Abstract
Background
Single dose (SD) nevirapine (NVP) at birth plus NVP to the infant up to 6 weeks of age is superior to SD NVP alone for prevention of HIV vertical transmission through breastfeeding. We analyzed NVP resistance in HIV-infected Ugandan infants who received either SD NVP or extended NVP prophylaxis.
Methods
We tested plasma HIV using a genotyping assay (ViroSeq), a phenotypic resistance assay (PhenoSense), and sensitive point mutation assay (LigAmp, for K103N, Y181C, G190A).
Results
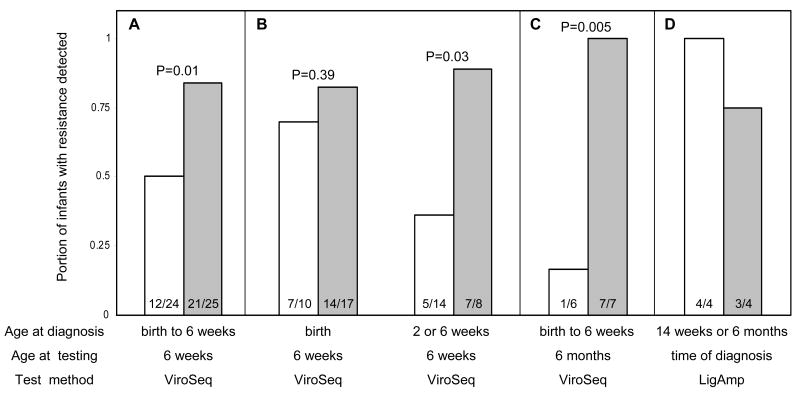
At 6 weeks, NVP resistance was detected by ViroSeq in a higher proportion of infants in the extended NVP arm than in the SD NVP arm (21/25=84% vs. 12/24=50%, p=0.01). Similar results were obtained with LigAmp and PhenoSense. Infants who were HIV-infected at birth had high rates of resistance in both study arms. In contrast, infants who were HIV-infected after birth were more likely to have resistance detected at 6 weeks in the extended NVP arm. Use of extended NVP prophylaxis was also associated with detection of NVP resistance by ViroSeq at 6 months (7/7=100% extended NVP arm vs. 1/6=16.7% SD NVP arm, p=0.005).
Conclusions
Use of extended NVP prophylaxis was associated with increased selection and persistence of NVP resistance in HIV-infected Ugandan infants.
Keywords: HIV-1, infant, mother-to-child transmission, nevirapine, resistance
Introduction
Single-dose (SD) nevirapine (NVP) can reduce the risk of HIV mother-to-child transmission (MTCT) [1, 2], but may lead to emergence of resistant variants in infants who are HIV-infected despite prophylaxis. After SD NVP exposure, 46% of Ugandan infants [3] and 87% of Malawian infants [4] who were HIV-infected had NVP resistance at 6-8 weeks of age. NVP resistance has also been observed in infants after SD NVP in South Africa [5] and India [6]. The most common NVP resistance mutation detected in infants after SD NVP is Y181C, although other mutations are also seen [3, 5, 6]. NVP resistance can persist in some infants for a year or more after SD NVP [3, 5]. In many resource-limited settings, more than half of HIV-infected infants die by two years of age [7]. Therefore, antiretroviral treatment may be indicated in NVP-exposed infants before NVP-resistant variants can fade. In women, clinical studies show that prior SD NVP use does not compromise the efficacy of subsequent antiretroviral treatment, provided that sufficient time elapses between SD NVP exposure and treatment initiation [8, 9]. In one study, HIV-infected infants with prior SD NVP exposure responded poorly to antiretroviral therapy given within 6 months of age, compared to infants without prior SD NVP exposure [9]. In another study, children who were NVP exposed (median age 1.7 years) and NVP unexposed (median age 7.8 years) were able to achieve virologic suppression on NVP-based highly active antiretroviral therapy [10].
MTCT during breastfeeding accounts for one-third to one-half of HIV MTCT [11, 12], and is associated with high maternal viral load, cell-free and cell-associated HIV in breast milk, low maternal CD4 cell count, mastitis, infant co-infections, mixed feeding, and length of breastfeeding [13-18]. SD NVP does not prevent the majority of postnatal HIV infections. NVP-resistant variants have been detected in the breast milk of some women after exposure to SD NVP [19-21]. Transmission of NVP-resistant HIV through the breast milk has been documented [3], but there are few data describing resistance in late-infected infants.
