Abstract
The Forkhead Box m1 (Foxm1 or Foxm1b) transcription factor (previously called HFH-11B, Trident, Win, or MPP2) is an important positive regulator of DNA replication and mitosis in a variety of cell types. Global deletion of Foxm1 in Foxm1−/− mice is lethal in the embryonic period, causing multiple abnormalities in the liver, heart, lung, and blood vessels. In the present study, Foxm1 was deleted conditionally in the respiratory epithelium (epFoxm1−/−). Surprisingly, deletion of Foxm1 did not alter lung growth, branching morphogenesis, or epithelial proliferation but inhibited lung maturation and caused respiratory failure after birth. Maturation defects in epFoxm1−/− lungs were associated with decreased expression of T1-α and aquaporin 5, consistent with a delay of type I cell differentiation. Expression of surfactant-associated proteins A, B, C, and D was decreased by deletion of Foxm1. Foxm1 directly bound and induced transcriptional activity of the mouse surfactant protein B and A (Sftpb and Sftpa) promoters in vitro, indicating that Foxm1 is a direct transcriptional activator of these genes. Foxm1 is critical for surfactant homeostasis and lung maturation before birth and is required for adaptation to air breathing.
Keywords: epithelial cells, Foxm1, lung development, surfactant proteins
Lung formation in mice begins at 9.5 days postcoitum (E9.5), when the foregut endoderm invades the splanchnic mesenchyme and undergoes dichotomous branching. Branching morphogenesis depends on mesenchymal–epithelial cell signaling mediated by a number of secreted molecules and their receptors, including fibroblast growth factor 10 (Fgf10), sonic hedgehog (Shh), Wnt/β-catenin, transforming growth factor-β, bone morphogenetic protein-4 (Bmp4), and hepatocyte growth factor (Hgf) (see refs. 1 and 2 for review). Before birth, the lung undergoes remarkable anatomic and biochemical changes that generate peripheral saccules and the pulmonary vascular bed required for gas exchange. When peripheral saccules dilate, the synthesis of surfactant proteins and lipids increases. Pulmonary capillaries grow in close apposition to the respiratory epithelium, creating an extensive surface area required for respiration. Pulmonary immaturity and accompanying lack of pulmonary surfactant cause respiratory distress syndrome (RDS), a common cause of morbidity and mortality in preterm infants (3). Mutations in the genes encoding proteins critical for surfactant function, including SP-B, SP-C, and ABCA3, cause respiratory failure or severe lung disease in newborn infants and mice (4). While the prevention and treatment of RDS in newborn infants has improved its clinical outcome, identification of transcriptional pathways regulating perinatal lung maturation and surfactant homeostasis will provide novel targets for genetic screening, diagnosis, and treatment of RDS.
The Forkhead Box (Fox) proteins are an extensive family of transcription factors that share homology in the Winged Helix/Forkhead DNA binding domain. Previous studies demonstrated that Foxa2 plays important roles in lung maturation and differentiation of goblet cells (4, 5), whereas Foxj1 is required for proper development of ciliated cells in the lung (6). Deletion of the Foxf1 gene in mice caused lung hypoplasia and defects in formation of pulmonary capillaries (7). Loss of Foxp2 leads to defective postnatal lung alveolarization (8).
Expression of the Foxm1 transcription factor (previously known as HFH-11B, Trident, Win, or MPP2) is induced during cellular proliferation in a variety of different cell types and extinguished in terminally differentiated cells (9). Foxm1 stimulates aberrant proliferation of tumor cells during progression of liver, lung, and prostate cancers (10–12). Recently, we demonstrated that most of the Foxm1xs−/− embryos die in utero between E13.5 and E16.5 because of defects in development of the embryonic liver, lung, heart, and blood vessels (13–15). Abnormal accumulation of polyploid cells, resulting from diminished DNA replication and failure to enter mitosis, was observed in both the livers and the hearts of Foxm1−/− embryos (13, 15, 16). Foxm1 is required for differentiation of hepatoblast precursor cells toward the biliary epithelial cell lineage, and Foxm1−/− livers fail to form intrahepatic bile ducts (13). Likewise, Foxm1−/− embryos exhibit defects in differentiation of pulmonary mesenchyme into mature capillary endothelial cells during the canalicular stage of lung development (14). Since Foxm1 is required for the differentiation and proliferation of many cell lineages during embryonic development (13, 14, 16, 17), the specific role of Foxm1 in the respiratory epithelium remains unknown.
To investigate the role of Foxm1 during lung development, we generated transgenic mice in which Foxm1 was conditionally deleted in the developing pulmonary epithelium. Surprisingly, the Foxm1 deletion did not influence branching morphogenesis or epithelial cell proliferation, suggesting that Foxm1 is dispensable for DNA replication and mitosis in developing pulmonary epithelial cells. However, deletion of Foxm1 inhibited the anatomic and biochemical maturation of the lung, causing respiratory failure at birth. Maturation defects in Foxm1-deficient lungs were associated with diminished numbers of type I epithelial cells and decreased expression of surfactant-associated proteins A, B, C, and D. In vitro studies demonstrated that Foxm1 binds to and activates promoters of the mouse surfactant protein B and A genes (Sftpb and Sftpa), indicating that Foxm1 controls surfactant homeostasis, at least in part, by directly regulating transcription of surfactant genes.
