Abstract
The complete sequence of the chloroplast genome of cassava (Manihot esculenta, Euphorbiaceae) has been determined. The genome is 161,453 bp in length and includes a pair of inverted repeats (IR) of 26,954 bp. The genome includes 128 genes; 96 are single copy and 16 are duplicated in the IR. There are four rRNA genes and 30 distinct tRNAs, seven of which are duplicated in the IR. The infA gene is absent; expansion of IRb has duplicated 62 amino acids at the 3′ end of rps19 and a number of coding regions have large insertions or deletions, including insertions within the 23S rRNA gene. There are 17 intron-containing genes in cassava, 15 of which have a single intron while two (clpP, ycf3) have two introns. The usually conserved atpF group II intron is absent and this is the first report of its loss from land plant chloroplast genomes. The phylogenetic distribution of the atpF intron loss was determined by a PCR survey of 251 taxa representing 34 families of Malpighiales and 16 taxa from closely related rosids. The atpF intron is not only missing in cassava but also from closely related Euphorbiaceae and other Malpighiales, suggesting that there have been at least seven independent losses. In cassava and all other sequenced Malphigiales, atpF gene sequences showed a strong association between C-to-T substitutions at nucleotide position 92 and the loss of the intron, suggesting that recombination between an edited mRNA and the atpF gene may be a possible mechanism for the intron loss.
Introduction
Cassava, Manihot esculenta Crantz subsp. Esculenta (Euphorbiaceae) is an ancient crop species and starch grains or radiocarbon-dated macroscopic remains are in the archeological record from 1800–7500 BP (Ugent et al. 1986; Dickau et al. 2007). Molecular evidence based on the haplotypes of the single-copy nuclear gene glyceraldehyde 3-phosphate dehydrogenase and genetic variation in five microsatellite loci strongly support the view that cultivated cassava is most likely derived from wild populations referred to M. esculenta subsp. flabellifolia, particularly from the populations occurring along the southern border of the Amazon basin (Olsen and Schaal 1999, 2001). Cassava, also called manioc or tapioca, is a perennial woody shrub, which is cultivated for its starchy storage roots throughout tropical and subtropical regions of the world, particularly in South America, Africa, and Asia, where it is the major source of dietary energy for more than 500 million people. Africa now produces more cassava than the rest of the world and it is one of that continent’s staple food crops (Okezie and Kosikowski 1982; Hillocks 2002). The great need for crop improvement coupled with limitations imposed on traditional methods due to polyploidy have made cassava an ideal candidate for genomic approaches. Nuclear genetic maps collected from restriction fragment length polymorphism (RFLP), randomly amplified polymorphic DNA (RAPD) and simple sequence repeats (SSR), are available (Fregene et al. 1997; Fregene 2000; Mba et al. 2001), and recently advocated whole genome sequencing (Raven et al. 2006) has been initiated (http://www.jgi.doe.gov/sequencing/cspseqplans2007.html).
Several arthropod pests negatively impact cassava roots, foliage, and/or stems, particularly members of Lepidoptera, Diptera, and Hemiptera. There is little or no genetic resistance to these pests and their management is commonly achieved through biological control (El-Sharkawy 2004). Chloroplast genetic engineering would be ideal to address this problem because of the high-dosage strategy that can kill Bacillus thuringiensis susceptible and resistant insects (Kota et al. 1999; DeCosa et al. 2001). In addition, multi-gene engineering (DeCosa et al. 2001; Quesada-Vargas et al. 2005) should facilitate the introduction of genes that code for insecticidal proteins. Several other agronomic traits engineered via the chloroplast genome including disease resistance (DeGray et al. 2001), salt tolerance (Kumar et al. 2004a), drought tolerance (Lee et al. 2003), and herbicide resistance (Daniell et al. 1998; Dufourmantel et al. 2007), are valuable for cassava biotechnology. Chloroplast genetic engineering should offer transgene containment via maternal inheritance of transgenes (Hagemann 2004; Daniell 2002, 2007) or engineering cytoplasmic male sterility (Ruiz and Daniell 2005). Other advantages of chloroplast genetic engineering have been reviewed elsewhere (Maliga 2004; Daniell et al. 2005; Grevich and Daniell 2005; Daniell 2006).
