [113]. We have previously observed that the binding of aminoglycoside antibiotics to an E. coli 16S rRNA A-site model oligonucleotide exhibits a similar behavior [104]. Recall that steady-state AP fluorescence and CD measurements were consistent with HXDV- and HXLV-AC-induced destacking of adenine residues in the edgewise loops of hTel24 (see section 2.3). We therefore suggest that these binding-linked destacking reactions contribute to the observed negative ΔCp values for the binding interactions.
2.5.3 Entropy Provides a Greater Driving Force than Enthalpy for the Binding of HXLV-AC and HXDV to hTel24-NMR, with the Extent of This Differential Diminishing with Increasing Temperature
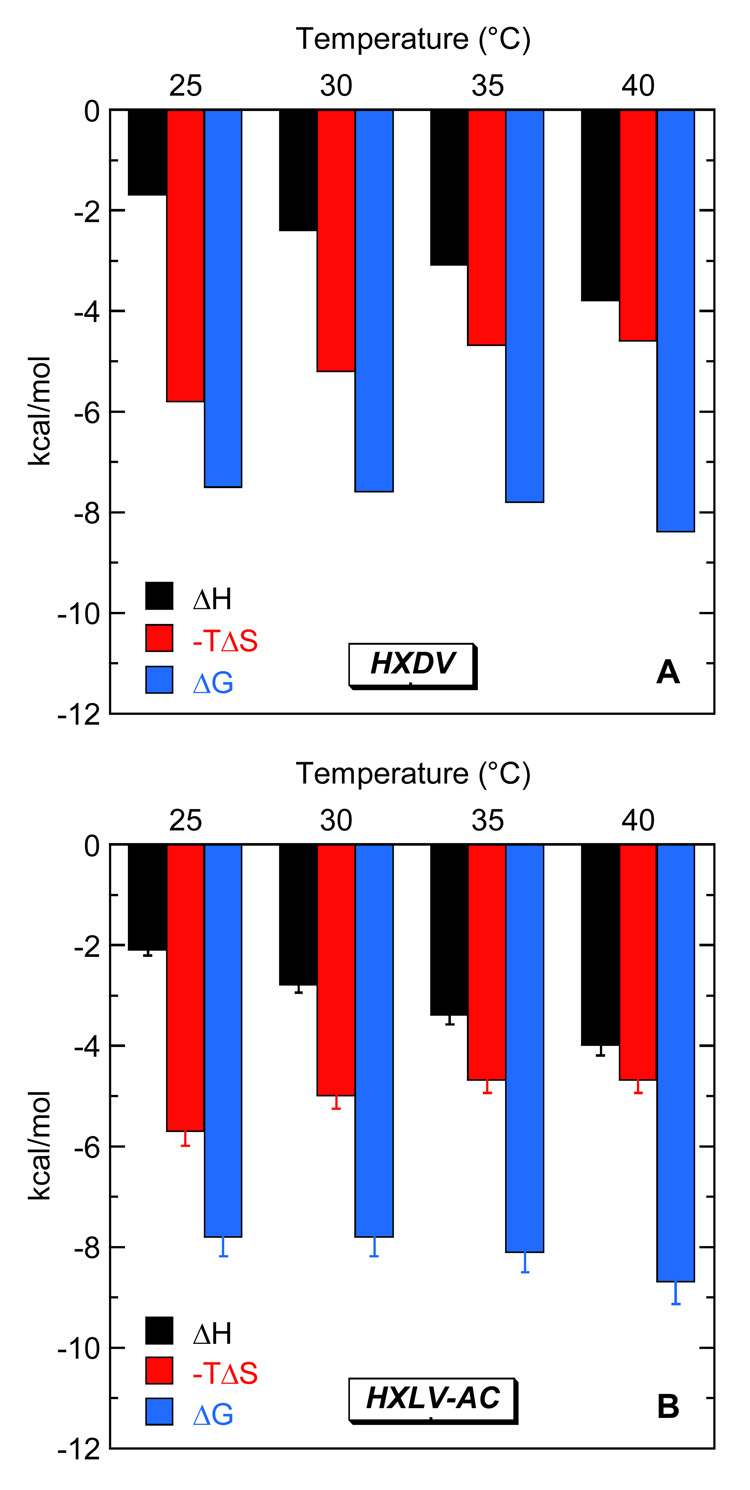
The ITC-derived values of K and ΔH listed in Table 3 allow us to complete the thermodynamic binding profiles by calculating corresponding values -TΔS and ΔG. The resulting thermodynamic profiles are listed in Table 4, with their temperature dependence being graphically depicted in Fig. 9. Inspection of these data reveals that the entropic contributions to the binding of both HXDV and HXLV-AC to hTel24-NMR are similar in magnitude. Moreover, these entropic contributions provide a greater driving force for the ligand-quadruplex binding reactions than enthalpy. However, the relative extents to which entropy drives the binding reactions diminish with increasing temperature. For example, the binding of HXDV is 77% entropy and 23% enthalpy driven at 25 °C, while being 57% entropy and 43% enthalpy driven at 40 °C. The binding of HXLV-AC exhibits a similar thermodynamic binding behavior.
Table 4.
Temperature Dependence of the Thermodynamic Parameters for the Binding of HXDV and HXLV-AC to hTel24-NMR in the presence of 50 mM K+
| Compound | Temperature (°C) | ΔH (kcal/mol)a | -TΔ S (kcal/mol)b | ΔG (kcal/mol)b |
|---|---|---|---|---|
| HXDV | 25 | −1.7 ± 0.1 | −5.8 ± 0.2 | −7.5 ± 0.1 |
| HXDV | 30 | −2.4 ± 0.1 | −5.2 ± 0.2 | −7.6 ± 0.1 |
| HXDV | 35 | −3.1 ± 0.1 | −4.7 ± 0.2 | −7.8 ± 0.1 |
| HXDV | 40 | −3.8 ± 0.1 | −4.6 ± 0.2 | −8.4 ± 0.1 |
| HXLV-AC | 25 | −2.1 ± 0.1 | −5.7 ± 0.2 | −7.8 ± 0.1 |
| HXLV-AC | 30 | −2.8 ± 0.1 | −5.0 ± 0.2 | −7.8 ± 0.1 |
| HXLV-AC | 35 | −3.4 ± 0.1 | −4.7 ± 0.2 | −8.1 ± 0.1 |
| HXLV-AC | 40 | −4.0 ± 0.1 | −4.7 ± 0.2 | −8.7 ± 0.1 |
Values of ΔH were determined as described in the legend to Table 3.
Values of -TΔS and ΔG were determined from the standard relationships -TΔS = ΔG - ΔH and ΔG = -RTlnK, respectively. The indicated uncertainties reflect the maximum possible errors as propagated through these relationships.
Fig. 9.
Temperature dependence of the ITC-derived thermodynamic profiles for the binding HXDV (A) and HXLV-AC (B) to hTel24-NMR. The solution conditions were as described in the legend to Fig. 3.
2.6 Defining the Molecular Origins of the Entropic Driving Force for the Binding of HXDV and HXLV-AC to Human Telomeric (3+1) Quadruplex DNA
2.6.1 Osmotic Stress Studies Reveal That HXDV Binding to hTel24-NMR Is Not Accompanied by a Net Change in Hydration
One commonly invoked explanation for entropically driven ligand-nucleic acid binding reactions is the coupled release of counterions. However, it is unlikely that the entropic driving force for the quadruplex binding reactions of HXDV and HXLV-AC reflects significant contributions from coupled counterion release, since neither compound is charged. A second commonly invoked explanation for entropically driven binding reactions is the coupled release of constrained water molecules. We sought to explore this possibility by using an osmotic stress approach to characterize the hydration changes, if any, adenine residues in the edgewise loops (at positions 14 and 20) are substituted with AP residues.