Results
Conditional Deletion of Foxm1 in the Respiratory Epithelium.
Consistent with previous studies (9), in situ hybridization demonstrated that Foxm1 is expressed in epithelium and mesenchyme of the embryonic lung and other organs [supporting information (SI) Fig. S1 A–G]. To determine whether Foxm1 is required for lung morphogenesis or function, we generated triple-transgenic mice (epFoxm1fl/fl) containing LoxP-flanked exons 4–7 of the Foxm1 gene (Foxm1fl/fl) and the SP-C–rtTAtg/− and the TetO-Cretg/− transgenes. To induce Cre expression in respiratory epithelium, doxycycline (Dox) was given to the dam in water from E7.5 to E14.5. Previous studies demonstrated that using this protocol, Cre-mediated recombination occurs in pulmonary epithelial cells from the initiation of lung development (18). In the presence of Dox, the reverse tetracycline transactivator (rtTA) binds to the TetO promoter and induces expression of Cre recombinase, deleting exons 4–7, which encode the DNA binding and transcriptional activation domains of the Foxm1 gene (Fig. S2 A–B), to produce lung epithelial-specific Foxm1 knockout mice (epFoxm1−/−).
Foxm1 is critical for perinatal survival and adaptation to air breathing.
In epFoxm1−/− newborn mice, the number of lung lobes, body weight, and lung to body weight ratio were unchanged (data not shown). However, deletion of Foxm1 caused respiratory failure and perinatal lethality in 82% of epFoxm1−/− mice within the first 24 h after birth (Table S1). No perinatal lethality was observed in control mice: Dox-treated single transgenic littermates, Dox-treated epFoxm1+/fl mice, or untreated epFoxm1fl/fl mice (data not shown). Histological examination of the epFoxm1−/− newborn mice revealed pulmonary congestion, focal atelectasis, bronchial occlusion, and hyaline membranes lining terminal airways (Figs. 1 A and B and S3 A–F), findings consistent with RDS in preterm infants (3, 4). Approximately 18% of epFoxm1−/− mice survived after birth and displayed mild RDS with focal atelectasis. Newborn epFoxm1−/− mice with severe RDS displayed a significantly decreased Foxm1 mRNA compared to either Foxm1fl/fl mice or epFoxm1−/− mice with mild RDS (Fig. S3G), indicating that a subset of epFoxm1−/− mice survives, most likely because of incomplete deletion of the Foxm1-floxed allele. When assessed before birth, Foxm1−/− mice were present as Mendelian inheritance (Table S1), indicating that Foxm1 was not required for fetal survival in utero, but was critical for survival immediately after birth. Interestingly, Foxm1 deletion during postnatal period (P)03–P30 did not influence overall lung morphology or expression of epithelial marker proteins (Fig. S4).
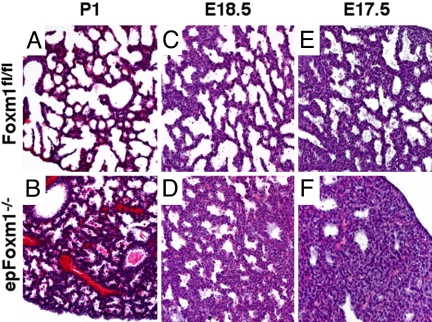
Fig. 1.
Diminished sacculation and delayed lung maturation in epFoxm1−/− mice. Embryonic lungs from epFoxm1−/− and control Foxm1fl/fl P1, E18.5, and E17.5 mice were fixed, paraffin embedded, sectioned, and stained with hematoxylin and eosin (H&E). Lung atelectasis and hyaline membranes were observed in epFoxm1−/− mice (B) with respiratory distress at birth (P1). Normal lung sacculation was observed in control Foxm1fl/fl mice at P1 (A), E18.5 (C), and E17.5 (E). Decreased sacculation and mesenchymal thickening were observed in epFoxm1−/− lungs (D and F). (Magnification, ×100.)
Foxm1 is Not Required for Epithelial Cell Proliferation During Lung Development.