Chloroplast genomes of bryophytes, gymnosperms, angiosperms, and their green algal relatives (Chlorophyta) share a basic set of introns which are believed to have been established prior to divergence of vascular and non-vascular plants. In land plant chloroplast genomes there are 17–20 group II introns within tRNA and protein-coding genes. Grasses have only 17 introns because they lack introns within the clpP and rpoC1 genes (Downie and Palmer 1992; Barkan 2004). A group I intron present within trnL-UAA is considered the most ancient because it is present in cyanobacteria (Xu et al. 1990) and in chloroplasts of algal lineages, as well as, land plants (Simon et al. 2003). The Chlamydomonas reinhardtii chloroplast genome contains five group I and two group II introns but none of these are conserved in land plants. The Euglena gracilis chloroplast genome contains 155 introns (groups II and III), accounting for almost 40% of the genome (Barkan 2004). These examples show that introns have been lost or gained during the course of chloroplast genome evolution. Group I and II introns have been considered mobile genetic elements exhibiting two modes of transposition: (1) intron homing (transmission to an intronless allele) or (2) transposition to ectopic sites, both involving the participation of maturases encoded within the introns (Barkan 2004). Functional group II intron maturases contain reverse transcriptase activity involved in intron homing, endonuclease activity involved in retrotransposition, and domains involved in RNA binding and splicing (Barkan 2004). The only maturase present in land plant chloroplast genomes, matK (an ORF in the trnK-UUU intron), lacks the reverse transcriptase domain and is therefore incapable of promoting intron mobility (Barkan 2004). Splicing of chloroplast group II introns, including the one in atpF, proceeds via lariat formation and depends on host-encoded splicing factors (Jenkins et al. 1997; Vogel et al. 1999; Vogel and Börner 2002; Barkan 2004). Intron presence/absence can be a useful marker for phylogenetic studies even though convergent losses have been documented in angiosperm chloroplast genomes (e.g., Downie et al. 1991a, 1994; McPherson et al. 2004). Detailed studies on the distribution of such structural changes have been sparse and each new fully sequenced chloroplast genome presents previously unsuspected structural variation that merits wider investigation to understand the evolutionary implications.
We report here on the complete sequence and organization of the chloroplast genome of M. esculenta, which is the first published genome sequence of a member of the family Euphorbiaceae. The most surprising structural change we uncovered in the cassava chloroplast genome is the loss of the atpF intron and the association of this loss with RNA editing. This is the first report of the absence of the atpF intron from a land plant chloroplast genome. We also present a detailed survey for this intron loss among members of Euphorbiaceae and in other families of Malpighiales. The phylogenetic implications of the distribution of mutiple losses of the atpF intron are also discussed.
Materials and methods
DNA sources
A bacterial artificial chromosome (BAC) library of M. esculenta cultivar TME3 was constructed by ligating size fractionated partial BstY1 digests of total cellular, high molecular weight DNA with the pINDIGOBAC536 vector. The average insert size of the cassava library was about 100 kb. Bacterial artificial chromosome-related resources for this public library (BAC library ME_TBa) can be obtained from the Clemson University Genomics Institute BAC/EST Resource Center (http://www.genome.clemson.edu). Bacterial artificial chromosome clone screening, selection and sequencing followed Daniell et al. (2006). Chloroplast genome inserts were identified with a soybean chloroplast DNA probe and the first 96 positive clones were pulled from the library, arrayed in a 96-well microtitre plate, copied, and archived. Selected clones were then subjected to Hind III fingerprinting and Not I digests. End-sequences were determined and localized on the chloroplast genome of Arabidopsis thaliana to deduce the relative positions of the clones; then a single clone that covered the entire chloroplast genome of cassava was chosen for sequencing.
DNA sequencing and genome assembly and annotation
The nucleotide sequence of the selected BAC clone was determined by the bridging shotgun method. The purified BAC DNA was subjected to hydroshearing, end repair, and then size-fractionated by agarose gel electrophoresis. Fractions of approximately 3.0–5.0 kb were eluted and ligated into the vector pBLUESCRIPT IIKS+. The shotgun libraries were plated and then arrayed into forty 96-well microtitre plates for the sequencing reactions. Sequencing was performed using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA). Sequence data from the forward and reverse priming sites of the shotgun clones were accumulated until equivalent to eight times the size of the genome and then assembled using Phred-Phrap programs (Ewing and Green 1998). Annotation of the cassava chloroplast genome was performed using DOGMA (Dual Organellar GenoMe Annotator, Wyman et al. 2004) with a custom database of previously published chloroplast genomes. Both tRNAs and rRNAs were identified by BLASTN searches against the same database of chloroplast genomes.