The Six Week Extended Nevirapine (SWEN) study, performed in Uganda, Ethiopia, and India, compared the efficacy of SD NVP alone to SD NVP plus an infant regimen of up to six weeks of NVP prophylaxis (daily NVP from day 8 to day 42) to prevent HIV transmission by breastfeeding [22]. In a modified intent-to-treat analysis excluding infants who were HIV infected at birth, the extended NVP regimen was 46% more effective at preventing MTCT by 6 weeks of age, and was associated with a significant 27% reduction in HIV transmission or infant mortality during the first 6 months of life, compared to SD NVP alone [22]. One concern with use of extended NVP prophylaxis was that it might enhance selection of NVP-resistant HIV, compared to use of SD NVP alone. In this report, we analyzed NVP resistance in infants who received either SD NVP alone or SD NVP plus up to six weeks of NVP to prevent HIV MTCT in the Ugandan component of the three-country SWEN trial. Three different laboratory methods were used for analysis of NVP resistance: (1) an FDA-cleared, population sequencing-based, HIV genotyping assay (ViroSeq), (2) a sensitive point mutation assay (LigAmp), and (3) a commercial phenotypic resistance assay (PhenoSense).
Methods
Source of Study Samples
Samples were obtained from infants in the study, “A phase III randomized clinical trial of the standard two dose nevirapine (NVP) regimen with the addition of HIV immune globulin (HIVIGLOB) or extended infant NVP dosing compared with the standard NVP regimen alone for the prevention of maternal-infant HIV transmission in Uganda” (NCT00639938) [22]. In this trial, pregnant HIV-infected women and their infants were randomized to one of three regimens for prevention of HIV MTCT: (1) SD NVP arm (SD NVP to the mother in labor and to the infant after birth, the HIVNET 012 regimen), (2) extended NVP arm (SD NVP to the mother and infant, plus daily NVP to the infant on days 8 to 42), and (3) HIVIGLOB arm (SD NVP to the mother and infant, plus antenatal HIV immune globulin given to the mother at 37-38 weeks gestation and to the infant shortly after birth). Infants in all three study arms received up to six weeks of daily multivitamins. Infants were tested for HIV infection at birth (by age 7 days), at 2 and 6 weeks of age, and at subsequent study visits. The intervention (daily NVP or daily multivitamins) was discontinued if HIV infection was confirmed in an infant before the 6-week visit. Some infants in the extended NVP arm who were HIV-infected at birth received multiple doses of NVP (median 14 doses) before HIV infection was confirmed and the regimen was discontinued. Analysis of NVP resistance in this report was limited to HIV-infected infants in SD NVP and extended NVP study arms.
Laboratory Methods
HIV genotyping was performed using the ViroSeq HIV Genotyping System v2.6 (Celera Diagnostics, Alameda, CA), as previously described [3]. Bidirectional sequence data was obtained at the positions of NVP resistance mutations for 47/49 infants tested. HIV subtyping was performed by phylogenetic analysis of pol region sequences [3]. PCR products generated in the ViroSeq system were also analyzed using the LigAmp assay to detect and quantify HIV variants with NVP resistance mutations (K103N=AAC, 0.5% assay cutoff; Y181C=TGT, 1% assay cutoff; G190A=GCA, 0.5% assay cutoff). The LigAmp assay was performed as previously described [23], with the following exception: for Y181C detection, two upstream oligonucleotides were used because of sequence diversity at the oligonucleotide binding site. Sequences of oligonucleotides used in the LigAmp assay are shown in Table 1. Assays for phenotypic drug susceptibility and HIV replication capacity were performed at a commercial laboratory (PhenoSense HIV assay, Monogram Biosciences, South San Francisco, CA), as described [24].
Table 1. Ligation oligonucleotides used in the LigAmp assay.
Different LigAmp oligonucleotides were used for HIV subtype A and D samples. For intersubtype recombinant samples, oligonucleotides were selected based on the HIV subtype in the region of interest. HIV subtype A oligonucleotides were used for analysis of one subtype G sample. Upstream olignucleotides contain a 5′ tail with the following sequence: 5′-ACTGTAAAACGACGGCCAGTGTTCCCCTCAAACTGGCAGATGCACG. Downstream oligonucleotides contain a 3′ tail with the following sequence: 5′-TGGTCATAGCTGTTTCCTGCA. The upstream oligonucleotide tail included binding sites for an M13 primer and a real-time PCR probe (underlined). The downstream oligonucleotide is phosphorylated at the 5′ end (Phos), and contains an M13 binding site. M13 primers and a real-time PCR probe are to detect the ligated oligonucleotides in the detection step of the LigAmp reaction [23]. For detection of Y181C, two different upstream oligonucleotides were used in the ligation reaction (upstream 1 and upstream 2, 1:1 mixture).