Histological studies were carried out to identify gross morphological defects in epFoxm1−/− lungs. At E15.5, lung structure was not altered by deletion of Foxm1 (Fig. S2C). Distal epithelial cells expressing TTF-1 and proSP-C were readily detected in epFoxm1−/− lungs (Fig. S2C). Consistent with previous studies (18), Cre recombinase protein was detected in 96.8 ± 3.0% of distal lung epithelial cells, but was absent from mesenchymal cells in epFoxm1−/− embryos (Fig. 2A, B, and G). In situ hybridization demonstrated that Foxm1 mRNA was selectively decreased in epithelial cells of epFoxm1−/− lungs (Figs. 2 C, D, H, and I and S1 E–J), confirming efficient deletion of Foxm1 from the distal respiratory epithelium by the Cre recombinase. Because previous studies demonstrated that Foxm1 induces proliferation of hepatocytes, cardiomyocytes, endothelial cells, and numerous neoplastic cells (10, 13, 14, 16, 17), we examined cellular proliferation in the periphery of the epFoxm1−/− lungs. Surprisingly, expression of proliferation-specific KI-67 antigen was unaltered in epFoxm1−/− lungs at E15.5 (Fig. 2 E and F) and E18.5 (Fig. 3C). Furthermore, no significant differences in total numbers of KI-67-positive cells were observed in epFoxm1−/− lungs compared to either control Foxm1fl/fl littermates or age-matched triple-transgenic embryos in the absence of Dox treatment (Fig. 2G). Foxm1 deficiency did not induce apoptosis in the lung as demonstrated by immunostaining of E15.5 sections with an antibody specific to the activated form of caspase 3 (Fig. S2C). Expression of proliferation-specific cyclin B1 mRNA was unaltered after deletion of Foxm1 (Fig. 3B). Thus, Foxm1 is not required for proliferation of distal respiratory epithelial cells during lung development. Significantly decreased mRNA levels of M-phase-promoting Cdc25B phosphatase (Fig. 3B), a known transcriptional target of Foxm1 (15), were observed in the lungs of epFoxm1−/− mice. Thus, reduced expression of Foxm1 and Cdc25B was not sufficient to inhibit proliferation in the Foxm1-deficient respiratory epithelium.
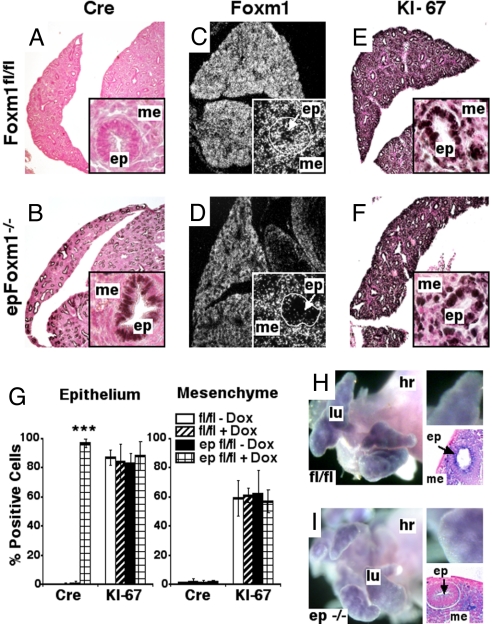
Fig. 2.
Normal proliferation rates in epFoxm1−/− lungs. (A–F) Foxm1 deletion did not influence KI-67 expression in epFoxm1−/− lungs. Paraffin sections from Dox-treated epFoxm1−/− and control Foxm1fl/fl E15.5 lungs were stained with either Cre antibody or KI-67 antibody (dark brown nuclei; slides counterstained with nuclear fast red). (A) Cre was not detected in control Foxm1fl/fl lungs. (B) Cre expression was observed in distal lung epithelium (ep), but not in mesenchymal cells (me) of Dox-treated epFoxm1−/− lungs. (C and D) In situ hybridization shows selective deletion of Foxm1 from epithelial cells of epFoxm1−/− lungs. (Magnifications, ×50 and ×400.) (G) Similar numbers of KI-67 positive cells were observed in epithelial and mesenchymal cellular compartments of Dox-treated epFoxm1−/− E15.5 lungs when compared to either untreated triple transgenic epFoxm1fl/fl embryos or Foxm1fl/fl embryos. Numbers of either Cre-positive or KI-67-positive cells were counted in 10 random 400× microscope fields. Mean ± SD was determined using five embryos in each group. ***, P < 0.001. (H and I) Whole-mount in situ hybridization of E13.5 lungs shows presence of Foxm1 mRNA in epithelium and mesenchyme of Foxm1fl/fl lungs. Foxm1 mRNA is decreased in epFoxm1−/− lung epithelium. lu, lung; hr, heart.
Fig. 3.