Analysis of the atpF intron
Genomic DNA extractions were initially screened with 15 μl PCR reactions using primers atpF-100F 5′-CTAAGTGTAGTSCTTGGTGTATTGA-3′ and atpF-465R 5′-AGTTCCTAGAGCTCCTTGTAA-3′, that are expected to yield a 367-bp intronless exonic fragment for taxa lacking an intron or a ca 1.1-kb fragment for taxa with an intron. If multiple copies of a gene are present which differ greatly in size (i.e., reflecting differences in intron content), then preferential amplification of the shorter fragments can occur. The intron-containing copy may also co-amplify or be supressed entirely as a PCR artifact (Wurdack, personal observation). To assess whether shorter intronless fragments might represent undesired paralogs, an intron/exon primer combination was tried on intronless taxa using atpF-465R and an internal intron primer, atpF-Intron-F 5′-TCGATATARAACACTCATRTCGATA-3′. Primers rooted in the flanking genes (atpH-162F 5′-GCTTAGTCTGGCTTTTATGGAAGC-3′ and atpA-152R 5′-CCTGCCATTACTTCATCAAGAC-3′) were used in combination with the original atpF primers or an alternative forward primer (atpF-1F 5′-ATGAAAAATRTAAC CGATTCTTTC-3′) to produce copies from the chloroplast genome for the purposes of sequencing and to establish whether intronless copies had the correct flanking chloroplast genes. Intronless copies could be processed paralogs from RNA intermediates and occur elsewhere in other genome (i.e., nuclear or mitochondrial) but these would not be expected to contain flanking chloroplast sequence. Finally, an intron insertion-site spanning primer atpF-ISP-R 5′-CGATTATCTAATAAATCACTTAACAC-3′ was used in combination with primers atpF-1F or atpF-100F to check for intronless copies in select intron-containing taxa. Amplification conditions were 35 cycles of 94°C for 1 min, 52°C for 1 min and 72°C for 2 min. Approximately 7% of the DNAs tested failed to yield bands, mostly due to degraded templates but some could be attributable to primer mismatches.
Phylogenetic analyses
Phylogenetic analyses of the atpF-coding region included 76 taxa of which 52 came from previously published, fully sequenced angiosperm chloroplast genomes and the remainder are newly generated, including cassava and 23 additional sequences from a carefully selected subset of the the taxa screened here for intron loss. Manual alignment was straightforward with no indels (insertions or deletions) in Malpighiales; among other angiosperms there were six autapomorphic insertions and six deletions (one informative, truncating the terminus of the gene by one codon). Indels were treated as missing data and not excluded or recoded. Analyses were performed with maximum parsimony (MP) using PAUP* ver. 4.0b10 (Swofford 2003), maximum likelihood (ML) using GARLI ver. 0.951 (Zwickl 2006), and Bayesian inference (BI) using MrBayes ver. 3.1.1 (Huelsenbeck and Ronquist 2001). The appropriate model (GTR + I + Γ) was determined with Modeltest ver. 3.7 (Posada and Crandall 1998). For MP, starting trees were generated with 1,000 random taxon addition TBR replicates (MulTrees off) and these were subjected to another round of searching with a limit of 50,000 trees (MulTrees on). Additional MP searches were conducted using the parsimony ratchet as implemented in PAUPRat ver. 1 (Sikes and Lewis 2001) and using five replicates of 200 interations each under default parameters. For ML, GARLI searches were run under the automated stopping criterion (20,000 generations); likelihood scores and parameter values were optimized in PAUP*. BI used four Markov chains run for 1.0 ×107 generations and sampled every 100 generations with a discarded burnin of 1.0 ×106 generations. Maximum likelihood and BI searches were done in replicate to determine the concordance between runs. Maximum parsimony non-parametric boostraps used 1,000 replicates, each with 10 random addition replicates but saving no more than 10 trees per iteration (see Wurdack et al. 2005). Maximum likelihood bootstraps in GARLI used 100 replicates and a 20,000-generation automated stopping criterion. We tested the hypothesis on correlated evolution between atpF intron status (present or absent) and RNA-editing (i.e., C or T at nucleotide position 92) using BayesDiscrete (part of BayesTraits; Pagel and Meade 2006). This Bayesian approach looks for correlated evolution between pairs of binary traits and accounts for phylogenetic uncertainty.
