Abstract
Despite their potential to regulate approximately one-third of the whole genome, relatively few microRNA (miRNA) targets have been experimentally validated, particularly in stratified squamous epithelia. Here we demonstrate not only that the lipid phosphatase SHIP2 is a target of miRNA-205 (miR-205) in epithelial cells, but, more importantly, that the corneal epithelial-specific miR-184 can interfere with the ability of miR-205 to suppress SHIP2 levels. This is the first example of a miRNA negatively regulating another to maintain levels of a target protein. Interfering with miR-205 function by using a synthetic antagomir, or by the ectopic expression of miR-184, leads to a coordinated damping of the Akt signaling pathway via SHIP2 induction. This was associated with a marked increase in keratinocyte apoptosis and cell death. Aggressive squamous cell carcinoma (SCC) cells exhibited elevated levels of miR-205. This was associated with a concomitant reduction in SHIP2 levels. Partial knockdown of endogenous miR-205 in SCCs markedly decreased phosphorylated Akt and phosphorylated BAD levels and increased apoptosis. We were able to increase SHIP2 levels in SCC cells after inhibition of miR-205. Therefore, miR-205 might have diagnostic value in determining the aggressivity of SCCs. Blockage of miR-205 activity with an antagomir or via ectopic expression of miR-184 could be novel therapeutic approaches for treating aggressive SCCs.
MicroRNAs (miRNAs) are small, 20- to 24-nucleotide, noncoding RNAs found in diverse organisms. In animals, most miRNAs mediate posttranscriptional silencing by binding with partial complementarity to the 3′ UTR of the target mRNA (1, 2). These endogenous, silencing RNAs have been shown to play important roles in development and differentiation (3–6), cellular stress responses (7), and cancer (8–11).
The role of miRNAs in stratified squamous epithelia remains poorly understood. Inactivation of Dicer in mouse skin caused hair follicles to evaginate into the epidermis rather than invaginating downward, thus forming cyst-like structures (12, 13). These results underscore the importance of miRNAs in the regulation of epidermal and follicular development. miRNAs have also been extensively profiled in the corneal epithelium and show expression patterns that are regionally restricted (14). For example, miR-184 was the most abundant miRNA in the corneal epithelium; however, it was conspicuously absent from the limbal epithelium, an area enriched in corneal epithelial stem cells (15–18). In contrast, miR-205 is broadly expressed throughout all viable cell layers in nearly all stratified squamous epithelia including the corneal, limbal, and conjunctival epithelia of the eye (12, 14). Thus, the corneal epithelium is unique in that it exhibits distinct as well as overlapping expression of miR-184 and miR-205 (14).
miRNAs have been predicted to regulate thousands of mammalian genes (19); however, few targets have been experimentally validated for the great majority of these miRNAs. With the exception of a recent demonstration that a p63-related family member is negatively regulated by miR-203 (20), little is known about stratified squamous epithelial miRNA targets. We report that miR-205 represses SH2-containing phosphoinositide 5′-phosphatase 2 (SHIP2). Our finding that miR-184 negatively modulates the activity of miR-205 to maintain SHIP2 levels is the first demonstration that a miRNA can interfere with another to ensure the expression of a target protein. We show (i) that SHIP2 levels can be modulated in a variety of epithelial cells using gain- and loss-of-function experiments with miR-184 and miR-205 and (ii) that manipulating SHIP2 levels through miRNAs diminishes Akt signaling leading to decreased keratinocyte survival. Finally, we find a reciprocal relationship between miR-205 and SHIP2 expression in squamous cell carcinoma (SCC) cell lines and suggest that miR-205 may be viewed as a tumor promoter in the context of SCCs.
Results
miR-205 Targets SHIP2.
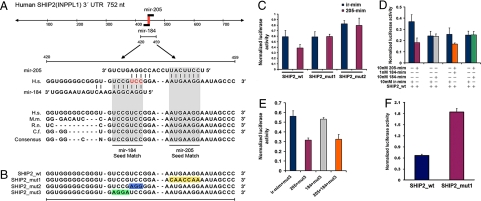
We found miR-205 in all squamous epithelium that we examined (14). We also reported that miR-184 and miR-205 are the most abundant miRNAs in corneal epithelium and that miR-184 expression was restricted to the corneal epithelium (14). Bioinformatic analysis suggested that, in humans, the SHIP2 (Inppl1) 3′ UTR is a putative target of both miR-184 and miR-205 (21) and is the only gene with overlapping binding sites to these two miRNAs. To test this prediction (Fig. 1A) we cotransfected HeLa cells with a miR-184 or miR-205 mimic and luciferase reporter constructs carrying the entire 3′ UTR of SHIP2 mRNA (Fig. 1B). In cells treated with a miR-205 mimic, we found a marked reduction (≈50%) in luciferase activity [Fig. 1 C and D and supporting information (SI) Fig. S1B]; however, no reduction in luciferase activity was seen in transfectants expressing miR-184 (Fig. S1A and Fig. 1D), suggesting that miR-184 does not inhibit SHIP2. To confirm this result, we mutated the miR-205 binding site on SHIP2 3′ UTR (Fig. 1B, SHIP2_mut1). The mutation prevented miR-205 from interfering with luciferase activity, indicating that the 3′ UTR of SHIP2 is indeed a target of miR-205 (Fig. 1C).