In the osmotic stress approach, one monitors the impact of neutral solutes or cosolvents (osmolytes) on the ligand-DNA association constants (K). An osmolyte-induced increase in K reflects a net release of water molecules in conjunction with binding, while an osmolyte-induced decrease in K reflects a net uptake of water molecules. We employed ethylene glycol (MW = 62.1 g/mol) and glycerol (MW = 92.1 g/mol) as the osmolytes in our studies, since their molecular weights are sufficiently small relative to that of HXDV and so as to minimize the potential for the introduction of volume exclusion (crowding) effects.
We monitored the fluorescence of hTel24-NMR-14,20AP as a function of added HXDV in the absence and presence of ethylene glycol or glycerol. Fig. 10 shows the resulting fluorescence titration profiles acquired at 25 °C in the presence of 50 mM K+. We determined ligand-quadruplex K values by analyzing the ligand-induced changes in the fluorescence of hTel24-NMR-14,20AP with the following formalism:
| (2) |
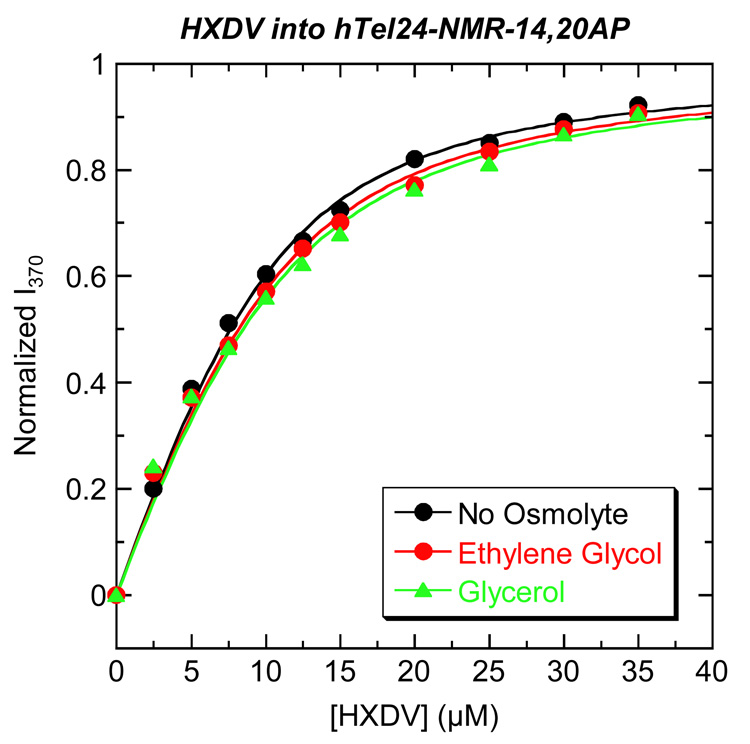
Fig. 10.
Fluorescence profiles at 25 °C for the titration of HXDV into a solution of hTel24-NMR-14,20AP in the absence (black) and presence of 2.0 osm ethylene glycol (red) or glycerol (green). The solid lines represent fits of the experimental data points with Eq. 2. The buffer conditions were as described in the legend to Fig. 3. For clarity of presentation, the fluorescence emission intensity at 370 nm (I370) was normalized by subtraction of the fluorescence intensity in the absence of drug (I0) and subsequent division by the total calculated binding-induced change in fluorescence (I∞ - I0).
In this relationship, I0 and I are the fluorescence emission intensities of the quadruplex in the absence and presence of ligand, respectively, I∞ is the fluorescence emission intensity of the quadruplex in the presence of infinite ligand concentration, and Ltot and Qtot are the total concentrations of ligand and quadruplex, respectively. Table 5 lists the K values derived from fits of the fluorescence titration profiles shown in Fig. 10 with Eq. 2. Note that in the absence of osmolyte, HXDV binds hTel24-NMR-14,20AP with a K value of (3.8 ± 0.5) × 105 M−1. This affinity constant is in excellent agreement with the corresponding value of (3.0 ± 0.4) × 105 M−1 independently derived by ITC using hTel24-NMR as the host quadruplex (see Table 2). Further note that the presence of neither ethylene glycol nor glycerol significantly alters the K value of HXDV, with any observed differences in K afforded by the osmolytes being within the experimental uncertainty. This observation is consistent with HXDV binding to hTel24-NMR-14,20AP being accompanied by no net change in hydration. In other words, hydration changes do not appear to provide a significant driving force for the quadruplex binding reactions of HXDV. Although not shown here, similar types of osmotic stress studies suggest that the same is true of the HXLV-AC-quadruplex binding reactions.
Table 5.
Osmolyte Dependence of the Affinity of HXDV for hTel24- NMR-14,20AP at 25 °C in the Presence of 50 mM K+a
| Osmolyte | Osmolality (osm) | K (M−1)b |
|---|---|---|
| None | 0 | (3.8 ± 0.5) × 105 |
| Ethylene Glycol | 2.0 | (3.2 ± 0.5) × 105 |
| Glycerol | 2.0 | (2.9 ± 0.7) × 105 |
2.6.2 The Entropic Driving Force for the Binding Reactions of Oxazole-Containing Macrocycles May Reflect Favorable Binding-Induced Alterations in the Configurational Entropies of the Edgewise Loops of the (3+1) DNA Quadruplex
Another potential explanation for the entropic nature of the primary driving force underlying the quadruplex binding reactions of HXDV and HXLV-AC is a concomitant increase in the configurational entropy of the host DNA. In this connection, our previously reported time-resolved fluorescence anisotropy studies probing the impact of HXDV binding on the conformational dynamics of AP-substituted hTel24 oligomers revealed that HXDV binding increased the mobility of the AP residue at position 21 in an edgewise loop, while exerting essentially no impact on the mobility of the AP residue at position 9 in the double-chain reversal loop [95]. These results are consistent with those described above (see section 2.3) showing the binding-induced destacking of the AP residue at position 21, but not at position 9. Recall that HXDV binding also induced the destacking of the AP residue at position 15 of hTel24. Thus, although the low fluorescence quantum yield of 15AP-substituted hTel24 precluded the use of this oligomer in fluorescence anisotropy studies, it is reasonable to suggest that HXDV binding would likely have increased the mobility of this residue as well. In the aggregate, the time-resolved fluorescence anisotropy results suggest that favorable binding-induced alterations in the configurational entropies of the edgewise loops of human telomeric (3+1) quadruplex DNA may contribute to the entropic driving force for the binding reactions oxazole-containing macrocyclic compounds.
3. Summary and Conclusions
The studies described here demonstrate that the macrocyclic hexaoxazoles HXDV and HXLV-AC exhibit cytotoxic potencies versus human cancer cells that are either similar to or exceed those previously reported for telomestatin. Both compounds bind solely to the G-quadruplex form of nucleic acids under physiological conditions, with no detectable binding to duplex or triplex forms. This behavior may prove to be a general feature of macrocyclic oxazole-containing ligands, since telomestatin (a heptaoxazole-containing macrocycle) appears to exhibit a similar degree of binding specificity [37, 79, 96, 97]. The bistriazoles are the only other class of compounds reported to date that approach this degree of quadruplex binding specificity [79]. Significantly, this degree of target specificity is likely to be an important determinant of the therapeutic utility of G-quadruplex-directed agents.
Our present and previously reported [95] spectroscopic studies described here and elsewhere suggest that HXDV and HXLV-AC bind human telomeric (3+1) G-quadruplex DNA via a nonintercalative “terminal capping” mode in which one ligand molecule binds to each end of the quadruplex. Hurley and coworkers have previously used docking and molecular dynamics simulations to propose such a binding model for the interaction of telomestatin with d(T2AG3)4 [36]. Our results lend support to this proposal, while also suggesting that terminal capping binding model may prove general for oxazole-containing macrocycles.
At temperatures ranging from 25 to 40 °C, the binding of both HXDV and HXLV-AC to human telomeric (3+1) G-quadruplex DNA is primarily entropy driven, with this thermodynamic driving force reflecting contributions from favorable binding-induced alterations in the configurational entropies of the edgewise loops of the quadruplex and not from changes in net hydration. These binding-linked changes in loop conformation also give rise to observed negative heat capacity changes that are associated with the ligand-quadruplex binding reactions.