| Subtype | Mutation | Olignucleotide | Oligonucleotide sequence |
|---|---|---|---|
| A | K103N | Upstream | 5′-(tail)- AGGAATACCACATCCAGCAGGTCTAAAAAAGGAC |
| Downstream | 5-Phos-AAATCAGTAACAGTACTAGATGTGGGGG | ||
| Y181C | Upstream 1 | 5′-(tail)-CCTTTAGATCACAAAATCCAGAAATAATTATATG | |
| Upstream 2 | 5′-(tail)-CCTTTAGATCACAAAATCCAGAAATGATTATATG | ||
| Downstream | 5′-Phos-TCAATACATGGATGACTTGTATGTAGGA-(tail) | ||
| G190A | Upstream | 5′-(tail)-TTATCTATCAATACATGGATGACTTGTATGTGGC | |
| Downstream | 5′-Phos-ATCTGATTTAGAAATAGGGCAGCATAGA-(tail) | ||
| D | K103N | Upstream | 5′-(tail)- AGGAATACCACATCCTGCAGGGCTAAAAAAGGAC |
| Downstream | 5-Phos-AAATCAGTAACAGTACTGGATGTGGGTG | ||
| Y181C | Upstream 1 | 5′(tail)-CTTTTAGAAAACAAAATCCAGAAATGGTTATATG | |
| Upstream 2 | 5′(tail)-CTTTTAGAAAACAAAATCCAGAAATAGTTATATG | ||
| Downstream | 5′-Phos-TCAATACATGGATGATTTGTATGTAGGA-(tail) | ||
| G190A | Upstream | 5′-(tail)-TTATCTATCAATACATGGATGATTTGTATGTGGC | |
| Downstream | 5′-Phos-ATCTGACTTAGAAATAGGGCAGCATAGA-(tail) |
Statistical Methods
Baseline characteristics of infants included in the primary analysis and proportions of infants with each mutation (or combination of mutations) were compared by study arm. Baseline characteristics were also compared between infants included in this study and infants whose samples were not available for analysis. Statistical significance for comparisons of proportions was assessed using Fisher's exact test. Mann-Whitney rank sum test was used for comparing medians. Analysis was performed in Stata Version 8 (StataCorp LP, College Station, TX). Associations were considered statistically significant at p<0.05.
Informed Consent
Guidelines of the U.S. Dept. of Health and Human Services and the authors' institutions were followed in the conduct of this research. Informed consent was obtained from all study subjects for participation in the SWEN trial in Uganda; the study was approved by Institutional Review Boards in Uganda and at Johns Hopkins University School of Medicine.
Results
Study Cohort
Samples were available from 49 (71.0%) of 69 infants who were diagnosed with HIV infection by 6 weeks of age (24 in the SD NVP arm and 25 in the extended NVP arm). We compared characteristics of the 49 infants included in this study with those of the 20 infants who did not have samples available for analysis (Table 2). There was no significant difference between these two groups in terms of study arm, maternal pre-NVP viral load, maternal pre-NVP CD4 cell count, median number of NVP doses received, or infant viral load. The proportion of infants who were HIV-infected at birth was higher among those included in this study (27/49=55.1%) than among those who did not have samples for resistance testing (6/20=30%). However, this difference was not statistically significant (p=0.07).
Table 2. Characteristics of infants who were HIV infected by 6 weeks of age.
Comparison of infants who did vs. did not have resistance results (left) and comparison of infants with resistance results who were in the SD NVP arm vs. the extended NVP arm (right).
| All HIV-infected infants | Infants with resistance results | ||||||
|---|---|---|---|---|---|---|---|
| Total | No sample | Sample tested | P valuea | SD NVP | Extended NVP | P value b | |
| (n=69) | (n=20) | (n=49) | (n=24) | (n=25) | |||
| SD NVP study arm (%) | 36 (52.2%) | 12 (60.0%) | 24 (49.0%) | 0.44 | -- | -- | -- |
|
| |||||||
| Median maternal log10 viral load at enrollment | 5.1 | 5.0 | 5.1 | 0.62 c | 5.0 | 5.1 | 0.53 c |
| Median maternal CD4 cell count at enrollment | 353 | 367.5 | 349 | 0.90 c | 299 | 392 | 0.53 c |
|
| |||||||
| Time of diagnosis of HIV infection | |||||||
| Birth | 33 (47.8%) | 6 (30%) | 27 (55.1%) | 0.07 d | 10 (41.7%) | 17 (68%) | 0.09 d |
| 2 weeks | 11 (15.9%) | 4 (20%) | 7 (14.3%) | 0.72 d | 2 (8.3%) | 5 (20%) | 0.42 d |
| 6 weeks | 25 (36.2%) | 10 (50%) | 15 (30.6%) | 0.17 d | 12 (50%) | 3 (12%) | 0.005 d |
|
| |||||||
| Median number of NVP doses received (range)e | 14 (0-36) | 18 (0-36) | 14 (3-35) | 0.48 c | -- | 14 (3-35) | |
|
| |||||||
| Median first infant log10 viral load after diagnosis of HIV infection | 5.5 | 5.6 | 5.5 | 0.89 | 5.6 | 5.3 | 0.19 |
|
| |||||||
| HIV subtype | |||||||
| A | 25 (51.0%) | 12 (50%) | 13 (52%) | 1.0 d | |||
| C | 1 (2.0%) | 1 (4.2%) | 0 (0%) | 0.49 d | |||
| D | 13 (26.5%) | 8 (33.3%) | 5 (20%) | 0.35 d | |||
| Recombinant | 10 (20.4%) | 3 (12.5%) | 7 (28.0%) | 0.29 d | |||
Infants with no sample vs. infants with resistance results (sample tested).
Infants in the SD NVP arm vs. the extended NVP arm.