Fig. 1.
miR-205 targets SHIP2 at 3′ UTR and can be regulated by miR-184. (A) Sequence of the miR-205 and miR-184 binding sites within the human SHIP2 (INPPL1) 3′ UTR. Red nucleotides are overlapping binding sites. Shaded areas represent conserved complementary nucleotides of miR-184 and miR-205 seed sequences in various mammals (H.s, human; M.m, mouse; R.n, rat; C.f, chicken). (B) Schematic of the reporter constructs showing entire 3′ UTR SHIP2 sequence (SHIP2 _wt) and the mutated 3′ UTR nucleotides (yellow) of the miR-205 binding site (SHIP2_mut1). SHIP2_mut2 represents the reporter construct containing mutated overlapping nucleotides (blue) of miR-184 and miR-205. SHIP2_mut3 represents the reporter construct containing nucleotides (green) predicted to be exclusively used for miR-184 binding to SHIP2 mRNA. (C) Luciferase activity of (i) SHIP2_wt in the presence of 10 nM of miR-205 showing the inhibitory activity of this reporter and (ii) the SHIP2_mut1 and mut2 reporters, showing that miR-205 mimic cannot inhibit the luciferase activity of these constructs compared with the wild-type construct. The overlapping nucleotides of miR-184 and miR-205 are required for miR-205 binding to SHIP2 3′ UTR as is the seed sequence of miR-205. Error bars (SEM) are derived from six experiments in triplicate. (D) Luciferase activity of SHIP2_wt reporter in the presence (+) or absence (−) of various concentrations of miR-205, miR-184, or nontargeting (irrelevant) mimics. Transfection (red) of miR-205 mimic inhibits luciferase activity, whereas transfection (gray) of miR-184 mimic has no effect. Cotransfection (orange) of 1 nM miR-184 and 10 nM miR-205 mimics cannot completely restore luciferase activity, whereas cotransfection (green) of equal amounts of miR-184 and miR-205 mimics completely rescued luciferase activity. Error bars (SEM) are derived from three experiments in triplicate. (E) Luciferase activity of SHIP2_mut3 reporter showing that (i) this mutation does not inhibit miR-205 binding to SHIP2 3′ UTR (blue and red columns); (ii) miR-184 does not inhibit this mutated reporter (gray column); and (iii) cotransfection of miR-184 and miR-205 cannot restore luciferase activity of 184 mut3 (orange column). Error bars (SEM) are derived from three experiments in triplicate. Controls for these experiments are shown in Fig. S1 C and D. (F) Luciferase activity of SHIP2_wt and SHIP2_mut1 in HEKs showing that endogenous miR-205 inhibits SHIP2. Positive controls (184/205_PER) are shown in Fig. S1E.
In an effort to confirm that endogenous miR-205 regulates SHIP2 expression, we transfected SHIP2_wt or SHIP2_mut1 reporters into primary human epidermal keratinocytes (HEKs), respectively. Endogenous miR-205 indeed inhibited the luciferase activity of the SHIP2_wt but did not affect the luciferase activity of the SHIP2_mut1 (Fig. 1F).
miR-184 Negatively Interferes with the Regulation of SHIP2 by miR-205.
Interestingly, when we cotransfected equal amounts of miR-184 and miR-205 into HeLa cells, miR-205 no longer inhibited luciferase activity of the SHIP2 reporter (Fig. 1D). This suggested that the binding of miR-184 through its seed sequence (the nucleotides on a miRNA that interact with a target) prevents full binding of miR-205 with its complementary nucleotides and that nucleotides upstream of the miR-205 seed match (the complementary nucleotides of the target) on SHIP2 3′ UTR are required for full miR-205 activity (Fig. 1B). To confirm this, we mutated the 3 nucleotides upstream of the seed match predicted for full binding of miR-205. Cotransfection of this mutated construct with a miR-205 mimic did not decrease luciferase activity (Fig. 1C, SHIP2_mut2), indicative that these nucleotides are required for miR-205 binding.
The observation that miR-184 interfered with the ability of miR-205 to regulate SHIP2 levels can most easily be explained by a competition for binding to the 3′ UTR. To test this idea, we mutated the nucleotides predicted to be exclusively used for miR-184 binding to SHIP2 mRNA (Fig. 1 A and B, SHIP2_mut3). miR-205 mimic was still able to suppress luciferase activity when cotransfected with SHIP2_mut3 (Fig. 1E, blue and red columns), indicative that this mutation did not affect the overall inhibitory activity of miR-205. Cotransfection of SHIP2_mut3 with miR-184 mimic had no effect on luciferase activity (Fig. 1E, gray column, and Fig. S1C), confirming that miR-184 does not directly inhibit SHIP2. However, when we cotransfected miR-205 plus miR-184 mimic with SHIP2_mut3, the luciferase activity was reduced by ≈60% (Fig. 1E, orange column, and Fig. S1D). This provided additional data in support of the idea that miR-184 negatively regulates miR-205 to maintain SHIP2 levels in HeLa cells.
SHIP2 Protein Is Diminished by miR-205.
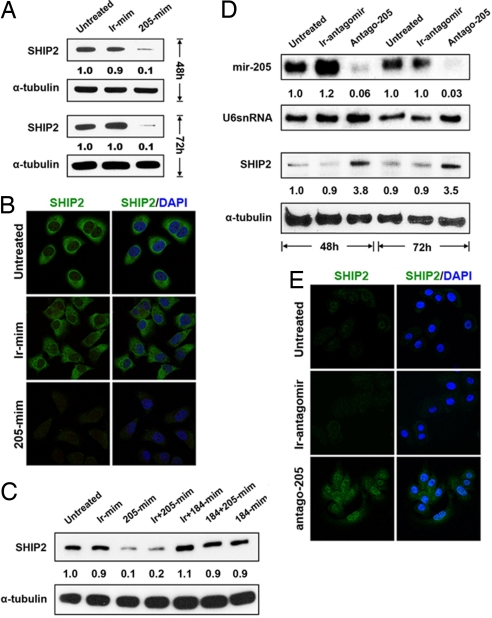
HeLa cells have negligible endogenous levels of miR-205 (22) and readily detectable levels of SHIP2 (23). The most straightforward prediction from our luciferase reporter assays would be that ectopic expression of miR-205 should reduce SHIP2 protein levels in HeLa cells. We found that treatment of HeLa cells with the miR-205 mimic indeed caused a marked reduction in SHIP2 expression, whereas treatment with an irrelevant (nontargeting) mimic caused no reduction in SHIP2 protein (Fig. 2A). Similarly, SHIP2 immunoreactivity was diminished after HeLa cells were transfected with the miR-205 mimic when compared with untreated HeLa cells or cells treated with the irrelevant mimic (Fig. 2B). Taken together, these findings indicate that, in HeLa cells, SHIP2 can be negatively regulated by miR-205.