Recently reported high-resolution structures of G-quadruplexes adopted by four-repeat human telomeric DNA model sequences [38, 40, 45–48] form a tempting database for structure-based drug design efforts. However, such efforts should be undertaken with caution, since the quadruplexes formed by the different model sequences are structurally heterogeneous, particularly with regard to their loop configurations. Furthermore, drug binding can be coupled to conformational changes in the host quadruplex that are difficult to mimic in computational studies, particularly docking methodologies in which the macromolecular target is typically fixed. It is important to note that drug-induced conformational changes in the host DNA need not be restricted to an intercalative mode of interaction, since the studies discussed here suggest that they can also occur in conjunction with a “terminal capping” mode of interaction.
Yang and coworkers have proposed a model for human telomeric DNA in which the (3+1) G-quadruplex structure is multiply repeated along the length of the 3′-terminal overhang region of telomeric DNA [43]. As noted above in section 1.1, this overhang region consists of approximately 16 to 35 repeats of the 5′-T2AG3-3′ sequence in human telomeric DNA [4, 6]. Thus, the 3′ -terminal overhang region of human telomeric DNA could potentially contain as many as eight repeats of the (3+1) G-quadruplex structure. The results described here imply that oxazole-containing macrocyclic ligands like HXDV, HXLV-AC, and telomestatin could conceivably bind to these repeating telomeric quadruplex structures with a stoichiometry of at least one and perhaps two ligand molecules per quadruplex structure.
4. Materials and Methods
4.1 Ligand and DNA Molecules
HXDV and HXLV-AC were synthesized as described previously [98, 116]. Stock solutions of these compounds were prepared in dimethyl sulfoxide (DMSO) and stored at −20 °C. All DNA oligomers were obtained in their HPLC-purified forms from Integrated DNA Technologies (Coralville, IA). The concentrations of all DNA solutions were determined spectrophotometrically using the following extinction coefficients at 260 nm and 90 °C (ε260-90°C) [in units of (mol strand/L)−1•cm−1]: 231,100 ± 2,300 for hTel24; 232,500 ± 1700 for hTel24-NMR; and 215,600 ± 1800 for hTel24-NMR-14,20AP. These extinction coefficients were determined by enzymatic digestion and subsequent colorimetric phosphate assay using previously established protocols [117]. Salmon testes (ST) DNA, as well as all DNA and RNA polynucleotides, were obtained from Sigma (St. Louis, MO), and used without further purification. Polynucleotide solution concentrations were determined using the following extinction coefficients in units of (mol nucleotide/L)−1•cm−1: ε260-25°C = 6,000 for p(dA)•p(dT); ε265-25°C = 8,700 for p(dT); ε258-25°C = 9,800 for p(rA); and ε260-25°C = 9,350 for p(rU). Prior to their use, all experimental nucleic acid solutions were preheated at 90 °C for 5 min and slowly cooled to room temperature over a period of 4 hr.
4.2 UV Melting Studies
Temperature-dependent absorption experiments were conducted on an AVIV Model 14DS Spectrophotometer (Aviv Biomedical, Lakewood, NJ) equipped with a thermoelectrically controlled cell holder. Quartz cells with a pathlength of 1.0 cm were used for all the absorbance studies. Temperature-dependent absorption profiles were acquired at either 260 (for duplex and triplex) or 295 (for quadruplex) nm with a 5 sec averaging time. The temperature was raised in 0.5 °C increments, and the samples were allowed to equilibrate for 1.5 min at each temperature setting. In the quadruplex melting studies, the concentration of hTel24-NMR was 5 µM in strand and, when present, the HXDV and HXLV-AC concentrations were 25 µM. In the duplex and triplex melting studies, the nucleic acid concentration was 15 µM in base pair or 15 µM in base triple and, when present, the HXDV and HXLV-AC concentrations were 15 µM. The buffer for all the UV melting experiments contained 10 mM EPPS (pH 7.5). In addition, sufficient KCl was added to each solution to bring the total K+ concentration to either 50 mM for hTel24-NMR and ST-DNA, 150 mM for p(rA)•p(dT), 250 mM for p(dA)•2p(dT), or 20 mM for p(rA)•2p(rU).
4.3 Isothermal Titration Calorimetry (ITC)
ITC measurements were conducted at temperatures ranging from 25 to 40 °C on a MicroCal VP-ITC (MicroCal, Inc., Northampton, MA). In these experiments, 10 µL aliquots of DNA (hTel24-NMR at a concentration of 200 µM in strand and ST-DNA at a concentration of 200 µM base pair) were injected from a 250 µL rotating syringe (300 RPM) into an isothermal sample chamber containing 1.42 ml of an HXDV or HXLV-AC solution that was 20 µM. Each experiment of this type was accompanied by the corresponding control experiment in which 10 µL aliquots of 200 µM DNA were injected into a solution of buffer alone. The duration of each injection was ten seconds and the initial delay prior to the first injection was 60 seconds. The delay between injections was 300 seconds. Each DNA injection generated a heat burst curve (µcal/sec versus sec), the area under which was determined by integration to obtain a measure of the heat associated with that injection. The heat associated with each DNA-buffer injection was subtracted from the corresponding heat associated with each DNA-compound injection to yield the heat, if any, of DNA binding for that injection. The buffer-corrected ITC profiles for the binding of hTel24-NMR to HXDV and HXLV-AC were fit with a model for one set of binding sites using the Origin version 7.0 software package (MicroCal, Inc., Northampton, MA) appropriately set to register the ligand as being in the cell rather than in the syringe. The buffer conditions were 10 mM EPPS (pH 7.5) and sufficient KCl to bring the total K+ concentration to 50 mM.
4.4 Circular Dichroism (CD) Spectroscopy
CD experiments were conducted at 25 °C on an AVIV Model 202 spectropolarimeter equipped with a thermoelectrically controlled cell holder. A quartz cell with a 1 cm pathlength was used for all the CD studies. CD spectra were recorded from 350 to 240 nm in 1 nm increments with an averaging time of 2 sec. The concentration of hTel24 and hTel24-NMR was 5 µM in strand and, when present, the HXDV and HXLV-AC concentrations were 25 µM. The buffer conditions were as described above for the ITC experiments (section 4.3).
4.5 Steady-State Fluorescence Spectroscopy
All steady-state fluorescence experiments were conducted on an AVIV model ATF105 spectrofluorometer equipped with a thermoelectrically controlled cell holder. In the HXDV titration experiments, which were conducted at 25 °C, 1 to 2 µL aliquots of 5 mM ligand in DMSO were sequentially added to solutions of hTel24-NMR-14,20AP that were 5 µM in strand. After each addition, the sample was left to equilibrate for 2 minutes, whereupon the emission spectrum from 400 to 320 nm was recorded. These emission spectra were corrected by subtraction of the corresponding spectra of buffer alone or buffer plus the appropriate amount of ligand. In these measurements, the excitation wavelength was set at 310 nm, with the excitation and emission bandwidths being set 3 nm. The pathlength of the quartz cell used in these measurements was 1 cm in both the excitation and emission directions. The buffer conditions were as described above for the ITC experiments (section 4.3). When present, ethylene glycol and glycerol were used at an osmolality of 2.0.
4.6 Cytotoxicity Assays
The cytotoxic activities of HXDV and HXLV-AC were determined using a standard MTT (tetrazolium-based) colorimetric assay [118–120]. Human lymphoblast RPMI 8402 cells were maintained in RPMI 1640 medium, while human oral carcinoma KB3-1 cells were maintained in DMEM medium. Both types of media were supplemented with 10% heat-inactivated fetal bovine serum, 2 mM l-glutamine, 100 U/mL penicillin, and 0.1 mg/mL streptomycin. Cytotoxicity assays were conducted using a 96-well microtiter plate containing either 2000–3000 cells/well (for RPMI 8402 cells) or 1000 cells/well (for KB3-1 cells) and 200 µL of growth medium per well. Cells were exposed continuously for 4 days to differing concentrations of compound, and MTT assays were performed at the end of the fourth day. Each assay was performed with a control that did not contain any compound. All assays were conducted at least four times in triplicate wells.