Mann-Whitney rank sum test.
Fisher's exact test.
Median number of NVP doses for infants in the extended arm only.
In the extended NVP arm, daily NVP dosing was discontinued as soon as a diagnosis of HIV infection was confirmed. The median number of NVP doses received was 14 doses for infants diagnosed with HIV infection at birth (range 3-33, 17 infants), 14 doses for infants diagnosed with HIV infection at 2 weeks of age (range 7-26, five infants), and 34 doses for infants diagnosed with HIV infection at 6 weeks of age (range 21-35, three infants).
We next compared the same clinical and laboratory characteristics described above, as well as HIV subtype, for the 24 infants in the SD NVP arm vs. the 25 infants in the extended NVP arm who were included in this study (Table 2). There was no significant difference in maternal pre-NVP viral load or maternal pre-NVP CD4 cell count, infant viral load, or infant HIV subtype distribution between these two groups. The proportion of infants diagnosed with HIV infection at birth was lower in the SD NVP arm than in the extended NVP arm (10/24=41.7% vs. 17/25=68%), but this difference was not statistically significant (p=0.09). The proportion of infants who were diagnosed with HIV infection at 6 weeks of age was higher in the SD NVP arm (12/24=50%) than in the extended NVP arm (3/25=12%, p=0.005), reflecting the reduced risk of post-natal HIV transmission in the extended NVP arm [22].
Analysis of NVP Resistance using the ViroSeq HIV Genotyping System
HIV genotyping results were obtained for all 49 infants who had 6-week samples. A higher proportion of infants in the extended NVP arm had at least one NVP resistance mutation detected compared to infants in the SD NVP arm (21/25=84% vs. 12/24=50%, p=0.01, Table 3 and Figure 1A). This difference was also observed in the subset of infants who were diagnosed with HIV infection at 2 or 6 weeks of age (7/8=88.9% of infants in the extended NVP arm had NVP resistance vs. 5/14=35.7% of infants in the SD NVP arm, p=0.03, Figure 1B). In contrast, in the subset of infants who were HIV-infected at birth, the proportion of infants with NVP resistance was similar in the two study arms (14/17=82.3% in the extended NVP arm vs. 7/10=70% in the SD NVP arm, p=0.39, Figure 1B). Among all 49 infants analyzed, detection of NVP resistance was not associated with subtype A vs. D (p=1.00). In the extended NVP arm, there was no association between detection of NVP resistance and the number of NVP doses received (median of 14 doses among infants with resistance vs. 17.5 doses among infants without resistance, p=0.85).
Table 3. Genotypic resistance results (ViroSeq and LigAmp assays).
| Total | SD NVP arm | Extended NVP arm | P value | |
|---|---|---|---|---|
| VIROSEQ RESULTS AT 6 WEEKS | 49 | 24 | 25 | |
| ≥ 1 NVP resistance mutation | 33 | 12 (50%) | 21 (84%) | 0.01 a |
| K101E only | 1 | 0 | 1 | |
| K103N only | 4 | 1 | 3 | |
| V106A only | 1 | 1 | 0 | |
| V106M only | 1 | 1 | 0 | |
| Y181C only | 12 | 4 | 8 | |
| Y188C only | 1 | 1 | 0 | |
| G190A only | 1 | 0 | 1 | |
| ≥ 2 NVP resistance mutations | 12 | 4 (16.7%) | 8 (32%) | 0.18 a |
| K103N + Y181C | 4 | 3 | 1 | |
| K103N + V106A + Y181C | 2 | 1 | 1 | |
| K103N + Y188C | 2 | 0 | 2 | |
| Y181C + G190A | 2 | 0 | 2 | |
| K103N + V106A | 1 | 0 | 1 | |
| V106A + Y181C | 1 | 0 | 1 | |
|
| ||||
| LIGAMP RESULTS AT 6 WEEKS b | 44 | 20 | 24 | |
| ≥ 1 NVP resistance mutation | 26 | 7 (35%) | 19 (79%) | 0.005 a |
| # (%) infants with ≥ 0.5% K103N | 13 | 3 (15%) | 10 (41.7%) | |
| Median % K103N | 4.7 | 4.2 | 5.2 | |
| # (%) infants with ≥ 1% Y181C | 22 | 7 (35%) | 15 (62.5%) | |
| Median %Y181C | 15.6 | 10.8 | 20.5 | |
| # (%) infants with ≥ 0.5% G190A | 9 | 3 (15%) | 6 (25%) | |
| Median % G190A | 3.2 | 1.6 | 5.6 | |
Fisher's exact test
LigAmp results over 100% were adjusted to 100%. Median values were calculated only for samples with the mutation detected.
Figure 1. Resistance results for infants in the SD NVP and Extended NVP arms.