Fig. 2.
SHIP2 levels are controlled by miR-205 and miR-184. (A) Immunoblotting of SHIP2 in HeLa cells that were treated with a miR-205 mimic, decrease protein 48 and 72 h after treatment. (B) Immunofluorescence microscopy of HeLa cells stained with anti-SHIP2 and anti-SHIP2/DAPI showing a marked decrease in staining 72 h after treatment with miR-205 mimic. Staining data at 48 h is presented in Fig. S2A. (C) Immunoblotting of SHIP2 in HeLa cells that were untreated (1), transfected with an irrelevant mimic (ir-mim; 2), miR-205 mimic (205-mim; 3), miR-205 mimic plus and irrelevant mimic (ir + 205-mim; 4), miR-184 mimic plus an irrelevant mimic (ir + 184-mim; 5), miR-184 plus miR-205 mimics (184 + 205-mim;6), and miR-184 mimic (184-mim; 7) for 48 h. miR-205 mimic reduces SHIP2 levels (3, 4) whereas miR-184 inhibits miR-205 from reducing SHIP2 levels (6). (D) Northern analysis using a miR-205 specific probe showing a marked decrease in miR-205 levels in HEKs treated with an antagomir to miR-205 (Antago-205) for 48 and 72 h. Immunoblotting of SHIP2 and α-tubulin in HEKs shows an increase in SHIP2 expression 48 and 72 h after treatment with Antago-205. (E) Immunofluorescence microscopy of HEKs stained with SHIP2 showing an increase in staining after 72 h of treatment with Antago-205. Staining data at 48 h are presented in Fig. S2B. Numbers below the panels represent the normalized expression signal of proteins and RNAs.
We next considered whether miR-184 had the capacity to maintain SHIP2 expression by antagonizing miR-205. If this was the case, we would expect to see an increase in endogenous SHIP2 protein after transfection with mimics to miR-184 and miR-205. As demonstrated previously, transfection of HeLa cells with miR-205 mimic led to a marked reduction in SHIP2 (Fig. 2 A and C, lane 3). In contrast, treatment with a miR-184 mimic (Fig. 2C, lane 7), a miR-184 mimic plus an irrelevant mimic (Fig. 2C, lane 5), or a miR-184 mimic plus a miR-205 mimic (Fig. 2C, lane 6) did not reduce the SHIP2 levels. These findings confirm the luciferase reporter data indicating that miR-184 blocks the ability of miR-205 to negatively regulate SHIP2.
To study this novel regulation of SHIP2 in squamous epithelia, we first used primary HEK cultures. These cells express miR-205 but do not express miR-184, thereby making the analysis of SHIP2 more straightforward. We reasoned that down-regulation of miR-205 should result in a rise in SHIP2 levels. We conducted such a miRNA loss-of-function study using an antagomir to miR-205 (Antago-205). Antagomirs are cholesterol-linked single-stranded RNAs that are complementary to a specific miRNA and cause the depletion of the miRNA (24). Endogenous miR-205 was markedly reduced at 48 and 72 h after treatment with Antago-205, whereas an irrelevant antagomir [Antago-124—a neuronal-specific miRNA (25)] had no effect (Fig. 2D). As predicted, HEKs treated with Antago-205 showed a marked increase in SHIP2 levels by Western (Fig. 2D) and immunohistochemical (Fig. 2E) analyses when compared with the irrelevant antagomir-treated or untreated HEKs. Thus, in HeLa cells and HEKs, SHIP2 levels are down-regulated by miR-205.
Down-Regulation of miR-205 Dampens Akt Signaling.
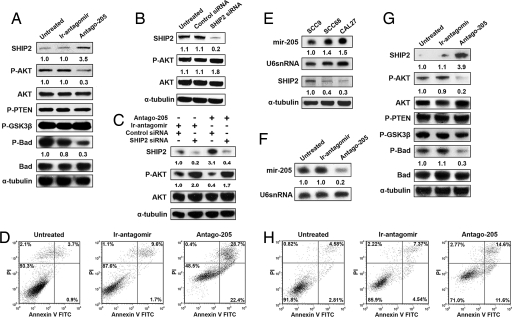
One of the roles ascribed to SHIP2 has been the negative regulation of the Akt pathway (26–28); however, this ability of SHIP2 has not been investigated in keratinocytes. Toward this aim, siRNA oligonucleotides specific for SHIP2 were transfected into HEKs and harvested for Western blot analysis after 72 h. Consistent with our previous experiments, reduced SHIP2 levels resulted in a concomitant increase in phosphorylated AKT (p-Akt) (Fig. 3B).
Fig. 3.