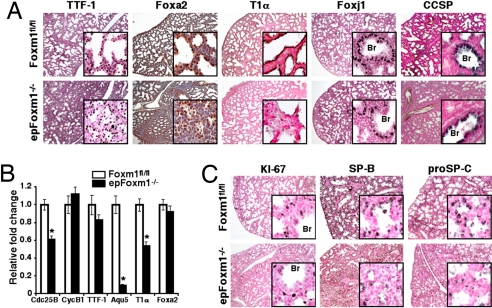
Defects in pulmonary surfactant protein expression and peripheral lung maturation in epFoxm1−/− embryos. (A) Diminished T1-α expression in epFoxm1−/− E18.5 lungs. Lung paraffin sections from Dox-treated epFoxm1−/− and Foxm1fl/fl E18.5 embryos were stained with antibodies against epithelial-specific marker proteins. Sections were counterstained with either hematoxylin (blue, Foxa2 staining) or nuclear fast red (red, A and C). T1-α (T1α) staining was decreased in epFoxm1−/− lungs, whereas TTF-1, Foxa2, Foxj1, and CCSP proteins were unaltered. (B) qRT–PCR of total RNA from epFoxm1−/− E17.5 lungs showed decreased T1α, Cdc25B, and Aqu5 mRNAs. *P < 0.05. (C) Decreased expression of SP-B and proSP-C in epFoxm1−/− lungs. SP-B and proSP-C proteins were readily detected at E18.5 in control Foxm1fl/fl lungs, but were decreased in epFoxm1−/− lungs. KI-67-positive cells are observed in both epFoxm1−/− and Foxm1fl/fl lungs. Br, bronchioles. (Magnifications, ×50 and ×400.)
Foxm1 Deficiency Delays Lung Maturation.
Because abnormal lung maturation contributes to perinatal lethality and respiratory distress (4, 5), we examined epFoxm1−/− embryos at E17.5 and E18.5 during the late canalicular–saccular phase of lung maturation. Decreased size of the peripheral saccules was observed in epFoxm1−/− embryos at E17.5 and E18.5 (Figs. 1 C–F and S3H). Consistent with delayed lung maturation, pulmonary mesenchyme failed to thin and peripheral lung tubules remained closed (Fig. 1F). No vascular abnormalities were detected in epFoxm1−/− lungs as demonstrated by Pecam-1 staining (Fig. S3 I and J). Numbers of squamous type I epithelial cells were decreased in the epFoxm1−/− lungs as demonstrated by immunostaining with antibodies against T1-α (Fig. 3A), a specific marker of type I cells in the mature lung (4). Consistent with diminished numbers of squamous type I cells in epFoxm1−/− lungs, mRNA levels of T1-α and aquaporin 5 were significantly decreased as demonstrated by quantitative real-time (RT-PCR) (qRT–PCR) analysis (Fig. 3B). In contrast, lungs of control Foxm1fl/fl littermates displayed dilated peripheral saccules with thinning of mesenchyme and differentiated squamous type I epithelial cells expressing the T1-α protein (Fig. 3A), a finding consistent with normal lung maturation in late gestation. Despite the reduced number of type I epithelial cells, no ultrastructural abnormalities were detected in type I cells by transmission electronic microscopy (Fig. 4 A and B). However, lamellar bodies in epFoxm1−/− type II cells were significantly smaller compared to those in type II cells in control lungs (Fig. S5 A, B, and E). Ultrastructure and immunohistochemical staining for Foxj1 and Clara cell secretory proteins (CCSP) was consistent with normal differentiation of ciliated and Clara cells in conducting airways of epFoxm1−/− mice (Figs. 3A and S5 C and D). Taken together, these findings indicate that deletion of the Foxm1 gene from respiratory epithelium did not influence differentiation of ciliated and Clara cells, but impaired lung sacculation and delayed differentiation of type I and type II epithelial cells.
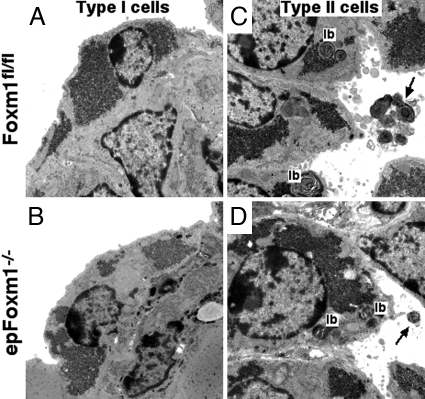
Fig. 4.
Ultrastructure of lung epithelial cells in epFoxm1−/− lungs: transmission electron microscopy of epFoxm1−/− E17.5 lungs. Similar ultrastructure was observed in type I (A and B) and type II (C and D) epithelial cells of epFoxm1−/− and Foxm1fl/fl lungs. Lamellar bodies (lb) and secreted surfactant (arrows) were observed in both epFoxm1−/− and Foxm1fl/fl type II cells. Lamellar body size was significantly decreased in epFoxm1−/− lungs (Fig. S5). (Magnification, ×15,000.)
Foxm1 Regulates Expression of Surfactant Proteins in Developing Lung.