Resistance results from infants in the SD NVP arm (white bars) and in the extended NVP arm (shaded bars) are shown. The p-values for comparisons of proportions (Fisher's exact test) are shown above the bars. The number of infants with resistance / the number of infants tested in each subgroup is shown in each bar. For each subgroup, the age of infant at diagnosis of HIV infection (Age at diagnosis), the age of the infant at the time of collection of the sample used for resistance testing (Age at testing), and the test method used for testing (Test method) are shown below the bar graph. Panels A and B: Portion of infants with NVP resistance detected by ViroSeq at 6 weeks of age. Panel A: Results for all infants tested. Panel B: Results for infants who were diagnosed with HIV infection at birth (left) and results for infants who were HIV-uninfected at birth and were diagnosed with HIV infection at 2 or 6 weeks of age (right). Panel C: Portion of infants with NVP resistance detected by ViroSeq at 6 month of age. Panel D: Portion of late-infected infants with NVP resistance mutations (K103N, Y181C, or G190A) detected by the LigAmp assay at the time of HIV diagnosis.
Analysis of K103N, Y181C, and G190A using the LigAmp assay
We used a point mutation assay, LigAmp, to detect and quantify HIV variants with the K103N, Y181C, and G190A NVP resistance mutations. Forty-four (89.7%) of the 49 6-week samples that had ViroSeq results (see above) were tested with the LigAmp assay. Two subtype C samples were not tested, and three infants did not have sufficient samples remaining for analysis with LigAmp. For the 44 samples tested with both ViroSeq and LigAmp, 31 (91.2%) of the 34 K103N, Y181C, and G190A mutations detected by ViroSeq were also detected by LigAmp; two K103N mutations and one Y181C mutation were encoded by unusual nucleotide codons and were not detected by LigAmp. LigAmp detected 13 additional NVP resistance mutations that were below the level of detection of the ViroSeq system.
We compared the portion of infants who had Y181C, K103N, or G190A detected by LigAmp in the two study arms. A higher proportion of infants in the extended NVP arm had one or more of these mutations detected, compared to infants in the SD NVP arm (19/24=79% vs. 7/20=35%, p=0.005, Table 3). In the extended NVP arm, detection of any NVP resistance mutation (K103N, Y181C, or G190A) was not associated with the number of NVP doses received (median of 14 doses among those with at least one mutation detected vs. 21 doses among those with no mutations detected, p=0.14). There was no significant difference in the median level of any of the three mutations in the two study arms (Table 3).
When data from infants in the two study arms were combined, a higher portion of infants with HIV subtype A had NVP resistance mutations detected by LigAmp than infants with subtype D (17/23=73.9% vs. 5/12=41.7%), but this difference was not statistically significant (p=0.08). Among infants who had one or more of these mutations detected, the median level of each of the three NVP resistance mutations (proportion of the viral population with the mutation) was as follows: for infants who were HIV-infected at birth: K103N=4.4%, Y181C=25%, and G190A=3%; for infants who were diagnosed with HIV infection at 2 or 6 weeks of age: K103N=5.4%, Y181C=2.6%, and G190A=18.7%.
Analysis of Phenotypic Drug Resistance
Phenotypic resistance results were obtained with the PhenoSense assay for 42 (85.7%) of 49 available samples. Phenotypic NVP resistance was more frequent among infants in the extended NVP arm than the SD NVP arm (19/22=86.3% vs. 9/20=45%, p=0.005). Results from the ViroSeq and PhenoSense assays (resistance detected vs. not detected) were the same for all but one of the 42 infants who had results from both assays; one infant had Y181C detected by ViroSeq as a mixture, but did not have phenotypic resistance detected. There was only one infant who had NVP resistance that was detected by LigAmp only (1.4% Y181C, with no resistance detected by ViroSeq or PhenoSense).
The PhenoSense assay also provides quantitative information about drug resistance. In this study, among infants with phenotypic NVP resistance, the median fold change in IC50 for NVP was higher in the extended NVP arm than in the SD NVP arm (143 vs. 66, respectively), but the result was not statistically significant (p=0.30). Many of the infants who had phenotypic NVP resistance also had cross resistance to other non-nucleoside reverse transcriptase inhibitors (NNRTI; 10/28=35.7% had cross resistance to delavirdine (DLV), 2/28=7.1% had cross resistance to efavirenz (EFV), and 12/28=42.8% had cross resistance to EFV and DLV,). Among the 42 infants who had PhenoSense results, we found no difference in the median HIV replication capacity for infants in the SD NVP vs. extended NVP study arms (both 33%) and no significant difference between the median replication capacity measured in infants who did vs. did not have NVP resistance mutations detected by ViroSeq (35% vs. 33%, p=0.74). This result suggests that presence of resistance mutations in some HIV variants does not significantly impact the HIV replication capacity of the viral population.
Interestingly, three infants in the extended NVP arm did not have NVP resistance detected with ViroSeq, PhenoSense, or LigAmp. Two of these infants were HIV-infected at birth; these infants received 5 and 14 doses of NVP. One of these infants was diagnosed with HIV infection at 6 weeks, and received 21 doses of NVP.