miR-205 affects the Akt pathway in keratinocytes directly through targeting of SHIP2 and is inversely correlated with SHIP2 in SCC cell lines. (A) Immunoblotting of SHIP2, phosphorylated Akt (p-Akt), total pan (1/2/5) Akt, phosphorylated BAD, total BAD, phosphorylated PTEN (p-PTEN), and phosphorylated GSK3β (p-GSK3β) in HEKs that were untreated (un-rx) or treated with an ir-antagomir or Antago-205 for 48 h. α-Tubulin serves as a loading control. (B) Immunoblots of SHIP2, p-Akt, AKT, and α-tubulin in HEKs 72 h after transfection with SHIP2 siRNA and control siRNA, showing decreases in SHIP2 and increases in p-Akt. (C) Immunoblots of SHIP2, p-Akt, AKT, and α-tubulin in HEKs 48 h after treatment with an antagomir to miR-205 or an irrelevant antagomir. HEKs were subsequently treated for another 72 h with combinations of siRNA to SHIP2, control siRNA, antagomir-205, and irrelevant antagomir. (D) Keratinocytes were stained with propidium iodide and annexin V 48 h after treatment with an ir-antagomir or Antago-205 and compared with untreated cells. Late apoptotic cells are seen in the top right quandrant. (E) Northern analysis of oral SCC cell lines using a miR-205-specific probe showing increases in miR-205 in SCC68 and CAL27 cells. Immunoblotting of SHIP2 in oral SCCs showing a marked decrease in SHIP2 in SCC68 and CAL27 cells. (F) Northern analysis with a miR-205-specific probe in SCC68 cells that were treated with an ir-antagomir or Antago-205 for 48 h. U6 serves as a loading control. (G) Immunoblotting of SHIP2, p-Akt, total Akt, p-PTEN, p-GSK3β, p-BAD, and BAD in SCC68 cells treated as described in F. α-Tubulin serves as loading control. (H) SCC68 cells were treated as described in F and G and then stained with propidium iodide and annexin V. Numbers below the panels represent the normalized expression signal of proteins and RNAs.
In view of these observations, we reasoned that increased levels of SHIP2 in HEKs after treatment with Antago-205 might decrease levels of p-Akt and phosphorylated BAD (p-BAD). Western blot analysis was used to measure the protein levels of SHIP2, pan (1/2/5)Akt, p-Akt, BAD, p-BAD, phosphorylated PTEN (p-PTEN), and phosphorylated GSK3β (p-GSK3β) in HEK cells after Antago-205 treatment. We observed an increase in SHIP2 and a coordinated decrease in p-Akt and p-BAD when compared with the irrelevant antagomir or untreated HEKs (Fig. 3A). However, no major change in total Akt, BAD, p-PTEN, or p-GSK3β levels was observed. Moreover, silencing of SHIP2 to prevent its induction by Antago-205 treatment led to an increase in p-Akt (Fig. 3C). Taken together, these studies demonstrate that SHIP2 is regulated by miR-205 and is required for the negative regulation of the Akt pathway in keratinocytes (Figs. 3A).
One of the outcomes of Akt signaling is to induce endogenous BAD phosphorylation, which ultimately leads to the inhibition of BAD-dependent death (29). To address whether the lower levels of p-BAD resulting from the down-regulation of miR-205 (Fig. 3A) would induce keratinocyte apoptosis and cell death, we determined the number of early and late apoptotic keratinocytes after treatment with Antago-205. As expected, there were few early apoptotic cells (1%) in the untreated and irrelevant antagomir-treated (2%) keratinocytes, whereas Antago-205 caused an ≈10-fold increase in early apoptotic cells as judged by annexin V staining (Fig. 3D). Similarly, there was a notable increase in propidium iodide staining, indicating elevated levels of cell death (Fig. 3D). This dramatic increase in apoptosis and cell death indicates that miR-205 may enhance keratinocyte survival by negatively regulating SHIP2.
miR-205 Is Abundant in SCC Cell Lines.
It has been reported that miR-205 is overexpressed in head and neck SCC cell lines (30, 31); however, no attempt has been made to validate potential targets of miR-205 in these cell lines. We postulated that if SHIP2 levels are controlled by miR-205, we would see a correlation between miR-205 and SHIP2 in oral SCC cell lines. We cultured SCC9 [tongue (32)], SCC68 [oral (32)], and CAL27 [tongue (33)] cell lines and observed a reciprocal relationship between the miR-205 levels and SHIP2 expression in these cells (Fig. 3E). SCC68 and CAL27, aggressive oral SCC lines (33–35), had high levels of miR-205 and low amounts of SHIP2. SCC9, which is minimally invasive (36), had lower amounts of miR-205 along with higher levels of SHIP2 (Fig. 3E).
Treatment of SCC68 cells with Antago-205 showed (i) a dramatic decrease in miR-205 levels (Fig. 3F), (ii) an increase in SHIP2 expression (Fig. 3G), (iii) a decrease in p-Akt and p-BAD expression (Fig. 3G), and (iv) an increase in apoptotic cells (Fig. 3H) paralleling our observation in normal HEKs (Fig. 3D). Taken together, these results provide additional evidence that SHIP2 levels are regulated by miR-205 and suggest that high levels of miR-205 may contribute to SCC pathogenesis via a SHIP2-mediated enhancement of Akt signaling and cell survival. The restoration of SHIP2 in SCCs via an antagomir to miR-205, which dampens Akt signaling and increases apoptosis, might be a novel use for this antagomir in the treatment of these neoplasias.
SHIP2 Regulation Is Unique in Corneal Keratinocytes.
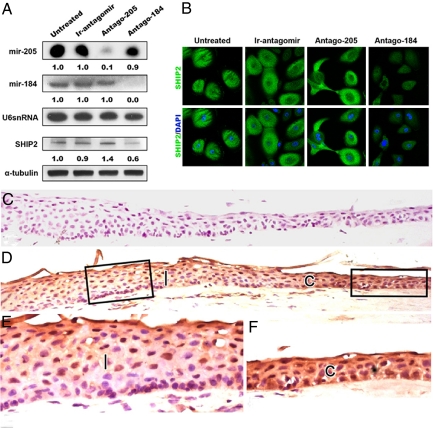
Having established that SHIP2 is a target of miR-205 in HEKs and SCC cell lines, we next examined the relationship between SHIP2 and miR-205 in human corneal epithelial keratinocytes (HCEKs). The situation in HCEKs is more complex because these cells express miR-184 and miR-205 (Fig. 4A), which interact to maintain SHIP2 levels in HeLa cells (Figs. 1 D and E and 2C). We reasoned that if miR-184 normally maintains SHIP2 levels by inhibiting the interaction of miR-205 with SHIP2, treatment of HCEKs with an antagomir to miR-184 would “release” miR-205 to down-regulate SHIP2. As expected, both SHIP2 expression and miR-184 levels decreased 72 h after treatment with Antago-184 (Fig. 4 A and B). In contrast, Antago-205 resulted in a down-regulation of miR-205 and an increase in SHIP2 levels compared with the untreated and control cells (Fig. 4 A and B).