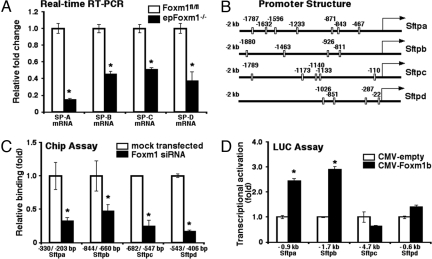
Surfactant-associated proteins (SP-A, SP-B, SP-C, and SP-D) are produced by type II epithelial cells and play important roles in surfactant structure, homeostasis, and function. Mutations in SFTPB and SFTPC genes cause respiratory failure or chronic pulmonary disease in full-term neonates and children, respectively (4). Although differentiated type II cells were observed in epFoxm1−/− mice at the ultrastructural level (Fig. 4 C and D), expression of the genes encoding the surfactant proteins was significantly decreased in the epFoxm1−/− lungs as demonstrated by qRT–PCR analysis of total lung RNA (Fig. 5A) and by immunostaining of lungs at E18.5 (Fig. 3C). Since SP-B is required for formation of lamellar bodies and adaptation to air breathing in mice and humans (4), decreased levels of SP-B may contribute to the respiratory failure in newborn epFoxm1−/− mice. Expression of TTF-1 and Foxa2 transcription factors, both of which are known transcriptional activators of the surfactant genes (2), was not altered by deletion of Foxm1 (Fig. 3 A and B).
Fig. 5.
Foxm1 induces expression of surfactant-associated proteins. (A) qRT–PCR of total lung RNA demonstrated decreased SP-A, SP-B, SP-C, and SP-D mRNAs in epFoxm1−/− E17.5 lungs. (B) Schematic drawings of –2-kb promoter regions of mouse Sftpa, Sftpb, Sftpc, and Sftpd genes are shown. Locations of potential Foxm1 DNA binding sites are indicated (open boxes). (C) ChIP assay demonstrated that Foxm1 protein binds to promoter regions of surfactant protein genes. Cross-linked chromatin from mock-transfected MLE-15 cells or MLE-15 cells transfected with Foxm1-specific siRNA was used for immunoprecipitation (IP) with either Foxm1 antibodies or IgG control. After IP, genomic DNA was analyzed for the amount of surfactant promoter DNA using qPCR. Foxm1 binding to genomic DNA was normalized to IgG control. (D) Foxm1 induced the transcriptional activity of Sftpa and Sftpb promoters. U2OS cells were transfected with CMV-Foxm1b expression vector and luciferase (LUC) reporters driven by the surfactant promoter regions. Cells were harvested at 24 h after transfection and processed for dual LUC assays to determine LUC activity. Transcriptional activities of Sftpa and Sftpb promoters were increased, while activities of Sftpc and Sftpd promoters were not altered. *, P < 0.05.
Foxm1 Induces the Transcription of Sftpb and Sftpa Promoters.
Promoter regions of the mouse surfactant protein genes contain potential Foxm1-binding sites (Fig. 5B). To determine whether Foxm1 protein directly binds to the surfactant promoter regions, we used chromatin immunoprecipitation (ChIP) assays. The cross-linked and sonicated chromatin from untransfected mouse lung epithelial (MLE)-15 cells or MLE-15 cells transfected with Foxm1-specific siRNA (15) was immunoprecipitated (IP) with either Foxm1 or control IgG antibodies. Binding of surfactant promoter DNA associated with the IP chromatin was determined by real-time PCR with primers specific for potential Foxm1-binding sites in the mouse surfactant promoters. Foxm1 protein specifically bound to regulatory regions of the surfactant protein genes (Sftpa, Sftpb, Sftpc, and Sftpd) as demonstrated by the ability of Foxm1 siRNA to reduce binding of Foxm1 protein to each of the gene promoters (Fig. 5C). To determine whether the Foxm1-binding sites were transcriptionally active, cotransfection experiments were performed using CMV-Foxm1b expression vector (14) and luciferase (LUC) reporter constructs driven by promoter regions of the surfactant protein genes. Cotransfection of the CMV-Foxm1b expression vector significantly increased expression of the –0.9-kb Sftpa and the –1.7-kb Sftpb reporter plasmids when compared to CMV-empty vector (Fig. 5D), indicating that Foxm1 is a transcriptional activator of Sftpa and Sftpb genes. In contrast, transcriptional activity of –4.7-kb Sftpc and –0.6-kb Sftpd promoters was not altered by CMV-Foxm1b transfection (Fig. 5D).
Discussion
Cell-selective deletion of exons 4–7 of the Foxm1 gene in the developing respiratory epithelium inhibited the morphological and biochemical maturation of the lung, causing respiratory failure at birth. These exons encode the Foxm1 DNA binding domain and C-terminal transcriptional activation domains, both of which are required for Foxm1 transcriptional activity (13). Despite extensive data supporting the important role of Foxm1 in cell proliferation, deletion of Foxm1 did not alter lung growth or proliferation but impaired lung sacculation and inhibited the expression of genes encoding pulmonary surfactant proteins. Foxm1 deficiency caused decreased numbers of type I cells, suggesting a delayed differentiation of type I cells from its precursors. Alternatively, the decreased number of type I cells may result from diminished numbers of its precursor cells in the epFoxm1−/− lungs.