Analysis of NVP Resistance in Infants at 6 Months of Age
We used the ViroSeq system and LigAmp assay to analyze the persistence of NVP resistance at 6 months of age. Of the 33 infants with ViroSeq results at 6 weeks of age, 17 infants were excluded because they initiated antiretroviral therapy before 6 months of age, one infant died before 6 months of age, one infant was lost to follow-up, and one infant had no sample available. Therefore, this analysis included 13 infants. In the SD NVP arm, one (16.7%) of six infants had a NVP resistance mutation detected by ViroSeq (Y181C), and two additional infants had a NVP resistance mutation detected by LigAmp only (both with Y181C, at 1.4% and 3.5%, Figure 1C). In contrast, all of the seven infants in the extended NVP arm had NVP resistance detected by ViroSeq at 6 months (1/6 vs. 7/7, p=0.005).
Analysis of NVP Resistance in Late-infected Infants
Sixteen infants were HIV-uninfected at 6 weeks of age, but were HIV-infected by either 14 weeks (n=15) or 6 months (n=1) of age. Samples collected from the time of HIV diagnosis were available for 8 (50%) of these 16 infants (4 in the SD NVP arm and 4 in the extended NVP arm). Only 1 (12.5%) of the 8 infants analyzed had NVP resistance detected by ViroSeq (one infant diagnosed at 14 weeks in the extended NVP arm had the K103N mutation). In contrast, 7 (87.5%) of the 8 late-infected infants had at least one mutation detected by LigAmp (4 had K103N, 1 had G190A, 1 had K103N+Y181C, and 1 had K103N+G190A, Figure 1D). The infant who did not have resistance detected by either assay was in the extended NVP arm, received 34 doses of NVP, and was diagnosed with HIV infection at 14 weeks of age.
Discussion
In this study, infants in the extended NVP arm were more likely to have genotypic and phenotypic NVP resistance at 6 weeks of age than infants in the SD NVP arm. Many of the infants with phenotypic resistance to NVP also had cross-resistance to DLV and/or EFV. The majority of HIV-infected infants who were exposed to SD NVP prophylaxis and require antiretroviral treatment by 6 months of age respond poorly to NNRTI-based antiretroviral regimens [9], but may respond better to NVP-based therapy if it is initiated later [10]. In this study, infants in the extended NVP arm were more likely to have resistance detected at 6 months compared to infants in the SD NVP arm. Although this study is limited by the small number of infants tested at 6 months of age (n=13), this finding suggests that infants who are HIV-infected despite extended NVP prophylaxis may be more likely to fail NVP-based antiretroviral therapy, than infants exposed to SD NVP alone.
The association of study arm with resistance at 6 weeks of age was only seen among infants who were HIV-uninfected at birth and were diagnosed with HIV infection at 2 or 6 weeks of age. Among that subset of infants, those who received extended NVP prophylaxis were more likely to develop NVP resistance. In contrast, infants who were HIV-infected prior to NVP exposure (i.e. HIV positive at birth) had high rates of NVP resistance at 6 weeks of age, whether they received the SD NVP regimen only (SD NVP arm), or the SD NVP regimen plus daily NVP until their HIV infection was confirmed (extended NVP arm; median 14 doses, range 3-33). These results suggest that infants who are HIV-infected in utero can receive several doses of NVP without significantly increasing their risk of resistance.
We also analyzed NVP resistance in late-infected infants (diagnosed with HIV infection at 14 weeks or 6 months of age, four infants in each study arm). All but one of the late-infected infants had NVP resistance mutations detected by the sensitive LigAmp assay at the time of HIV diagnosis. The four late-infected infants in the extended NVP arm received 34-36 doses of NVP. For infants in the SD NVP arm, and for the one infant in the extended NVP arm who was diagnosed with HIV infection at 6 months, NVP would have been cleared from the infant by the time of HIV infection. Therefore, in these cases, one can assume that the resistant strain was transmitted to the infant by breastfeeding. In the other three infants (those in the extended NVP arm who were diagnosed with HIV infection at 14 weeks), it is possible that a NVP-susceptible strain was transmitted to the infant shortly after the 6-week visit (before NVP had cleared), with subsequent selection of NVP-resistant HIV in the infant.
The SWEN trial demonstrated that extended NVP prophylaxis is more effective than SD NVP for prevention of MTCT. This study shows that use of extended NVP prophylaxis is associated with increased emergence and persistence of NVP-resistant HIV variants in Ugandan infants. The SWEN trial also included trials performed in India and Ethopia that used the same NVP-based regimens for prevention of HIV MTCT. Even though the trial design (e.g. timing of infant HIV testing and sample collection) and resistance testing methods were different in the Ugandan and Indian components of the SWEN study, HIV-infected infants in the Indian trial who were HIV-uninfected at birth but diagnosed with HIV infection by 6 weeks of age were also more likely to have NVP resistance if they had received extended NVP prophylaxis [25]. Clinical studies are needed to determine the impact of extended NVP prophylaxis on subsequent antiretroviral treatment in HIV-infected infants.