Fig. 4.
miR-184 alters the ability of miR-205 to affect SHIP2 in corneal keratinocytes in vitro and in vivo. (A) Northern analysis of primary human corneal epithelial (HCEKs) cells using specific probes for miR-184 and miR-205, showing expression of both of these miRNAs in untreated and control (Ir-antagomir) cells. After 72 h of treatment, each antagomir abrogates the respective miRNA. U6 serves as a loading control. Shown is immunoblotting of SHIP2 and α-tubulin in HCEKs that were untreated or were treated with Ir-antagomir, Antago-205, or an antagomir to miR-184 (Antago-184) for 72 h. (B) Immunofluorescence microscopy of HCEKs stained for SHIP2 showing a marked decrease in staining after a 72-h treatment with antagomir-184, whereas treatment with antagomir to miR-205 resulted in an increase in SHIP2 staining. (C and D) Serial frozen sections of human limbal and corneal epithelium immunohistochemically stained with an antibody that recognizes IgG (control, C) or SHIP2 (D). (E and F) Higher magnification of the boxed areas of the limbal (l, E) and corneal (c, F) epithelia, showing a decrease in SHIP2 staining in the limbal epithelium compared with the corneal epithelium. Numbers below the panels represent the normalized expression signal of proteins and RNAs.
Our previous in situ hybridization studies demonstrated that miR-184 was expressed in the corneal epithelium but not in the limbal epithelium, whereas miR-205 was expressed in both the corneal and limbal epithelia (14). If the function of miR-184 in corneal epithelium is to maintain SHIP2 levels by antagonizing miR-205, SHIP2 staining should be more intense in corneal versus limbal epithelium. Indeed, SHIP2 was detected immunohistochemically in normal human corneal epithelium (Fig. 4 D and F) whereas much less SHIP2 staining was observed in the limbal region (Fig. 4 D and E). These in vivo data strongly supports our in vitro findings that miR-184 antagonizes miR-205 to maintain SHIP2 levels.
We propose that a balance exists between miR-184 and miR-205 and that this maintains SHIP2 levels (Fig. 5A); however, abrogation of miR-205 elevates SHIP2 because the miR-184/205 balance is altered and miR-184 alone has no inhibitory effect on SHIP2 (Fig. 5B). Similar to the HeLa cell transfections, miR-184 antagonizes miR-205 to maintain SHIP2 levels in corneal keratinocytes and corneal epithelium, and this highlights the uniqueness of the corneal epithelium with respect to SHIP2 regulation (Figs. 4 and 5B).
Fig. 5.
Proposed regulatory effects of miR-205 and miR-184 on SHIP2 levels in various epithelial contexts. (A) Epidermal keratinocytes. Decreasing miR-205 via antagomir-205 increases SHIP2 levels resulting in the dampening of Akt signaling and an increase in apoptosis and cell death. (B) Corneal keratinocytes. Decreasing miR-184 via antagomir-184 “releases” miR-205 to reduce SHIP2 levels augmenting the Akt pathway, with increased cell survival and angiogenesis as possible outcomes. Because miR-184 does not inhibit SHIP2, decreasing miR-205 via antagomir-205 disturbs the normal balance between SHIP2 and miR-184/205. (C) SCC. Ectopic expression of miR-184 or treatment with an antagomir to miR-205 represents potential therapeutic modalities for the treatment of SCCs by increasing SHIP2 levels, which might act as a tumor suppressor in these neoplasias.
Discussion
A chief impediment to understanding miRNA function has been the relative lack of experimentally validated targets. We demonstrate that SHIP2 mRNA is a target of miR-205 in HEKs and that, in HCEKs, miR-184 antagonizes miR-205, thereby maintaining SHIP2 levels. To our knowledge, this is the first example in a vertebrate system where one miRNA abrogates the inhibitory function of another. Our mutation analyses indicate that miR-205 binds to SHIP2 mRNA leading to translational repression. This has been proposed as the “classical” manner in which miRNAs affect protein synthesis in mammalian systems (1). The mechanism by which miR-184 negatively regulates miR-205 is unique. Binding of miR-184 to its seed sequence has no direct effect on SHIP2 translation, but instead prevents miR-205 from interacting with SHIP2 mRNA. This neutralizes the inhibitory activity of miR-205 on SHIP2, a situation special to the corneal epithelium because this is the only known epithelium that exhibits overlapping expression of miR-184 and miR-205 (14). Previously, investigators have considered the regulation of proteins or mRNAs by miRNAs as a one-to-one event; however, our findings indicate that in some instances the situation is more complex and that cross-talk between individual miRNAs can occur.
The need for maintaining SHIP2 levels, which down-regulate the Akt pathway, may relate to the requirement of corneal avascularity so that light required for vision can be transmitted to the lens. Inhibition of Akt can lead to the down-regulation of VEGF, which can repress angiogenesis. We suggest that SHIP2, via its ability to negatively regulate the Akt pathway, could suppress corneal angiogenesis through inhibition of VEGF (37). In this scenario, SHIP2 would be functioning similarly to inhibitory PAS domain protein, which has been shown to maintain an avascular phenotype in corneal epithelium via the negative regulation of VEGF (38).