Acknowledgements
This work was supported by NIH grants CA097123 (D.S.P.) and CA098127 (E.J.L.) and American Cancer Society grant RSG-99-153-04-CDD (D.S.P.). C.M.B. was supported in part by an NIH training grant (T32 CA108455) in Cancer Pharmacology. The authors thank Angela Liu of UMDNJ for conducting the cytotoxicity assays.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Blackburn EH. Switching and Signaling at the Telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 2.De Cian A, Lacroix L, Douarre C, Temime-Smaali N, Trentesaux C, Riou JF, Mergny J-L. Targeting Telomeres and Telomerase. Biochimie. 2008;90:131–155. doi: 10.1016/j.biochi.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 3.Moyzis RK, Buckingham JM, Cram LS, Dani M, Deaven LL, Jones MD, Meyne J, Ratliff RL, Wu JR. A Highly Conserved Repetitive DNA Sequence, (TTAGGG)n, Present at the Telomeres of Human Chromosomes. Proc. Natl. Acad. Sci. USA. 1988;85:6622–6626. doi: 10.1073/pnas.85.18.6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makarov VL, Hirose Y, Langmore JP. Long G Tails at Both Ends of Human Chromosomes Suggest a C Strand Degradation Mechanism for Telomere Shortening. Cell. 1997;88:657–666. doi: 10.1016/s0092-8674(00)81908-x. [DOI] [PubMed] [Google Scholar]
- 5.McElligott R, Wellinger RJ. The Terminal DNA Structure of Mammalian Chromosomes. EMBO J. 1997;16:3705–3714. doi: 10.1093/emboj/16.12.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wright WE, Tesmer VM, Huffman KE, Levene SD, Shay JW. Normal Human Chromosomes Have Long G-Rich Telomeric Overhangs at One End. Genes Dev. 1997;11:2801–2809. doi: 10.1101/gad.11.21.2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chai W, Shay JW, Wright WE. Human Telomeres Maintain Their Overhang Length at Senescence. Mol. Cell. Biol. 2005;25:2158–2168. doi: 10.1128/MCB.25.6.2158-2168.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chai W, Du Q, Shay JW, Wright WE. Human Telomeres Have Different Overhang Sizes at Leading versus Lagging Strands. Mol. Cell. 2006;21:427–435. doi: 10.1016/j.molcel.2005.12.004. [DOI] [PubMed] [Google Scholar]
- 9.Harley CB, Futcher AB, Greider CW. Telomeres Shorten During Ageing of Human Fibroblasts. Nature. 1990;345:458–460. doi: 10.1038/345458a0. [DOI] [PubMed] [Google Scholar]
- 10.Allsopp RC, Vaziri H, Patterson C, Goldstein S, Younglai EV, Futcher AB, Greider CW, Harley CB. Telomere Length Predicts Replicative Capacity of Human Fibroblasts. Proc. Natl. Acad. Sci. USA. 1992;89:10114–10118. doi: 10.1073/pnas.89.21.10114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanahan D, Weinberg RA. The Hallmarks of Cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 12.Masutomi K, Yu EY, Khurts S, Ben-Porath I, Currier JL, Metz GB, Brooks MW, Kaneko S, Murakami S, DeCaprio JA, Weinberg RA, Stewart SA, Hahn WC. Telomerase Maintains Telomere Structure in Normal Human Cells. Cell. 2003;114:241–253. doi: 10.1016/s0092-8674(03)00550-6. [DOI] [PubMed] [Google Scholar]
- 13.Autexier C, Lue NF. The Structure and Function of Telomerase Reverse Transcriptase. Annu. Rev. Biochem. 2006;75:493–517. doi: 10.1146/annurev.biochem.75.103004.142412. [DOI] [PubMed] [Google Scholar]
- 14.Bertuch AA, Lundblad V. The Maintenance and Masking of Chromosome Termini. Curr. Opin. Cell Biol. 2006;18:247–253. doi: 10.1016/j.ceb.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 15.Morin GB. The Human Telomere Terminal Transferase Enzyme Is a Ribonucleoprotein That Synthesizes TTAGGG Repeats. Cell. 1989;59:521–529. doi: 10.1016/0092-8674(89)90035-4. [DOI] [PubMed] [Google Scholar]
- 16.Masutomi K, Possemato R, Wong JM, Currier JL, Tothova Z, Manola JB, Ganesan S, Lansdorp PM, Collins K, Hahn WC. The Telomerase Reverse Transcriptase Regulates Chromatin State and DNA Damage Responses. Proc. Natl. Acad. Sci. USA. 2005;102:8222–8227. doi: 10.1073/pnas.0503095102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu D, O'Connor MS, Qin J, Songyang Z. Telosome, a Mammalian Telomere-Associated Complex Formed by Multiple Telomeric Proteins. J. Biol. Chem. 2004;279:51338–51342. doi: 10.1074/jbc.M409293200. [DOI] [PubMed] [Google Scholar]
- 18.de Lange T. Shelterin: The Protein Complex That Shapes and Safeguards Human Telomeres. Genes Dev. 2005;19:2100–2110. doi: 10.1101/gad.1346005. [DOI] [PubMed] [Google Scholar]
- 19.Hastie ND, Dempster M, Dunlop MG, Thompson AM, Green DK, Allshire RC. Telomere Reduction in Human Colorectal Carcinoma and with Ageing. Nature. 1990;346:866–868. doi: 10.1038/346866a0. [DOI] [PubMed] [Google Scholar]
- 20.Counter CM, Avilion AA, LeFeuvre CE, Stewart NG, Greider CW, Harley CB, Bacchetti S. Telomere Shortening Associated with Chromosome Instability Is Arrested in Immortal Cells which Express Telomerase Activity. EMBO J. 1992;11:1921–1929. doi: 10.1002/j.1460-2075.1992.tb05245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Counter CM, Botelho FM, Wang P, Harley CB, Bacchetti S. Stabilization of Short Telomeres and Telomerase Activity Accompany Immortalization of Epstein-Barr Virus- Transformed Human B Lymphocytes. J. Virol. 1994;68:3410–3414. doi: 10.1128/jvi.68.5.3410-3414.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim NW, Piatyszek MA, Prowse KR, Harley CB, West MD, Ho PL, Coviello GM, Wright WE, Weinrich SL, Shay JW. Specific Association of Human Telomerase Activity with Immortal Cells and Cancer. Science. 1994;266:2011–2015. doi: 10.1126/science.7605428. [DOI] [PubMed] [Google Scholar]
- 23.Nakajima A, Tauchi T, Sashida G, Sumi M, Abe K, Yamamoto K, Ohyashiki JH, Ohyashiki K. Telomerase Inhibition Enhances Apoptosis in Human Acute Leukemia Cells: Possibility of Antitelomerase Therapy. Leukemia. 2003;17:560–567. doi: 10.1038/sj.leu.2402825. [DOI] [PubMed] [Google Scholar]
- 24.Sumi M, Tauchi T, Sashida G, Nakajima A, Gotoh A, Shin-Ya K, Ohyashiki JH, Ohyashiki K. A G-Quadruplex-Interactive Agent, Telomestatin (SOT-095), Induces Telomere Shortening with Apoptosis and Enhances Chemosensitivity in Acute Myeloid Leukemia. Int. J. Oncol. 2004;24:1481–1487. [PubMed] [Google Scholar]