Deletion of either Foxm1 or Foxa2 transcription factor from respiratory epithelium was sufficient to decrease lung maturation and disrupt surfactant homeostasis (ref. 5 and this article). Thus, both of these Fox transcription factors are required for proper development of lung epithelial cells and secretion of surfactant proteins. Interestingly, Foxm1 and Foxa2 proteins share homologous winged helix DNA binding domains and recognize similar consensus DNA binding sites, raising the possibility that these Fox proteins act through the same DNA binding sequences and regulate similar target genes in developing epithelial cells. However, differences in biochemical and ultrastructural findings in the two models indicate that Foxm1 and Foxa2 play distinct roles in lung morphogenesis. Although Foxa2 did not activate the Sftpa promoter in cotransfection experiments (5), Foxm1 directly induced Sftpa promoter activity, suggesting that Foxm1 and Foxa2 may also regulate unique epithelial target genes during lung development.
While deletion of the Sftpb gene in mice caused pulmonary failure at birth (19), defects in surfactant structure and function caused by lack of Sftpa, Sftpc, or Sftpd are not lethal (reviewed in refs. 2 and 4). In this study, we found that SP-B mRNA in epFoxm1−/− mice was reduced to 40% of that of controls. Since heterozygous Sftpb+/− mice (50% reduction) are susceptible to pulmonary disease under stress (20), it is likely that decreased SP-B contributes to the respiratory failure seen in the epFoxm1−/− mice. Alternatively, it is also possible that the respiratory distress in epFoxm1−/− mice is secondary to an impaired gas exchange resulting from decreased numbers of type I epithelial cells and sacculation defects.
The reduction in SP-B and proSP-C proteins seen by immunohistochemistry was consistent with the mRNA data wherein the intensity of epithelial cell staining was decreased. Decreased expression of all of the surfactant proteins may have influenced pulmonary function at birth. Interestingly, a combined surfactant deficiency was associated with perinatal lethal phenotype in mice deficient for Foxa2, TTF-1, calcineurin b1, ABCA 3, β-catenin, and other genes (reviewed in refs. 2 and 4), indicating a complexity of surfactant regulation in the lung. Although diminished surfactant protein levels could be a consequence of delayed maturation in epFoxm1−/− mice, Foxm1 may directly influence transcription of the surfactant protein genes in vivo. Consistent with this hypothesis, cotransfection studies with MLE-15 cells demonstrated that Foxm1 directly bound and induced the transcriptional activity of Sftpa and Sftpb promoter regions. However, Sftpc and Sftpd promoter constructs were not responsive to Foxm1 in vitro, suggesting that Foxm1 regulates Sftpc and Sftpd expression by indirect mechanisms. Alternatively, DNA regulatory sequences that were not included in the promoter constructs may contain Foxm1-responsive elements.
Foxm1 appears to be dispensable for cell cycle progression in the developing respiratory epithelium. This finding was surprising, considering that Foxm1 directly activates transcription of multiple cell cycle regulators (13, 17, 21). Previous studies from our laboratory demonstrated that mice with global deletion of the Foxm1 gene (Foxm1−/−) exhibited an embryonic lethal phenotype between E13.5 and E16.5 because of severe abnormalities in liver and heart morphogenesis, hypertrophy of blood vessels, and inability of lung mesenchyme to proliferate and to form peripheral pulmonary capillaries (13–15). In support of the role of Foxm1 in cell proliferation, Foxm1 accelerated cellular proliferation in a variety of cell types during liver regeneration, tumor formation, and lung repair in response to lung injury (10–12, 22). In contrast to the previous findings, we demonstrated that deletion of Foxm1 in respiratory epithelial cells throughout lung morphogenesis did not influence epithelial proliferation and branching lung morphogenesis. While expression of M-phase-promoting Cdc25B phosphatase was reduced in epFoxm1−/− mouse lungs, expression of other proliferation-specific Foxm1 targets was not altered. Thus, reduced expression of Cdc25B alone appears to be insufficient to cause proliferation defects in Foxm1-deficient epithelial cells.
In summary, deletion of Foxm1 from respiratory epithelium did not influence branching lung morphogenesis or epithelial proliferation, but impaired lung sacculation, delayed type I cell differentiation, and reduced expression of surfactant-associated proteins, causing respiratory failure after birth. The identification of critical regulators of lung maturation, such as Foxm1, may provide novel strategies for diagnosis, prevention, and treatment of respiratory distress syndrome in preterm infants.
Methods
Mouse Strains.
We previously described the generation of Foxm1flox/flox (Foxm1fl/fl) mice, which contain LoxP sequences flanking DNA binding and transcriptional activation domains (exons 4–7; Fig. S1A) of the Foxm1 gene (13). The Foxm1fl/fl mice were bred with SP-C–rtTAtg/−/TetO-Cretg/tg mice (18) to generate the SP-C–rtTAtg/−/TetO-Cretg/− Foxm1fl/fl triple transgenic mice. Dox (1% in drinking water) was given to pregnant mice during embryonic period E7.5–E14.5. Foxm1fl/fl littermates lacking the SP-C–rtTA, the TetO-Cre, or both transgenes were used as controls. Further controls included SP-C–rtTAtg/−/TetO-Cretg/− Foxm1fl/fl embryos without Dox treatment and Dox-treated SP-C–rtTAtg/−/TetO-Cretg/− Foxm1fl/+ embryos. No lung abnormalities were observed in control mice. Animal studies were reviewed and approved by the Animal Care and Use Committee of Cincinnati Children's Hospital Research Foundation.