Despite the ubiquitous distribution of SHIP2 in vertebrate tissues (28, 39), little attention has been directed toward this lipid phosphatase in stratified squamous epithelia, and consequently the function(s) of endogenous SHIP2 in these tissues remain poorly understood. Antago-205 increased keratinocyte SHIP2 levels, which was coordinated with a dampening of Akt signaling (Fig. 5A). Moreover, the down-regulation of miR-205 markedly increased keratinocyte apoptosis and cell death. This is consistent with the report that SHIP2 overexpression in MDCK epithelial cells resulted in cytotoxicity (40). We believe that one of the functions of miR-205, which is broadly expressed in epithelia, is to control SHIP2 levels and maintain cell survival through the Akt pathway.
It is becoming increasingly clear that alterations in miRNAs may adversely impact on cancer (10, 41–43). Of particular relevance to the present study are observations that miR-205 is up-regulated in a variety of carcinomas (8, 9, 30, 31, 44, 45). Our finding that elevated levels of miR-205 markedly reduce SHIP2 in aggressive SCC cell lines provides some insight into a potential role of miR-205 in SCCs. PTEN (phosphatase and tensin homologue deleted on chromosome 10) is a lipid phosphatase similar to SHIP2 in that PIP3 is a common lipid substrate (for review see ref. 46). PTEN is more widely regarded as a tumor suppressor than SHIP2; however, PTEN mutations are rarely found in head and neck, oral, and skin SCCs (47–49), suggestive that another tumor suppressor gene may be associated with the development of these neoplasias (49). Our observations indicate that SHIP2 might fulfill this role through its negative regulation of the Akt pathway, which is frequently deregulated in many types of cancer (for review see ref. 50). Because down-regulation of miR-205 in an aggressive SCC cell line restores SHIP2, we suggest that miR-205 may be viewed as a tumor promoter in the context of SCCs (Fig. 5C). Therefore (i) miR-205 might have diagnostic value in determining the aggressivity of SCCs, and (ii) an antagomir to miR-205 or ectopic expression of miR-184 could be novel therapeutic approaches for treating aggressive SCCs (Fig. 5C).
The idea that SHIP2 might function as a tumor suppressor in keratinocytes makes excellent biological sense from the perspective of corneal epithelial SCCs. These tumors develop from limbal rather than corneal epithelium (51). It is noteworthy that the stem cell compartment, the primary site for malignant transformations (52, 53), is localized to the limbus (15–17). We suggest that an additional factor for a limbal origin of corneal epithelial SCCs may be the absence of miR-184 in the limbal epithelium; because miR-184 is present in the corneal epithelium, this helps preserve SHIP2 levels (Fig. 4 D and F), thereby maintaining the presence of a potential tumor suppressor (Fig. 5B). Conversely, the abrupt absence of miR-184 in the limbal epithelium enables miR-205 to negatively regulate SHIP2 levels (Fig. 4 D and E), decreasing its potential tumor suppressor function in a stem-cell enriched region. As many neoplasias result from underexpressed tumor suppressor genes, down-regulation of SHIP2 in limbal basal cells could contribute to the neoplastic transformation of these cells.
Materials and Methods
Constructs, Transfections, and Assays.
The 3′ UTR of the human SHIP2 mRNA was cloned in between the SpeI and HinDIII sites of pMIR-Report. Mutants 1–3 of the SHIP2 sequence were created by using a QuikChange Site-Directed Mutagenesis Kit (Stratagene). miRNA mimics specifying miR-184 and miR-205 and nontargeting control were obtained from Dharmacon.
HeLa cells and HEKs were seeded onto 24-well plates (1 × 105 cells per well) the day before transfections were performed. Cells (≈70% confluent) were transfected with pMIR-Report constructs (50 ng per well), pRL-SV40 Renilla luciferase (10 ng per well), and miRNA mimics (10 nM). All transfections were carried out in triplicate with Lipofectamine 2000 (Invitrogen). Cell lysates were prepared with Passive Lysis Buffer (Promega) 48 h after transfection, and luciferase activities were measured by using the Dual Luciferase Reporter Assay (Promega). For Western and immunohistochemical analyses, HeLa cells were serum-starved for 48 h after transfection.
Antagomirs directed against miR-184, miR-205, and miR-124 were synthesized by Dharmacon and were added to culture media to a final concentration of 1,000 pmol/mL. Cells were grown in normal culture media to a 70% confluent state and were then treated with antagomir-containing culture media for 48–72 h.
SHIP2 siRNA and control siRNA pools were synthesized by Dharmacon and transfected into HEKs (200 nM) by using Lipofectamine 2000. Medium was changed every 24 h, and cells were cultured for 72 h and harvested for Western blot analyses as described below. For the siRNA and antagomir combination experiments, HEKs were grown in normal culture media containing 1,000 pmol/mL antagomir-205 or an irrelevant antagomir for 48 h. These cells were then cotreated with 200 nM siRNA and 1,000 pmol/mL antagomir-205 for another 72 h. Cells were harvested for Western blot analyses. Please see SI Materials and Methods for details.
Cell Culture.
Primary human epidermal keratinocytes (HEKs) were grown in keratinocyte serum-free media (154 media; Cascade Biologicals) containing HKGS growth supplements and 70 μM CaCl2. HCEKs were cultured in CnT20 with supplements (CellnTech). SCC9 and CAL27 were grown in DMEM/F12 (Gibco) containing 10% FBS. SCC68 was cultured in Keratinocyte SFM (Gibco) with recommended supplements. HeLa cells were obtained from American Type Culture Collection and grown in F12 Ham's media with 10% FBS.
Northern and Western Blots.
Northern blots were performed as described previously (14). For Western blots the following antibodies were used: SHIP2, p-Akt, Akt, p-BAD, BAD, p-PTEN, p-GSK-3β, and α-tubulin. Please see SI Materials and Methods for details.
Immunohistochemistry and Light Microscopy.
We conducted immunocytochemistry on HeLa, HEK, and HCEK cultures and immunohistochemistry on normal human corneal epithelium as is routinely performed in our laboratory. Please see SI Materials and Methods for details.
Apoptosis Assays.