- 25.Tauchi T, Shin-ya K, Sashida G, Sumi M, Okabe S, Ohyashiki JH, Ohyashiki K. Telomerase Inhibition with a Novel G-Quadruplex-Interactive Agent, Telomestatin: In Vitro and in Vivo Studies in Acute Leukemia. Oncogene. 2006;25:5719–5725. doi: 10.1038/sj.onc.1209577. [DOI] [PubMed] [Google Scholar]
- 26.Blackburn EH, Greider CW, Szostak JW. Telomeres and Telomerase: The Path from Maize, Tetrahymena and Yeast to Human Cancer and Aging. Nat. Med. 2006;12:1133–1138. doi: 10.1038/nm1006-1133. [DOI] [PubMed] [Google Scholar]
- 27.Komata T, Kanzawa T, Kondo Y, Kondo S. Telomerase As a Therapeutic Target for Malignant Gliomas. Oncogene. 2002;21:656–663. doi: 10.1038/sj.onc.1205072. [DOI] [PubMed] [Google Scholar]
- 28.Cech TR. Beginning to Understand the End of the Chromosome. Cell. 2004;116:273–279. doi: 10.1016/s0092-8674(04)00038-8. [DOI] [PubMed] [Google Scholar]
- 29.Gomez D, Paterski R, Lemarteleur T, Shin-Ya K, Mergny J-L, Riou JF. Interaction of Telomestatin with the Telomeric Single-Strand Overhang. J. Biol. Chem. 2004;279:41487–41494. doi: 10.1074/jbc.M406123200. [DOI] [PubMed] [Google Scholar]
- 30.Karlseder J, Broccoli D, Dai Y, Hardy S, de Lange T. p53- and ATM-Dependent Apoptosis Induced by Telomeres Lacking TRF2. Science. 1999;283:1321–1325. doi: 10.1126/science.283.5406.1321. [DOI] [PubMed] [Google Scholar]
- 31.Karlseder J, Smogorzewska A, de Lange T. Senescence Induced by Altered Telomere State, Not Telomere Loss. Science. 2002;295:2446–2449. doi: 10.1126/science.1069523. [DOI] [PubMed] [Google Scholar]
- 32.Biroccio A, Rizzo A, Elli R, Koering CE, Belleville A, Benassi B, Leonetti C, Stevens MF, D'Incalci M, Zupi G, Gilson E. TRF2 Inhibition Triggers Apoptosis and Reduces Tumourigenicity of Human Melanoma Cells. Eur. J. Cancer. 2006;42:1881–1888. doi: 10.1016/j.ejca.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 33.Garvik B, Carson M, Hartwell L. Single-Stranded DNA Arising at Telomeres in cdc13 Mutants May Constitute a Specific Signal for the RAD9 Checkpoint. Mol. Cell. Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qi H, Li TK, Kuo D, Nur EKA, Liu LF. Inactivation of Cdc13p Triggers MEC1- Dependent Apoptotic Signals in Yeast. J. Biol. Chem. 2003;278:15136–15141. doi: 10.1074/jbc.M212808200. [DOI] [PubMed] [Google Scholar]
- 35.Shin-ya K, Wierzba K, Matsuo K, Ohtani T, Yamada Y, Furihata K, Hayakawa Y, Seto H. Telomestatin, a Novel Telomerase Inhibitor from Streptomyces anulatus. J. Am. Chem. Soc. 2001;123:1262–1263. doi: 10.1021/ja005780q. [DOI] [PubMed] [Google Scholar]
- 36.Kim MY, Vankayalapati H, Shin-Ya K, Wierzba K, Hurley LH. Telomestatin, a Potent Telomerase Inhibitor That Interacts Quite Specifically with the Human Telomeric Intramolecular G-Quadruplex. J. Am. Chem. Soc. 2002;124:2098–2099. doi: 10.1021/ja017308q. [DOI] [PubMed] [Google Scholar]
- 37.Kim M-Y, Gleason-Guzman M, Izbicka E, Nishioka D, Hurley LH. The Different Biological Effects of Telomestatin and TMPyP4 Can Be Attributed to Their Selectivity for Interaction with Intramolecular or Intermolecular G-Quadruplex Structures. Cancer Res. 2003;63:3247–3256. [PubMed] [Google Scholar]
- 38.Wang Y, Patel DJ. Solution Structure of the Human Telomeric Repeat d[AG3(T2AG3)3] G-Tetraplex. Structure. 1993;1:263–282. doi: 10.1016/0969-2126(93)90015-9. [DOI] [PubMed] [Google Scholar]
- 39.Mergny JL, Phan AT, Lacroix L. Following G-Quartet Formation by UV-Spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 40.Parkinson GN, Lee MP, Neidle S. Crystal Structure of Parallel Quadruplexes from Human Telomeric DNA. Nature. 2002;417:876–880. doi: 10.1038/nature755. [DOI] [PubMed] [Google Scholar]
- 41.Ren J, Qu X, Trent JO, Chaires JB. Tiny Telomere DNA. Nucleic Acids Res. 2002;30:2307–2315. doi: 10.1093/nar/30.11.2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li J, Correia JJ, Wang L, Trent JO, Chaires JB. Not So Crystal Clear: The Structure of the Human Telomere G-Quadruplex in Solution Differs from That Present in a Crystal. Nucleic Acids Res. 2005;33:4649–4659. doi: 10.1093/nar/gki782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ambrus A, Chen D, Dai J, Bialis T, Jones RA, Yang D. Human Telomeric Sequence Forms a Hybrid-Type Intramolecular G-Quadruplex Structure with Mixed Parallel/Antiparallel Strands in Potassium Solution. Nucleic Acids Res. 2006;34:2723–2735. doi: 10.1093/nar/gkl348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu Y, Noguchi Y, Sugiyama H. The New Models of the Human Telomere d[AGGG(TTAGGG)3] in K+ solution. Bioorg. Med. Chem. 2006;14:5584–5591. doi: 10.1016/j.bmc.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 45.Luu KN, Phan AT, Kuryavyi V, Lacroix L, Patel DJ. Structure of the Human Telomere in K+ Solution: An Intramolecular (3 + 1) G-Quadruplex Scaffold. J. Am. Chem. Soc. 2006;128:9963–9970. doi: 10.1021/ja062791w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dai J, Punchihewa C, Ambrus A, Chen D, Jones RA, Yang D. Structure of the Intramolecular Human Telomeric G-Quadruplex in Potassium Solution: A Novel Adenine Triple Formation. Nucleic Acids Res. 2007;35:2440–2450. doi: 10.1093/nar/gkm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Phan AT, Kuryavyi V, Luu KN, Patel DJ. Structure of Two Intramolecular G-Quadruplexes Formed by Natural Human Telomere Sequences in K+ Solution. Nucleic Acids Res. 2007;35:6517–6525. doi: 10.1093/nar/gkm706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Phan AT, Luu KN, Patel DJ. Different Loop Arrangements of Intramolecular Human Telomeric (3+1) G-Quadruplexes in K+ Solution. Nucleic Acids Res. 2006;34:5715–5719. doi: 10.1093/nar/gkl726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Paeschke K, Simonsson T, Postberg J, Rhodes D, Lipps HJ. Telomere End-Binding Proteins Control the Formation of G-Quadruplex DNA Structures In Vivo. Nat. Struct. Biol. 2005;12:847–854. doi: 10.1038/nsmb982. [DOI] [PubMed] [Google Scholar]
- 50.Chang C-C, Chu J-F, Kao F-J, Chiu Y-C, Lou P-J, Chen H-C, Chang T-C. Verification of Antiparallel G-Quadruplex Structure in Human Telomeres by Using Two-Photon Excitation Fluorescence Lifetime Imaging Microscopy of the 3,6-Bis(1-methyl-4-vinylpyridinium)carbazole Diiodide Molecule. Anal. Chem. 2006;78:2810–2815. doi: 10.1021/ac052218f. [DOI] [PubMed] [Google Scholar]