Acknowledgments
The authors thank the SWEN study team for providing the samples and data used in this report, the JHU and Makerere lab staff for assistance with sample processing, and the Monogram Biosciences laboratory staff for performing phenotypic resistance and replication capacity testing. Finally, the authors would like to thank Prof. Francis Mmiro for his life-long dedication to improving the health of pregnant women and children, and for his substantial contributions in the field of HIV prevention. Sadly, Prof. Mmiro passed away while this paper was In Revision.
Sources of Support: This work was supported by (1) the HIV Prevention Trials Network (HPTN) sponsored by the National Institutes of Allergy and Infectious Diseases (NIAID), National Institutes of Child Health and Human Development (NICH/HD), National Institute on Drug Abuse, National Institute of Mental Health, and Office of AIDS Research, of the NIH, DHHS (U01-AI-46745, U01-AI-48054, and U01-AI068613), (2) the International Maternal Pediatric and Adolescent AIDS Clinical Trials Group subcontract BRS-IMPCT-Q-06-00100-T007 from U01-AI-068632 (NIAID, NICHD), (3) R01-AI-34235-04 (NIAID), and (4) U01-AI-038576-07 (NIAID).
Footnotes
Conflict of Interest Statement None of the authors have a commercial or other association that might pose a conflict of interest with the following exception: (1) Dr. Susan Eshleman is a co-inventor of the LigAmp assay and Johns Hopkins University has filed a patent application with the US-Patent and Trademark Office. The inventors may receive royalty payments if the patent is awarded and licensed. (2) Wei Huang is an employee and stockholder of Monogram Biosciences, Inc. (3) Susan Eshleman is a member of the Clinical Advisory Board of Monogram Biosciences.
References
- 1.Guay LA, Musoke P, Fleming T, et al. Intrapartum and neonatal single-dose nevirapine compared with zidovudine for prevention of mother-to-child transmission of HIV-1 in Kampala, Uganda: HIVNET 012 randomised trial. Lancet. 1999;354:795–802. doi: 10.1016/S0140-6736(99)80008-7. [DOI] [PubMed] [Google Scholar]
- 2.Jackson JB, Musoke P, Fleming T, et al. Intrapartum and neonatal single-dose nevirapine compared with zidovudine for prevention of mother-to-child transmission of HIV-1 in Kampala, Uganda: 18 months follow-up of the HIVNET 012 randomised trial. Lancet. 2003;362:859–868. doi: 10.1016/S0140-6736(03)14341-3. [DOI] [PubMed] [Google Scholar]
- 3.Eshleman SH, Mracna M, Guay LA, et al. Selection and fading of resistance mutations in women and infants receiving nevirapine to prevent HIV-1 vertical transmission (HIVNET 012) AIDS. 2001;15:1951–1957. doi: 10.1097/00002030-200110190-00006. [DOI] [PubMed] [Google Scholar]
- 4.Eshleman SH, Hoover DR, Chen S, et al. Resistance after single dose nevirapine prophylaxis emerges in a high portion of Malawian newborns. AIDS. 2005;19:2167–2168. doi: 10.1097/01.aids.0000194800.43799.94. [DOI] [PubMed] [Google Scholar]
- 5.Martinson NA, Morris L, Gray G, et al. Selection and persistence of viral resistance in HIV-infected children after exposure to single-dose nevirapine. J Acquir Immune Defic Syndr. 2006;44:148–53. doi: 10.1097/QAI.0b013e31802b920e. [DOI] [PubMed] [Google Scholar]
- 6.Kurle SN, Gangakhedkar RR, Sen S, Hayatnagarkar SS, Tripathy SP, Paranjape RS. Emergence of NNRTI drug resistance mutations after single-dose nevirapine exposure in HIV type 1 subtype C-infected infants in India. AIDS Res Hum Retroviruses. 2007;23:682–5. doi: 10.1089/aid.2006.0167. [DOI] [PubMed] [Google Scholar]
- 7.Brahmbhatt H, Kigozi G, Wabwire-Mangen F, et al. Mortality in HIV-infected and uninfected children of HIV-infected and uninfected mothers in rural Uganda. J Acquir Immune Defic Syndr. 2006;41:504–8. doi: 10.1097/01.qai.0000188122.15493.0a. [DOI] [PubMed] [Google Scholar]
- 8.Jourdain G, Ngo-Giang-Huong N, Le Coeur S, et al. Intrapartum exposure to nevirapine and subsequent maternal responses to nevirapine-based antiretroviral therapy. N Engl J Med. 2004;351:229–40. doi: 10.1056/NEJMoa041305. [DOI] [PubMed] [Google Scholar]
- 9.Lockman S, Shapiro RL, Smeaton LM, et al. Response to antiretroviral therapy after a single, peripartum dose of nevirapine. N Engl J Med. 2007;356:135–147. doi: 10.1056/NEJMoa062876. [DOI] [PubMed] [Google Scholar]
- 10.Barlow-Mosha L, Ajunua P, Mubiru M, et al. Early effectiveness of a NVP-based HAART regimen among HIV-infected children with and without prior single-dose NVP exposure. 15th Conference on Retroviruses and Opportunistic Infections (Boston, MA) Foundation for Retrovirology and Human Health; 2008. Abstract #358. [Google Scholar]