Apoptosis assay was performed on HEKs and the SCC68 cell line 48 h after treatment with either an antagomir directed against miR-205 or an irrelevant antagomir using the Annexin V-FITC Apoptosis Detection Kit I (BD Biosciences) according to the manufacturer's protocols and analyzed by using the FACSCalibur Flow Cytometer (BD Biosciences).
Supplementary Material
Acknowledgments.
We thank P. Stein and T.-T. Sun for critical reading of the manuscript and useful discussions; Q. Ruan for assistance with immunohistochemistry; and A. Paller and K. Green (Northwestern University, Chicago, IL) for the SCC cell lines. This research is supported by National Institutes of Health Grant EY017536 (to R.M.L.) and a Dermatology Foundation grant (to S.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803992105/DCSupplemental.
References
- 1.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 2.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 3.Esau C, et al. MicroRNA-143 regulates adipocyte differentiation. J Biol Chem. 2004;279:52361–52365. doi: 10.1074/jbc.C400438200. [DOI] [PubMed] [Google Scholar]
- 4.Hornstein E, et al. The microRNA miR-196 acts upstream of Hoxb8 and Shh in limb development. Nature. 2005;438:671–674. doi: 10.1038/nature04138. [DOI] [PubMed] [Google Scholar]
- 5.Sempere LF, et al. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13.11–R13.11. doi: 10.1186/gb-2004-5-3-r13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214–220. doi: 10.1038/nature03817. [DOI] [PubMed] [Google Scholar]
- 7.Leung AK, Sharp PA. MicroRNAs: A safeguard against turmoil? Cell. 2007;130:581–585. doi: 10.1016/j.cell.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Iorio MV, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 9.Iorio MV, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 10.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 11.Ma L, Teruya-Feldstein J, Weinberg RA. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 12.Yi R, et al. Morphogenesis in skin is governed by discrete sets of differentially expressed microRNAs. Nat Genet. 2006;38:356–362. doi: 10.1038/ng1744. [DOI] [PubMed] [Google Scholar]
- 13.Andl T, et al. The miRNA-processing enzyme Dicer is essential for the morphogenesis and maintenance of hair follicles. Curr Biol. 2006;16:1041–1049. doi: 10.1016/j.cub.2006.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ryan DG, Oliveira-Fernandes M, Lavker RM. MicroRNAs of the mammalian eye display distinct and overlapping tissue specificity. Mol Vision. 2006;12:1175–1184. [PubMed] [Google Scholar]
- 15.Cotsarelis G, Cheng SZ, Dong G, Sun T-T, Lavker RM. Existence of slow-cycling limbal epithelial basal cells that can be preferentially stimulated to proliferate: Implications on epithelial stem cells. Cell. 1989;57:201–209. doi: 10.1016/0092-8674(89)90958-6. [DOI] [PubMed] [Google Scholar]
- 16.Lavker RM, Tseng SC, Sun TT. Corneal epithelial stem cells at the limbus: Looking at some old problems from a new angle. Exp Eye Res. 2004;78:433–446. doi: 10.1016/j.exer.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 17.Schermer A, Galvin S, Sun T-T. Differentiation-related expression of a major 64K corneal keratin in vivo and in culture suggests limbal location of corneal epithelial stem cells. J Cell Biol. 1986;103:49–62. doi: 10.1083/jcb.103.1.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tseng SC, Tsai RJ. Limbal transplantation for ocular surface reconstruction—A review. Fortsch Ophthalmol. 1991;88:236–242. [PubMed] [Google Scholar]
- 19.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 20.Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing “stemness.”. Nature. 2008;452:225–229. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Krek A, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 22.Barad O, et al. MicroRNA expression detected by oligonucleotide microarrays: System establishment and expression profiling in human tissues. Genome Res. 2004;14:2486–2494. doi: 10.1101/gr.2845604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prasad NK, Decker SJ. SH2-containing 5′-inositol phosphatase, SHIP2, regulates cytoskeleton organization and ligand-dependent down-regulation of the epidermal growth factor receptor. J Biol Chem. 2005;280:13129–13136. doi: 10.1074/jbc.M410289200. [DOI] [PubMed] [Google Scholar]
- 24.Krutzfeldt J, et al. Silencing of microRNAs in vivo with “antagomirs.”. Nature. 2005;438:685–689. doi: 10.1038/nature04303. [DOI] [PubMed] [Google Scholar]
- 25.Lagos-Quintana M, et al. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- 26.Sasaoka T, et al. Dual role of SRC homology domain 2-containing inositol phosphatase 2 in the regulation of platelet-derived growth factor and insulin-like growth factor I signaling in rat vascular smooth muscle cells. Endocrinology. 2003;144:4204–4214. doi: 10.1210/en.2003-0190. [DOI] [PubMed] [Google Scholar]
- 27.Sharrard RM, Maitland NJ. Regulation of protein kinase B activity by PTEN and SHIP2 in human prostate-derived cell lines. Cell Signalling. 2007;19:129–138. doi: 10.1016/j.cellsig.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 28.Sleeman MW, et al. Absence of the lipid phosphatase SHIP2 confers resistance to dietary obesity. Nat Med. 2005;11:199–205. doi: 10.1038/nm1178. [DOI] [PubMed] [Google Scholar]
- 29.Datta SR, et al. 14-3-3 proteins and survival kinases cooperate to inactivate BAD by BH3 domain phosphorylation. Mol Cell. 2000;6:41–51. [PubMed] [Google Scholar]