Immunohistochemical Staining, in Situ Hybridization, and Transmission Electron Microscopy.
Embryos were harvested, fixed overnight with 10% buffered formalin, and then embedded into paraffin blocks. Paraffin 5-μm sections were stained with hematoxylin and eosin (H&E) for morphological examination. Paraffin sections were also used for immunostaining as described (23). Information about antibodies is provided in SI Text. Paraffin E15.5 sections were also used for in situ hybridization with 35S-labeled antisense riboprobe specific to the 1649- to 1947-bp region of the mouse Foxm1 mRNA as described (9). Whole-mount hybridization of E13.5 lungs was performed as described (24). Transmission electron microscopy was performed on E17.5 lungs as previously described (5).
qRT–PCR.
Total lung RNA was prepared from epFoxm1−/− or Foxm1fl/fl lungs and then analyzed by qRT–PCR using a StepOnePlus Real-Time PCR system (Applied Biosystems). Samples were amplified with TaqMan Gene Expression Master Mix (Applied Biosystems) combined with inventoried TaqMan gene expression assays for the gene of interest (Table S2). Reactions were analyzed in triplicates and expression levels were normalized to β-actin.
Cotransfection Studies and ChIP Assays.
We transfected human osteosarcoma U2OS cells with either CMV-Foxm1b or control CMV-empty expression plasmids and with LUC reporters driven by –0.9-kb mouse Sftpa promoter, –1.7-kb Sftpb promoter, –4.7-kb Sftpc promoter, or –0.6-kb Sftpd promoter. CMV-Renilla was used as an internal control to normalize transfection efficiency. Dual luciferase assay (Promega) was performed 24 h after transfection as described (14, 23).
Nuclear extracts from untransfected or siRNA-transfected mouse lung epithelial MLE-15 cells were cross-linked by addition of formaldehyde, sonicated, and used for the immunoprecipitation with Foxm1 rabbit polyclonal antibodies (H-300, Santa Cruz) as described previously (21). DNA fragments were 500–1000 bp. Reverse cross-linked ChIP DNA samples were subjected to real-time PCR, using the oligonucleotides specific to promoter regions of mouse surfactant genes (Table S3). DNA binding was normalized to control ChIP DNA samples, which were immunoprecipitated using control rabbit serum.
Statistical Analysis.
Student's T-test was used to determine statistical significance. P values ≤0.05 were considered significant. Values for all measurements were expressed as the mean ± SD.
Supplementary Material
Acknowledgments.
We thank Ms. Ann Maher for secretarial support and D. Loudy for technical assistance. This work was supported by March of Dimes Birth Defects Foundation Grant 6-FY2005–1266 (to V.V.K.), Research Scholar Grant RSG-06–187-01-MGO from the American Cancer Society (to V.V.K.), and National Institutes of Health Grants HL61646, HL90156 (to J.A.W.), and HL 84151 (to V.V.K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806748105/DCSupplemental.
References
- 1.Warburton D, et al. The molecular basis of lung morphogenesis. Mech Dev. 2000;92(1):55–81. doi: 10.1016/s0925-4773(99)00325-1. [DOI] [PubMed] [Google Scholar]
- 2.Maeda Y, Dave V, Whitsett JA. Transcriptional control of lung morphogenesis. Physiol Rev. 2007;87(1):219–244. doi: 10.1152/physrev.00028.2006. [DOI] [PubMed] [Google Scholar]
- 3.Avery ME, Mead J. Surface properties in relation to atelectasis and hyaline membrane disease. J Am Med Assoc. 1959;97(5):517–523. doi: 10.1001/archpedi.1959.02070010519001. Part 1. [DOI] [PubMed] [Google Scholar]
- 4.Whitsett JA, Wert SE, Trapnell BC. Genetic disorders influencing lung formation and function at birth. Hum Mol Genet. 2004;13:R207–R215. doi: 10.1093/hmg/ddh252. Spec No 2. [DOI] [PubMed] [Google Scholar]
- 5.Wan H, et al. Foxa2 is required for transition to air breathing at birth. Proc Natl Acad Sci USA. 2004;101(40):14449–14454. doi: 10.1073/pnas.0404424101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brody SL, Yan XH, Wuerffel MK, Song SK, Shapiro SD. Ciliogenesis and left-right axis defects in forkhead factor HFH-4-null mice. Am J Resp Cell Mol Biol. 2000;23(1):45–51. doi: 10.1165/ajrcmb.23.1.4070. [DOI] [PubMed] [Google Scholar]
- 7.Kalinichenko VV, et al. Defects in pulmonary vasculature and perinatal lung hemorrhage in mice heterozygous null for the Forkhead Box f1 transcription factor. Dev Biol. 2001;235:489–506. doi: 10.1006/dbio.2001.0322. [DOI] [PubMed] [Google Scholar]