- 51.Tauchi T, Shin-Ya K, Sashida G, Sumi M, Nakajima A, Shimamoto T, Ohyashiki JH, Ohyashiki K. Activity of a Novel G-Quadruplex-Interactive Telomerase Inhibitor, Telomestatin (SOT-095), Against Human Leukemia Cells: Involvement of ATM-Dependent DNA Damage Response Pathways. Oncogene. 2003;22:5338–5347. doi: 10.1038/sj.onc.1206833. [DOI] [PubMed] [Google Scholar]
- 52.Tahara H, Shin-Ya K, Seimiya H, Yamada H, Tsuruo T, Ide T. G-Quadruplex Stabilization by Telomestatin Induces TRF2 Protein Dissociation from Telomeres and Anaphase Bridge Formation Accompanied by Loss of the 3' Telomeric Overhang in Cancer Cells. Oncogene. 2006;25:1955–1966. doi: 10.1038/sj.onc.1209217. [DOI] [PubMed] [Google Scholar]
- 53.De Cian A, Cristofari G, Reichenbach P, De Lemos E, Monchaud D, Teulade-Fichou MP, Shin-Ya K, Lacroix L, Lingner J, Mergny JL. Reevaluation of Telomerase Inhibition by Quadruplex Ligands and Their Mechanisms of Action. Proc. Natl. Acad. Sci. USA. 2007;104:17347–17352. doi: 10.1073/pnas.0707365104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gomez D, O'Donohue MF, Wenner T, Douarre C, Macadre J, Koebel P, Giraud-Panis M-J, Kaplan H, Kolkes A, Shin-ya K, Riou J-F. The G-Quadruplex Ligand Telomestatin Inhibits POT1 Binding to Telomeric Sequences In Vitro and Induces GFP-POT1 Dissociation from Telomeres in Human Cells. Cancer Res. 2006;66:6908–6912. doi: 10.1158/0008-5472.CAN-06-1581. [DOI] [PubMed] [Google Scholar]
- 55.Gomez D, Wenner T, Brassart B, Douarre C, O'Donohue MF, El Khoury V, Shin-Ya K, Morjani H, Trentesaux C, Riou J-F. Telomestatin-Induced Telomere Uncapping Is Modulated by POT1 Through G-Overhang Extension in HT1080 Human Tumor Cells. J. Biol. Chem. 2006;281:38721–38729. doi: 10.1074/jbc.M605828200. [DOI] [PubMed] [Google Scholar]
- 56.Han H, Hurley LH. G-Quadruplex DNA: A Potential Target for Anti-Cancer Drug Design. Trends Pharmacol. Sci. 2000;21:136–142. doi: 10.1016/s0165-6147(00)01457-7. [DOI] [PubMed] [Google Scholar]
- 57.Hurley LH, Wheelhouse RT, Sun D, Kerwin SM, Salazar M, Fedoroff OY, Han FX, Han H, Izbicka E, Von Hoff DD. G-Quadruplexes As targets for drug design. Pharmacol. Ther. 2000;85:141–158. doi: 10.1016/s0163-7258(99)00068-6. [DOI] [PubMed] [Google Scholar]
- 58.Neidle S, Read MA. G-Quadruplexes As Therapeutic Targets. Biopolymers. 2000;56:195–208. doi: 10.1002/1097-0282(2000)56:3<195::AID-BIP10009>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 59.Hurley LH. Secondary DNA Structures As Molecular Targets for Cancer Therapeutics. Biochem. Soc. Trans. 2001;29:692–696. doi: 10.1042/0300-5127:0290692. [DOI] [PubMed] [Google Scholar]
- 60.Perry PJ, Jenkins TC. DNA Tetraplex-Binding Drugs: Structure-Selective Targeting Is Critical for Antitumour Telomerase Inhibition. Mini Rev. Med. Chem. 2001;1:31–41. doi: 10.2174/1389557013407304. [DOI] [PubMed] [Google Scholar]
- 61.Helder MN, Wisman GB, van der Zee GJ. Telomerase and Telomeres: From Basic Biology to Cancer Treatment. Cancer Invest. 2002;20:82–101. doi: 10.1081/cnv-120000370. [DOI] [PubMed] [Google Scholar]
- 62.Perry PJ, Reszka AP, Wood AA, Read MA, Gowan SM, Dosanjh HS, Trent JO, Jenkins TC, Kelland LR, Neidle S. Human Telomerase Inhibition by Regioisomeric Disubstituted Amidoanthracene-9,10-diones. J. Med. Chem. 1998;41:4873–4884. doi: 10.1021/jm981067o. [DOI] [PubMed] [Google Scholar]
- 63.Sun D, Thompson B, Cathers BE, Salazar M, Kerwin SM, Trent JO, Jenkins TC, Neidle S, Hurley LH. Inhibition of Human Telomerase by a G-Quadruplex- Interactive Compound. J. Med. Chem. 1997;40:2113–2116. doi: 10.1021/jm970199z. [DOI] [PubMed] [Google Scholar]
- 64.Gowan SM, Heald R, Stevens MF, Kelland LR. Potent Inhibition of Telomerase by Small-Molecule Pentacyclic Acridines Capable of Interacting with G-Quadruplexes. Mol. Pharmacol. 2001;60:981–988. doi: 10.1124/mol.60.5.981. [DOI] [PubMed] [Google Scholar]
- 65.Gowan SM, Harrison JR, Patterson L, Valenti M, Read MA, Neidle S, Kelland LR. A G-Quadruplex-Interactive Potent Small-Molecule Inhibitor of Telomerase Exhibiting In Vitro and In Vivo Antitumor Activity. Mol. Pharmacol. 2002;61:1154–1162. doi: 10.1124/mol.61.5.1154. [DOI] [PubMed] [Google Scholar]
- 66.Harrison RJ, Cuesta J, Chessari G, Read MA, Basra SK, Reszka AP, Morrell J, Gowan SM, Incles CM, Tanious FA, Wilson WD, Kelland LR, Neidle S. Trisubstituted Acridine Derivatives As Potent and Selective Telomerase Inhibitors. J. Med. Chem. 2003;46:4463–4476. doi: 10.1021/jm0308693. [DOI] [PubMed] [Google Scholar]
- 67.Harrison RJ, Reszka AP, Haider SM, Romagnoli B, Morrell J, Read MA, Gowan SM, Incles CM, Kelland LR, Neidle S. Evaluation of Disubstituted Acridone Derivatives As Telomerase Inhibitors: The Importance of G-Quadruplex Binding. Bioorg. Med. Chem. Lett. 2004;14:5845–5849. doi: 10.1016/j.bmcl.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 68.Moore MJ, Schultes CM, Cuesta J, Cuenca F, Gunaratnam M, Tanious FA, Wilson WD, Neidle S. Trisubstituted Acridines as G-Quadruplex Telomere Targeting Agents. Effects of Extensions of the 3,6- and 9-Side Chains on Quadruplex Binding, Telomerase Activity, and Cell Proliferation. J. Med. Chem. 2006;49:582–599. doi: 10.1021/jm050555a. [DOI] [PubMed] [Google Scholar]
- 69.Anantha NV, Azam M, Sheardy RD. Porphyrin Binding to Quadruplexed T4G4. Biochemistry. 1998;37:2709–2714. doi: 10.1021/bi973009v. [DOI] [PubMed] [Google Scholar]
- 70.Arthanari H, Basu S, Kawano TL, Bolton PH. Fluorescent Dyes Specific for Quadruplex DNA. Nucleic Acids Res. 1998;26:3724–3728. doi: 10.1093/nar/26.16.3724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Arthanari H, Bolton PH. Porphyrins Can Catalyze the Interconversion of DNA Quadruplex Structural Types. Anticancer Drug Des. 1999;14:317–326. [PubMed] [Google Scholar]
- 72.Han FX, Wheelhouse RT, Hurley LH. Interactions of TMPyP4 and TMPyP2 with Quadruplex DNA. Structural Basis for the Differential Effects on Telomerase Inhibition. J. Am. Chem. Soc. 1999;121:3561–3570. [Google Scholar]