Results
Size, gene content, order, and organization of the cassava chloroplast genome
The complete nucleotide sequence of the chloroplast genome of M. esculenta has been determined (Fig. 1, Genbank accession number EU117376). This genome is 161,453 bp in length and includes a pair of inverted repeats (IR) of 26,954 bp separated by small and large single copy (SSC, LSC) regions of 18,250 and 89,295 bp, respectively. The genome has 128 genes; 96 are single copy and 16 are duplicated in the IR. There are four rRNA genes and 30 distinct tRNAs, seven of which are duplicated in the IR. The genome consists of 49.82% protein-coding, 1.7% tRNA, and 5.6% rRNA genes, and 42.87% non-coding sequence. The G + C and A + T contents in the cassava chloroplast genome are 35.87 and 64.13%, respectively. The overall A + T content is similar to poplar (63.26%), tobacco (62.2%), citrus (61.52%), maize (61.5%), and rice (61.1%). The A + T content of LSC and SSC regions are 66.73 and 70.36%, respectively, whereas the IR has 57.70% A + T due to the presence of the rRNA gene cluster.
Fig. 1.

Map of the M. esculenta plastid genome represented in its circular monomeric form. Large and small single copy regions (LSC, SSC) are separated by the inverted repeats (IRa and IRb, 26,954 bp, respectively). Genes illustrated inside the circle are transcribed in the clockwise direction and genes illustrated outside the circle are transcribed in the counter-clockwise direction. Split genes or genes with introns are marked with asterisks
The infA gene, coding for translation initiation factor 1, is absent in the cassava chloroplast genome. The expansion of IRb has duplicated 62 amino acids of the 3′ end of rps19. There are indels within certain coding sequences. For example, between coordinates 540–560 of the 23S rRNA gaps are required to align the cassava sequences with sequences from other species. Moreover, the 23S rRNA (2,822 bp) is 11–12 bp longer than other core eudicots including members of the Solanaceae, Atropa (2,811 bp, Schmitz-Linneweber et al. 2002), Lycopersicon (2,810 bp, Kahlau et al. 2006), Nicotiana (2,810 bp, Shinozaki et al. 1986), Solanum (2,810 bp, Daniell et al. 2006), and species from other families, including Arabidopsis (2,810 bp, Sato et al. 1999), Citrus (2,809 bp, Bausher et al. 2006), Coffea (2,810 bp, Samson et al. 2007), Cucumis (2,810 bp, Plader et al. 2007), Daucus (2,806 bp, Ruhlman et al. 2006), Glycine (2,811 bp, Saski et al. 2005), Gossypium (2,810 bp, Lee et al. 2006), Populus (2,809 bp, Okumura et al. 2006), and Spinacia (2,810 bp, Schmitz-Linneweber et al. 2001).