- 8.Shu W, et al. Foxp2 and Foxp1 cooperatively regulate lung and esophagus development. Development (Cambridge, UK) 2007;134(10):1991–2000. doi: 10.1242/dev.02846. [DOI] [PubMed] [Google Scholar]
- 9.Ye H, et al. Hepatocyte nuclear factor 3/fork head homolog 11 is expressed in proliferating epithelial and mesenchymal cells of embryonic and adult tissues. Mol Cell Biol. 1997;17(3):1626–1641. doi: 10.1128/mcb.17.3.1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kalinichenko VV, et al. Forkhead Box m1b transcription factor is essential for development of hepatocellular carcinomas and is negatively regulated by the p19ARF tumor suppressor. Genes Dev. 2004;18:830–850. doi: 10.1101/gad.1200704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim IM, et al. The Forkhead Box m1 transcription factor stimulates the proliferation of tumor cells during development of lung cancer. Cancer Res. 2006;66(4):2153–2161. doi: 10.1158/0008-5472.CAN-05-3003. [DOI] [PubMed] [Google Scholar]
- 12.Kalin TV, et al. Increased levels of the FoxM1 transcription factor accelerate development and progression of prostate carcinomas in both TRAMP and LADY transgenic mice. Cancer Res. 2006;66(3):1712–1720. doi: 10.1158/0008-5472.CAN-05-3138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krupczak-Hollis K, et al. The mouse Forkhead Box m1 transcription factor is essential for hepatoblast mitosis and development of intrahepatic bile ducts and vessels during liver morphogenesis. Dev Biol. 2004;276:74–88. doi: 10.1016/j.ydbio.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 14.Kim IM, et al. The forkhead box M1 transcription factor is essential for embryonic development of pulmonary vasculature. J Biol Chem. 2005;280:22278–22286. doi: 10.1074/jbc.M500936200. [DOI] [PubMed] [Google Scholar]
- 15.Ramakrishna S, et al. Myocardium defects and ventricular hypoplasia in mice homozygous null for the Forkhead Box M1 transcription factor. Dev Dyn. 2007;236(4):1000–1013. doi: 10.1002/dvdy.21113. [DOI] [PubMed] [Google Scholar]
- 16.Korver W, et al. Uncoupling of S phase and mitosis in cardiomyocytes and hepatocytes lacking the winged-helix transcription factor trident. Curr Biol. 1998;8(24):1327–1330. doi: 10.1016/s0960-9822(07)00563-5. [DOI] [PubMed] [Google Scholar]
- 17.Laoukili J, et al. FoxM1 is required for execution of the mitotic programme and chromosome stability. Nat Cell Biol. 2005;7(2):126–136. doi: 10.1038/ncb1217. [DOI] [PubMed] [Google Scholar]
- 18.Perl AK, Wert SE, Nagy A, Lobe CG, Whitsett JA. Early restriction of peripheral and proximal cell lineages during formation of the lung. Proc Natl Acad Sci USA. 2002;99(16):10482–10487. doi: 10.1073/pnas.152238499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Clark JC, et al. Targeted disruption of the surfactant protein B gene disrupts surfactant homeostasis, causing respiratory failure in newborn mice. Proc Natl Acad Sci USA. 1995;92(17):7794–7798. doi: 10.1073/pnas.92.17.7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tokieda K, et al. Surfactant protein-B-deficient mice are susceptible to hyperoxic lung injury. Am J Resp Cell Mol Biol. 1999;21(4):463–472. doi: 10.1165/ajrcmb.21.4.3436. [DOI] [PubMed] [Google Scholar]
- 21.Wang IC, et al. Forkhead box M1 regulates the transcriptional network of genes essential for mitotic progression and genes encoding the SCF (Skp2-Cks1) ubiquitin ligase. Mol Cell Biol. 2005;25(24):10875–10894. doi: 10.1128/MCB.25.24.10875-10894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kalinichenko VV, et al. Ubiquitous expression of the forkhead box M1B transgene accelerates proliferation of distinct pulmonary cell-types following lung injury. J Biol Chem. 2003;278:37888–37894. doi: 10.1074/jbc.M305555200. [DOI] [PubMed] [Google Scholar]
- 23.Kim IM, et al. Functional characterization of evolutionary conserved DNA regions in forkhead box f1 gene locus. J Biol Chem. 2005;280(45):37908–37916. doi: 10.1074/jbc.M506531200. [DOI] [PubMed] [Google Scholar]
- 24.Lim L, Kalinichenko VV, Whitsett JA, Costa RH. Fusion of right lung lobes and pulmonary vessels in mice heterozygous for the Forkhead Box f1 targeted allele. Am J Physiol Lung Cell Mol Physiol. 2002;282:L1012–L1022. doi: 10.1152/ajplung.00371.2001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.