- 73.Haq I, Trent JO, Chowdhry BZ, Jenkins TC. Intercalative G-Tetraplex Stabilization of Telomeric DNA by a Cationic Porphyrin. J. Am. Chem. Soc. 1999;121:1768–1779. [Google Scholar]
- 74.Izbicka E, Wheelhouse RT, Raymond E, Davidson KK, Lawrence RA, Sun D, Windle BE, Hurley LH, Von Hoff DD. Effects of Cationic Porphyrins as G-Quadruplex Interactive Agents in Human Tumor Cells. Cancer Res. 1999;59:639–644. [PubMed] [Google Scholar]
- 75.Han H, Langley DR, Rangan A, Hurley LH. Selective Interactions of Cationic Porphyrins with G-Quadruplex Structures. J. Am. Chem. Soc. 2001;123:8902–8913. doi: 10.1021/ja002179j. [DOI] [PubMed] [Google Scholar]
- 76.Shi DF, Wheelhouse RT, Sun D, Hurley LH. Quadruplex-Interactive Agents As Telomerase Inhibitors: Synthesis of Porphyrins and Structure-Activity Relationship for the Inhibition of Telomerase. J. Med. Chem. 2001;44:4509–4523. doi: 10.1021/jm010246u. [DOI] [PubMed] [Google Scholar]
- 77.Grand CL, Han H, Munoz RM, Weitman S, Von Hoff DD, Hurley LH, Bearss DJ. The Cationic Porphyrin TMPyP4 Down-Regulates c-MYC and Human Telomerase Reverse Transcriptase Expression and Inhibits Tumor Growth In Vivo. Mol. Cancer Ther. 2002;1:565–573. [PubMed] [Google Scholar]
- 78.Yamashita T, Uno T, Ishikawa Y. Stabilization of Guanine Quadruplex DNA by the Binding of Porphyrins with Cationic Side Arms. Bioorg. Med. Chem. 2005;13:2423–2430. doi: 10.1016/j.bmc.2005.01.041. [DOI] [PubMed] [Google Scholar]
- 79.Moorhouse AD, Santos AM, Gunaratnam M, Moore M, Neidle S, Moses JE. Stabilization of G-Quadruplex DNA by Highly Selective Ligands via Click Chemistry. J. Am. Chem. Soc. 2006;128:15972–15973. doi: 10.1021/ja0661919. [DOI] [PubMed] [Google Scholar]
- 80.Fedoroff OY, Salazar M, Han H, Chemeris VV, Kerwin SM, Hurley LH. NMR-Based Model of a Telomerase-Inhibiting Compound Bound to G-Quadruplex DNA. Biochemistry. 1998;37:12367–12374. doi: 10.1021/bi981330n. [DOI] [PubMed] [Google Scholar]
- 81.Han H, Cliff CL, Hurley LH. Accelerated Assembly of G-Quadruplex Structures by a Small Molecule. Biochemistry. 1999;38:6981–6986. doi: 10.1021/bi9905922. [DOI] [PubMed] [Google Scholar]
- 82.Mazzitelli CL, Brodbelt JS, Kern JT, Rodriguez M, Kerwin SM. Evaluation of Binding of Perylene Diimide and Benzannulated Perylene Diimide Ligands to DNA by Electrospray Ionization Mass Spectrometry. J. Am. Soc. Mass Spectrom. 2006;17:593–604. doi: 10.1016/j.jasms.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 83.Guo Q, Lu M, Marky LA, Kallenbach NR. Interaction of the Dye Ethidium Bromide with DNA Containing Guanine Repeats. Biochemistry. 1992;31:2451–2455. doi: 10.1021/bi00124a002. [DOI] [PubMed] [Google Scholar]
- 84.Koeppel F, Riou JF, Laoui A, Mailliet P, Arimondo PB, Labit D, Petitgenet O, Helene C, Mergny JL. Ethidium Derivatives Bind to G-Quartets, Inhibit Telomerase and Act As Fluorescent Probes for Quadruplexes. Nucleic Acids Res. 2001;29:1087–1096. doi: 10.1093/nar/29.5.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Perry PJ, Read MA, Davies RT, Gowan SM, Reszka AP, Wood AA, Kelland LR, Neidle S. 2,7-Disubstituted Amidofluorenone Derivatives As Inhibitors of Human Telomerase. J. Med. Chem. 1999;42:2679–2684. doi: 10.1021/jm990084q. [DOI] [PubMed] [Google Scholar]
- 86.Mergny J-L, Lacroix L, Teulade-Fichou M-P, Hounsou C, Guittat L, Hoarau M, Arimondo PB, Vigneron J-P, Lehn J-M, Riou J-F, Garestier T, Helene C. Telomerase Inhibitors Based on Quadruplex Ligands Selected by a Fluorescence Assay. Proc. Natl. Acad. Sci. USA. 2001;98:3062–3067. doi: 10.1073/pnas.051620698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Leonetti C, Amodei S, D'Angelo C, Rizzo A, Benassi B, Antonelli A, Elli R, Stevens MF, D'Incalci M, Zupi G, Biroccio A. Biological Activity of the G-Quadruplex Ligand RHPS4 (3,11-Difluoro-6,8,13-trimethyl-8H-quino[4,3,2-kl]acridinium Methosulfate) Is Associated with Telomere Capping Alteration. Mol. Pharmacol. 2004;66:1138–1146. doi: 10.1124/mol.104.001537. [DOI] [PubMed] [Google Scholar]
- 88.Cookson JC, Dai F, Smith V, Heald RA, Laughton CA, Stevens MF, Burger AM. Pharmacodynamics of the G-Quadruplex-Stabilizing Telomerase Inhibitor 3,11-difluoro-6,8,13-trimethyl-8H-quino[4,3,2-kl]acridinium Methosulfate (RHPS4) In Vitro: Activity in Human Tumor Cells Correlates with Telomere Length and Can Be Enhanced, or Antagonized, with Cytotoxic Agents. Mol. Pharmacol. 2005;68:1551–1558. doi: 10.1124/mol.105.013300. [DOI] [PubMed] [Google Scholar]
- 89.Duan W, Rangan A, Vankayalapati H, Kim MY, Zeng Q, Sun D, Han H, Fedoroff OY, Nishioka D, Rha SY, Izbicka E, Von Hoff DD, Hurley LH. Design and Synthesis of Fluoroquinophenoxazines That Interact with Human Telomeric G-Quadruplexes and Their Biological Effects. Mol. Cancer Ther. 2001;1:103–120. [PubMed] [Google Scholar]
- 90.Kim MY, Duan W, Gleason-Guzman M, Hurley LH. Design, Synthesis, and Biological Evaluation of a Series of Fluoroquinoanthroxazines with Contrasting Dual Mechanisms of Action Against Topoisomerase II and G-Quadruplexes. J. Med. Chem. 2003;46:571–583. doi: 10.1021/jm0203377. [DOI] [PubMed] [Google Scholar]
- 91.Binz N, Shalaby T, Rivera P, Shin-ya K, Grotzer MA. Telomerase Inhibition, Telomere Shortening, Cell Growth Suppression and Induction of Apoptosis by Telomestatin in Childhood Neuroblastoma Cells. Eur. J. Cancer. 2005;41:2873–2881. doi: 10.1016/j.ejca.2005.08.025. [DOI] [PubMed] [Google Scholar]
- 92.Chamberlin MJ. Comparative Properties of DNA, RNA, and Hybrid Homopolymer Pairs. Federation Proceedings. 1965;24:1446–1457. [PubMed] [Google Scholar]
- 93.Riley M, Maling B, Chamberlin MJ. Physical and Chemical Characterization of Two- and Three-Stranded Adenine-Thymine and Adenine-Uracil Homopolymer Complexes. J. Mol. Biol. 1966;20:359–389. doi: 10.1016/0022-2836(66)90069-6. [DOI] [PubMed] [Google Scholar]
- 94.Shafer RH. Stability and Structure of Model DNA Triplexes and Quadruplexes and Their Interactions with Small Ligands. Prog. Nucleic Acid Res. Mol. Biol. 1998;59:55–94. doi: 10.1016/s0079-6603(08)61029-6. [DOI] [PubMed] [Google Scholar]