Evolution of atpF and loss of its intron
Seventeen genes in cassava have introns, 15 of which contain a single intron including six in tRNAs, while two (clpP, ycf3) have two introns. The alignment of the atpF sequences of cassava and Populus (i.e., the closest relative with a completely sequenced chloroplast genome) indicates that the intron is missing from the former (Fig. 2). The presence/absence of the atpF intron in 251 taxa representing 34 of the ~38 families of Malpighiales and 16 taxa from the closely related sister groups, Celastrales and Oxalidales (including Huaceae), was determined by a PCR survey (Appendix, Fig. 3). This screening determined the intron was missing not only in cassava but also from closely related members of Euphorbiaceae, some Phyllanthaceae, and Picrodendraceae, and all sampled members of Elatinaceae, Lophopyxidaceae, Malesherbiaceae, Passifloraceae, and Turneraceae. PCR product size variation probably due to small intron indels was evident on agarose gels for some taxa but large size differences (>ca 50 bp) indicating that larger indels were not detected. Secondary PCR screening on intronless taxa with the intron/exon primer pair did not detect intron-containing copies. Screening using the exon/intron-insertion-site spanning primers did not detect intronless copies in intron-containing taxa. Amplification using primers in the flanking genes atpH and atpA and sequencing of representative intronless taxa (see Appendix) verified intron absence at the same splice site. The length of the atpF protein-coding sequence was found to be 555 bp in all fully sequenced Malpighiales (some additional taxa sampled here have truncated sequences due to the use of internal atpF primers). Phylogenetic analyses (MP, ML, BI) were performed using atpF sequences from 76 taxa including 23 Malpighiales representing intron-containing and closely related intronless taxa. The data set had 576 aligned coding positions of which 380 were variable, and 267 were parsimony informative. Maximum parsimony searches reached the set tree limit of 50,000 (actual number of optimal trees is greater) most parsimonious trees (length 1,626 steps; retention index of 0.589; consistency index of 0.385/0.325 including/excluding uninformative characters, respectively) that in strict consensus are moderately resolved, except among rosids, Solanaceae, and basal angiosperms (results not shown). The parsimony ratchet found no shorter trees. The ML analyses resulted in an optimal tree with −lnL = 8741.21059 (Fig. 4). The topologies and relative levels of support were similar among trees from the three methods of analysis. Fifteen nodes (four in Malpighiales) had strong support (≥95 posterior probability, PP; ≥85% bootstrap) across all analyses. Malpighiales were resolved as monophyletic with ML and BI analyses (PP = 98) but not with MP (i.e., unresolved mostly with other rosids). Euphorbiaceae were monophyletic with high to moderate support. Phyllanthaceae were paraphyletic due to the inclusion of Picrodendraceae but this has weak support. In cassava and all other sequenced Malphigiales, the atpF gene sequences showed a strong association between C-to-T substitutions at nucleotide position 92 and losses of the intron (mapped in Fig. 4).
Fig. 2.
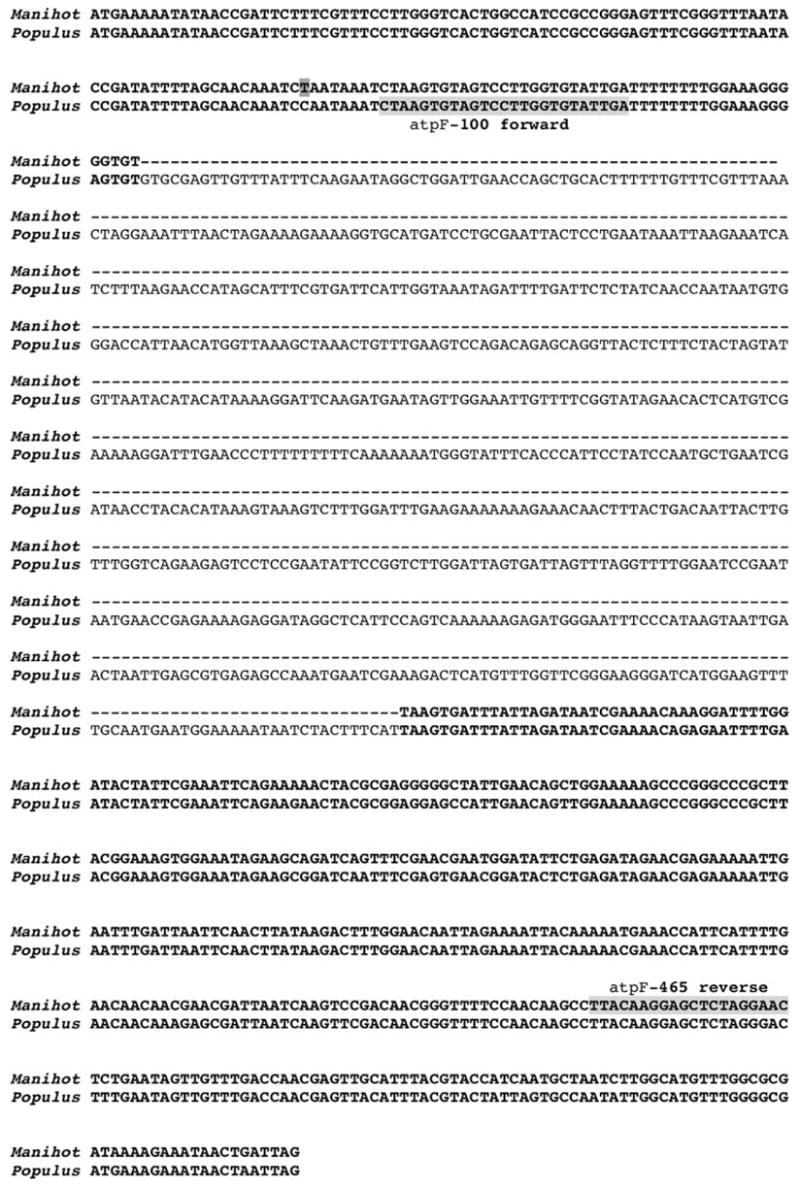
Alignment of atpF genes from cassava and Populus alba showing that there is 94.1% sequence identity in the exon regions and the precise loss of the intron in cassava. The nucleotide position 92 is highlighted in cassava, where C-U editing occurs. Primers used for DNA sequencing are also highlighted
Fig. 3.