- 11.Kourtis AP, Lee FK, Abrams EJ, Jamieson DJ, Bulterys M. Mother-to-child transmission of HIV-1: timing and implications for prevention. Lancet Infect Dis. 2006;6:726–32. doi: 10.1016/S1473-3099(06)70629-6. [DOI] [PubMed] [Google Scholar]
- 12.Taha TE, Hoover DR, Kumwenda NI, et al. Late postnatal transmission of HIV-1 and associated factors. J Infect Dis. 2007;196:10–4. doi: 10.1086/518511. [DOI] [PubMed] [Google Scholar]
- 13.John GC, Nduati RW, Mbori-Ngacha DA, et al. Correlates of mother-to-child human immunodeficiency virus type 1 (HIV-1) transmission: association with maternal plasma HIV-1 RNA load, genital HIV-1 DNA shedding, and breast infections. J Infect Dis. 2001;183:206–212. doi: 10.1086/317918. [DOI] [PubMed] [Google Scholar]
- 14.Rousseau CM, Nduati RW, Richardson BA, et al. Association of levels of HIV-1-infected breast milk cells and risk of mother-to-child transmission. J Infect Dis. 2004;190:1880–8. doi: 10.1086/425076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mbori-Ngacha D, Nduati R, John G, et al. Morbidity and mortality in breastfed and formula-fed infants of HIV-1-infected women: A randomized clinical trial. JAMA. 2001;286:2413–20. doi: 10.1001/jama.286.19.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kourtis AP, Jamieson DJ, de Vincenzi I, et al. Prevention of human immunodeficiency virus-1 transmission to the infant through breastfeeding: new developments. Am J Obstet Gynecol. 2007;197:S113–22. doi: 10.1016/j.ajog.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 17.Coovadia HM, Rollins NC, Bland RM, et al. Mother-to-child transmission of HIV-1 infection during exclusive breastfeeding in the first 6 months of life: an intervention cohort study. Lancet. 2007;369:1107–16. doi: 10.1016/S0140-6736(07)60283-9. [DOI] [PubMed] [Google Scholar]
- 18.Sinkala M, Kuhn L, Kankasa C, et al. No benefit of early cessation of breastfeeding at 4 months on HIV-free survival of infants born to HIV-infected mothers in Zambia: The Zambia exclusive breastfeeding study. 14th Conference on Retroviruses and Opportunistic Infections (Los Angeles, CA) Foundation for Retrovirology and Human Health; 2007. abstract # 74. [Google Scholar]
- 19.Andreotti M, Guidotti G, Galluzzo CM, et al. Resistance mutation patterns in plasma and breast milk of HIV-infected women receiving highly-active antiretroviral therapy for mother-to-child transmission prevention. Aids. 2007;21:2360–2. doi: 10.1097/QAD.0b013e3282f190a6. [DOI] [PubMed] [Google Scholar]
- 20.Kassaye S, Lee E, Kantor R, et al. Drug resistance in plasma and breast milk after single-dose nevirapine in subtype C HIV type 1: population and clonal sequence analysis. AIDS Res Hum Retroviruses. 2007;23:1055–61. doi: 10.1089/aid.2007.0045. [DOI] [PubMed] [Google Scholar]
- 21.Lee EJ, Kantor R, Zijenah L, et al. Breast-milk shedding of drug-resistant HIV-1 subtype C in women exposed to single-dose nevirapine. J Infect Dis. 2005;192:1260–4. doi: 10.1086/444424. [DOI] [PubMed] [Google Scholar]
- 22.The Six Week Extended Dose Nevirapine (SWEN) Study Team. Extended dose nevirapine to six weeks of age for infants in Ethiopia, India, and Uganda: a randomized trial for prevention of HIV transmission through breastfeeding. 15th Conf. on Retroviruses and Opportunistic infections (Boston, MA) Foundation for Retrovirology and Human Health; 2008. Abstract # S-135. [Google Scholar]
- 23.Church JD, Jones D, Flys T, et al. Sensitivity of the ViroSeq HIV-1 Genotyping System for detection of the K103N resistance mutation in HIV-1 subtypes A, C, and D. J Mol Diagnostics. 2006;8:420–2. doi: 10.2353/jmoldx.2006.050148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Petropoulos CJ, Parkin NT, Limoli KL, et al. A novel phenotypic drug susceptibility assay for human immunodeficiency virus type 1. Antimicrob Agents Chemother. 2000;44:920–8. doi: 10.1128/aac.44.4.920-928.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moorthy A, Gupta A, Sastry J, et al. Timing of infection is critical for nevirapine resistance outcomes among breastfed subtype C HIV-1-infected infants exposed to extended vs single-dose nevirapine prophylaxis: The India SWEN study. 15th Conference on Retroviruses and Opportunistic Infections (Boston, MA) Foundation for Retrovirology and Human Health; 2008. abstract# 44. [Google Scholar]