- 30.Jiang J, Lee EJ, Gusev Y, Schmittgen TD. Real-time expression profiling of microRNA precursors in human cancer cell lines. Nucleic Acids Res. 2005;33:5394–5403. doi: 10.1093/nar/gki863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tran N, et al. MicroRNA expression profiles in head and neck cancer cell lines. Biochem Biophys Res Commun. 2007;358:12–17. doi: 10.1016/j.bbrc.2007.03.201. [DOI] [PubMed] [Google Scholar]
- 32.Rheinwald JG, Beckett MA. Tumorigenic keratinocyte lines requiring anchorage and fibroblast support cultures from human squamous cell carcinomas. Cancer Res. 1981;41:1657–1663. [PubMed] [Google Scholar]
- 33.Magne N, et al. Influence of epidermal growth factor receptor (EGFR), p53 and intrinsic MAP kinase pathway status of tumour cells on the antiproliferative effect of ZD1839 (“Iressa”) Br J Cancer. 2002;86:1518–1523. doi: 10.1038/sj.bjc.6600299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ghosh S, et al. Loss of adhesion-regulated proteinase production is correlated with invasive activity in oral squamous cell carcinoma. Cancer. 2002;95:2524–2533. doi: 10.1002/cncr.10997. [DOI] [PubMed] [Google Scholar]
- 35.Sprague LD, et al. Effect of reoxygenation on the hypoxia-induced up-regulation of serine protease inhibitor PAI-1 in head and neck cancer cells. Oncology. 2006;71:282–291. doi: 10.1159/000106789. [DOI] [PubMed] [Google Scholar]
- 36.Liu SC, et al. Overexpression of cyclin D2 is associated with increased in vivo invasiveness of human squamous carcinoma cells. Mol Carcinog. 2002;34:131–139. doi: 10.1002/mc.10057. [DOI] [PubMed] [Google Scholar]
- 37.Jiang BH, Zheng JZ, Aoki M, Vogt PK. Phosphatidylinositol 3-kinase signaling mediates angiogenesis and expression of vascular endothelial growth factor in endothelial cells. Proc Natl Acad Sci USA. 2000;97:1749–1753. doi: 10.1073/pnas.040560897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Makino Y, et al. Inhibitory PAS domain protein is a negative regulator of hypoxia-inducible gene expression. Nature. 2001;414:550–554. doi: 10.1038/35107085. [DOI] [PubMed] [Google Scholar]
- 39.Schurmans S, et al. The mouse SHIP2 (Inppl1) gene: Complementary DNA, genomic structure, promoter analysis, and gene expression in the embryo and adult mouse. Genomics. 1999;62:260–271. doi: 10.1006/geno.1999.5995. [DOI] [PubMed] [Google Scholar]
- 40.Koch A, Mancini A, El Bounkari O, Tamura T. The SH2-domain-containing inositol 5-phosphatase (SHIP)-2 binds to c-Met directly via tyrosine residue 1356 and involves hepatocyte growth factor (HGF)-induced lamellipodium formation, cell scattering and cell spreading. Oncogene. 2005;24:3436–3447. doi: 10.1038/sj.onc.1208558. [DOI] [PubMed] [Google Scholar]
- 41.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 42.Kumar MS, Lu J, Mercer KL, Golub TR, Jacks T. Impaired microRNA processing enhances cellular transformation and tumorigenesis. Nat Genet. 2007;39:673–677. doi: 10.1038/ng2003. [DOI] [PubMed] [Google Scholar]
- 43.Zhang B, Pan X, Cobb GP, Anderson TA. MicroRNAs as oncogenes and tumor suppressors. Dev Biol. 2007;302:1–12. doi: 10.1016/j.ydbio.2006.08.028. [DOI] [PubMed] [Google Scholar]
- 44.Gottardo F, et al. Micro-RNA profiling in kidney and bladder cancers. Urol Oncol. 2007;25:387–392. doi: 10.1016/j.urolonc.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 45.Yanaihara N, et al. Unique microRNA molecular profiles in lung cancer diagnosis and prognosis. Cancer Cell. 2006;9:189–198. doi: 10.1016/j.ccr.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 46.Hawkins PT, Anderson KE, Davidson K, Stephens LR. Signaling through class I PI3Ks in mammalian cells. Biochem Soc Trans. 2006;34:647–662. doi: 10.1042/BST0340647. [DOI] [PubMed] [Google Scholar]
- 47.Chen Q, Samaranayake LP, Zhou H, Xiao L. Homozygous deletion of the PTEN tumor-suppressor gene is not a feature in oral squamous cell carcinoma. Oral Oncol. 2000;36:95–99. doi: 10.1016/s1368-8375(99)00068-8. [DOI] [PubMed] [Google Scholar]
- 48.Kubo Y, Urano Y, Hida Y, Arase S. Lack of somatic mutation in the PTEN gene in squamous cell carcinomas of human skin. J Dermatol Sci. 1999;19:199–201. doi: 10.1016/s0923-1811(98)00058-9. [DOI] [PubMed] [Google Scholar]
- 49.Snaddon J, Parkinson EK, Craft JA, Bartholomew C, Fulton R. Detection of functional PTEN lipid phosphatase protein and enzyme activity in squamous cell carcinomas of the head and neck, despite loss of heterozygosity at this locus. Br J Cancer. 2001;84:1630–1634. doi: 10.1054/bjoc.2001.1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Altomare DA, Testa JR. Perturbations of the AKT signaling pathway in human cancer. Oncogene. 2005;24:7455–7464. doi: 10.1038/sj.onc.1209085. [DOI] [PubMed] [Google Scholar]
- 51.Waring GO, Roth AM, Ekins MB. Clinical and pathologic description of 17 cases of corneal intraepithelial neoplasia. Am J Ophthalmol. 1984;97:547–559. doi: 10.1016/0002-9394(84)90371-4. [DOI] [PubMed] [Google Scholar]
- 52.Miller SJ, Lavker RM, Sun T-T. Interpreting epithelial cancer biology in the context of stem cells: Tumor properties and therapeutic implications. Biochim Biophys Acta. 2005;1756:25–52. doi: 10.1016/j.bbcan.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 53.Perryman SV, Sylvester KG. Repair and regeneration: Opportunities for carcinogenesis from tissue stem cells. J Cell Mol Med. 2006;10:292–308. doi: 10.1111/j.1582-4934.2006.tb00400.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.