Ethidium bromide stained 1.5% agarose gel showing amplification products for selected Malpighiales taxa screened for the presence/absence of the atpF intron. Intron present, lanes 1 Putranjiva roxburghii, 2 Hybanthus concolor, 3 Bischofia javanica, 4 Ricinus communis, 5 Suregada glomerulata, 6 Tetrorchidium cf. macrophyllum. Intron absent, 7 Elateriospermum tapos, 8 Manihot grahamii, 9 Micrandra inundata, 10 Savia bahamensis, 11 Lophopyxis maingayi 12 Malesherbia weberbaueri. L Invitrogen 1 Kb Plus DNA Ladder. Underlined taxa have been sequenced from these PCR products. See Appendix for sample details
Fig. 4.

Phylogenetic tree of the 76-taxon atpF dataset. Tree shown is from a ML analysis with −lnL = 8741.21059. Number series above or below branches indicate support from BI, ML, and MP analyses, respectively; single numbers indicate all 3 values are identical. Asterisk indicates <50% support in that analysis. Intron absence is indicated with an open square; all other taxa possess the atpF intron. A filled gray square indicates the presence of a thymine at nucleotide 92; all other taxa possess a cytosine at that site, which is potentially RNA edited as a C-U conversion. GenBank numbers are indicated for published data, all other taxa are newly generated (see Appendix)
Discussion
Implications for homologous recombination of transgenes in chloroplast genomes
Complete chloroplast genome sequences provide information on intergenic spacer regions for homologous recombination of transgenes in chloroplast vectors. Transformations of the Arabidopsis, potato, and tomato chloroplast genomes were achieved using the tobacco chloroplast genome flanking sequences but efficiencies were much lower than in tobacco because of less than 100% sequence identities (Sikdar et al. 1998; Sidorov et al. 1999; Ruf et al. 2001). In another study, Petunia flanking sequences were used to transform the tobacco plastid genome and transformation efficiency was lower than was observed with endogenous flanking sequences (DeGray et al. 2001). Furthermore, when Solanum nigrum flanking sequence was used in plastid transformation of tobacco, the efficiency of homologous recombination was very low even though flanking sequence identity was 98% (Zubkot et al. 2004). These examples emphasize the advantages of species-specific vectors for efficient homologous recombination within the intergenic spacer regions of chloroplast genomes. Species-specific chloroplast vectors were successfully employed in developing plastid transformants in carrot, cotton, and lettuce (Kumar 2007, 2004a, b; Kanamoto et al. 2006; Ruhlman et al. 2007). In addition to intergenic spacer regions, regulatory elements like promoters, 5′-UTRs and 3′-UTRs are important for expression of transgenes in plastids. Recently, high-level GFP expression was achieved in lettuce using endogenous regulatory elements (Kanamoto et al. 2006). Therefore, it is important to obtain complete genome sequences of crop plants for various biotechnology applications (Daniell et al. 2006; http://www.ncbi.nlm.nih.gov/genomes/ORGANELLES/plastids_tax.html).