- 95.Barbieri CM, Srinivasan AR, Rzuczek SG, Rice JE, Lavoie EJ, Pilch DS. Defining the Mode, Energetics and Specificity with Which a Macrocyclic Hexaoxazole Binds to Human Telomeric G-Quadruplex DNA. Nucleic Acids Res. 2007;35:3272–3286. doi: 10.1093/nar/gkm188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.De Cian A, Guittat L, Shin-Ya K, Riou J-F, Mergny J-L. Affinity and Selectivity of G4 Ligands Measured by FRET. Nucleic Acids Symp. Ser. 2005;49:235–236. doi: 10.1093/nass/49.1.235. [DOI] [PubMed] [Google Scholar]
- 97.Rosu F, Gabelica V, Shin-ya K, De Pauw E. Telomestatin-Induced Stabilization of the Human Telomeric DNA Quadruplex Monitored by Electrospray Mass Spectrometry. Chem. Comm. 2003;21:2702–2703. doi: 10.1039/b309394h. [DOI] [PubMed] [Google Scholar]
- 98.Minhas GS, Pilch DS, Kerrigan JE, Lavoie EJ, Rice JE. Synthesis and G-Quadruplex Stabilizing Properties of a Series of Oxazole-Containing Macrocycles. Bioorg. Med. Chem. Lett. 2006;16:3891–3895. doi: 10.1016/j.bmcl.2006.05.038. [DOI] [PubMed] [Google Scholar]
- 99.Fisher HF, Singh N. Calorimetric Methods for Interpreting Protein-Ligand Interactions. Methods Enzymol. 1995;259:194–221. doi: 10.1016/0076-6879(95)59045-5. [DOI] [PubMed] [Google Scholar]
- 100.Haq I, Ladbury JE, Chowdhry BZ, Jenkins TC, Chaires JB. Specific Binding of Hoechst 33258 to the d(CGCAAATTTGCG)2 Duplex: Calorimetric and Spectroscopic Studies. J. Mol.Biol. 1997;271:244–257. doi: 10.1006/jmbi.1997.1170. [DOI] [PubMed] [Google Scholar]
- 101.Chaires JB. Energetics of Drug-DNA Interactions. Biopolymers Nucleic Acid Sci. 1998;44:201–215. doi: 10.1002/(SICI)1097-0282(1997)44:3<201::AID-BIP2>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 102.Haq I, Jenkins TC, Chowdhry BZ, Ren J, Chaires JB. Parsing Free Energies of Drug-DNA Interactions. Methods Enzymol. 2000;323:373–405. doi: 10.1016/s0076-6879(00)23374-0. [DOI] [PubMed] [Google Scholar]
- 103.Mazur S, Tanious FA, Ding D, Kumar A, Boykin DW, Simpson IJ, Neidle S, Wilson WD. A Thermodynamic and Structural Analysis of DNA Minor -Groove Complex Formation. J. Mol. Biol. 2000;300:321–337. doi: 10.1006/jmbi.2000.3869. [DOI] [PubMed] [Google Scholar]
- 104.Barbieri CM, Srinivasan AR, Pilch DS. Deciphering the Origins of Observed Heat Capacity Changes for Aminoglycoside Binding to Prokaryotic and Eukaryotic Ribosomal RNA A-Sites: A Calorimetric, Computational, and Osmotic Stress Study. J. Am. Chem. Soc. 2004;126:14380–14388. doi: 10.1021/ja0457516. [DOI] [PubMed] [Google Scholar]
- 105.Murphy KP, Freire E. Thermodynamics of Structural Stability and Cooperative Folding Behavior in Proteins. Adv. Protein Chem. 1992;43:313–361. doi: 10.1016/s0065-3233(08)60556-2. [DOI] [PubMed] [Google Scholar]
- 106.Spolar RS, Livingstone JR, Record MTJ. Use of Liquid Hydrocarbon and Amide Transfer Data to Estimate Contributions to Thermodynamic Functions of Protein Folding from the Removal of Nonpolar and Polar Surface from Water. Biochemistry. 1992;31:3947–3955. doi: 10.1021/bi00131a009. [DOI] [PubMed] [Google Scholar]
- 107.Spolar RS, Record MT., Jr. Coupling of Local Folding to Site-Specific Binding of Proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- 108.Gómez J, Hilser VJ, Xie D, Freire E. The Heat Capacity of Proteins. Proteins. 1995;22:404–412. doi: 10.1002/prot.340220410. [DOI] [PubMed] [Google Scholar]
- 109.Habermann SM, Murphy KP. Energetics of Hydrogen Bonding in Proteins: a Model Compound Study. Protein Sci. 1996;5:1229–1239. doi: 10.1002/pro.5560050702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ren J, Jenkins TC, Chaires JB. Energetics of DNA Intercalation Reactions. Biochemistry. 2000;39:8439–8447. doi: 10.1021/bi000474a. [DOI] [PubMed] [Google Scholar]
- 111.Sturtevant JM. Heat Capacity and Entropy Changes in Processes Involving Proteins. Proc. Natl. Acad. Sci. USA. 1977;74:2236–2240. doi: 10.1073/pnas.74.6.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Holbrook JA, Capp MW, Saecker RM, Record MT., Jr. Enthalpy and Heat Capacity Changes for Formation of an Oligomeric DNA Duplex: Interpretation in Terms of Coupled Processes of Formation and Association of Single-Stranded Helices. Biochemistry. 1999;38:8409–8422. doi: 10.1021/bi990043w. [DOI] [PubMed] [Google Scholar]
- 113.Kozlov AG, Lohman TM. Adenine Base Unstacking Dominates the Observed Enthalpy and Heat Capacity Changes for the Escherichia coli SSB Tetramer Binding to Singe-Stranded Oligoadenylates. Biochemistry. 1999;38:7388–7397. doi: 10.1021/bi990309z. [DOI] [PubMed] [Google Scholar]
- 114.Kozlov AG, Lohman TM. Large Contributions of Coupled Protonation Equilibria to the Observed Enthalpy and Heat Capacity Changes for ssDNA Binding to Escherichia coli SSB Protein. Proteins: Structure, Function, and Genetics. 2000;41(S4):8–22. doi: 10.1002/1097-0134(2000)41:4+<8::aid-prot20>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 115.Kaul M, Pilch DS. Thermodynamics of Aminoglycoside-rRNA Recognition: The Binding of Neomycin-Class Aminoglycosides to the A Site of 16S rRNA. Biochemistry. 2002;41:7695–7706. doi: 10.1021/bi020130f. [DOI] [PubMed] [Google Scholar]
- 116.Rzuczek SG, Pilch DS, LaVoie EJ, Rice JE. Lysinyl Macrocyclic Hexaoxazoles: Synthesis and Selective G-Quadruplex Stabilizing Properties. Bioorg. Med. Chem. Lett. 2008;18:913–917. doi: 10.1016/j.bmcl.2007.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Plum GE. Optical Methods. In: Beaucage SL, Bergstrom DE, Glick GD, Jones RA, editors. Current Protocols in Nucleic Acid Chemistry. New York: John Wiley & Sons; 2000. pp. 7.3.1–7.3.17. [Google Scholar]
- 118.Mosmann T. Rapid Colorometric Assay for Cellular Growth and Survival: Application to Proliferation and Cytotoxicity Assays. J. Immunol. Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 119.Denizot F, Lang R. Rapid Colorimetric Assay for Cell Growth and Survival. Modifications to the Tetrazolium Dye Procedure Giving Improved Sensitivity and Reliability. J. Immunol. Methods. 1986;89:271–277. doi: 10.1016/0022-1759(86)90368-6. [DOI] [PubMed] [Google Scholar]
- 120.Carmichael J, DeGraff WG, Gazdar AF, Minna JD, Mitchell JB. Evaluation of a Tetrazolium-Based Semiautomated Colorimetric Assay: Assessment of Chemosensitivity Testing. Cancer Res. 1987;47:936–942. [PubMed] [Google Scholar]