Genome organization
The plastid genome of M. esculenta has the ancestral angiosperm genome organization (Raubeson et al. 2007) and in particular is co-linear with Populus, its closest fully sequenced relative. Major structural differences include the presence of rps16 and rpl32 (both absent in Populus) and the loss of infA (present in Populus) in cassava. The last is an unusually unstable gene, having been lost from the chloroplast genome of angiosperms on many separate occasions and transferred multiple times to the nuclear genome. Using two infA gene probes from Antirrhinum and a Southern hybridization approach accompanied by selective sequencing, Millen et al. (2001) ascertained that infA was lost or is a pseudogene in at least 24 lineages of angiosperms, including most rosids and five Euphorbiaceae sampled. Hevea, a close relative of cassava in Euphorbiaceae, was definitively determined in that study to possess a pseudogene based on sequencing; poor hybridization suggests other Malpighiales contain pseudogenes (Millen et al. 2001). In Populus and 17 other sequenced chloroplast genomes sequenced, infA is also pseudogene.
Evolution of atpF and loss of its intron
With the exception of the loss in cassava reported here, a group II intron in atpF at nucleotide position 148 (atpFi148) has been found in all previously sequenced land plant chloroplast genomes. Most green algae (Chlorophyta and charophycean green algae which are early-branching members of the streptophytes and sister to land plants) also appear to lack this intron. Among charophytes, however, Staurastrum does possess atpFi148, but its sister group Zygnema does not (Turmel et al. 2005, 2006). A single gain within the streptophytes before the origin of land plants followed by losses in charophytes has been suggested (Turmel et al. 2006). Details on exactly where the atpFi148 was gained and where losses have occurred must await detailed sampling for this intron and a more robust phylogeny of the early-branching lineages of the streptophytes. Another charaphyte, Chara, has a non-homologous atpF intron at a different insertion site (atpFi1380) (Turmel et al. 1999, 2006). Splicing of the atpF intron is well studied and specific host-encoded (nuclear) protein cofactors are required. Among assayed chloroplast introns in Arabidopsis and maize, crs1 is a required specific cofactor for the atpF intron where it promotes intron folding (Jenkins et al. 1997; Vogel et al. 1999; Ostersetzer et al. 2005; Asakura and Barkan 2006). Intron recognition by crs1 has been mapped to elements of specific intron domains in maize (Ostersetzer et al. 2005), although these are not highly conserved either across angiosperms or in the Malpighiales sequenced here. Lack of splicing of the spinach atpF intron in transgenic Chlamydomonas suggests host factors are also taxon specific (Deshpande et al. 1995). Mutations causing the loss of specific splicing factors such as crs1 could drive the concomitant evolutionary loss of the atpF intron since splicing defects are lethal by preventing biogenesis of the ATP synthase complex (Jenkins et al. 1997).
RNA editing and loss of atpF intron
Copies (processed paralogs sensu Bowe and dePamphilis 1996) of organellar genes lacking otherwise canonical introns can be the products of reverse-transcription of RNA intermediates. These may be integrated elsewhere (i.e., mitochondrial or nuclear genomes) or replace the native intron-containing copy via recombination (Bonen and Vogel 2001; Itchoda et al. 2002). RNA editing has been documented for atpF (Corneille et al. 2000; Tillich et al. 2006) and in particular for codon 31, where C-U editing occurs to correct a non-synonymous second position substitution (nucleotide 92) and conserve a leucine residue. Processed paralogs from RNA intermediates would be expected to show an editing signature. Cassava and all other intronless taxa sequenced for atpF appear by bioinformatic prediction to have lost this RNA-editing in codon number 31 (i.e., the conversion of CCA → CTA and the resulting amino acid change of P → L), which may be evidence of replacement via an RNA intermediate. The association of intron absence and this substitution is absolute among sequenced Malpighiales but not elsewhere in the tree where editing occurs but the intron is present (mapped in Fig. 4). This observation suggests recombination between an edited mRNA and the cognate chloroplast genome allele may be a possible mechanism for the loss of the atpF intron. Although this mechanism has been previously suggested for mitochondrial genes (e.g., nad4, Itchoda et al. 2002), it has not yet been reported for chloroplast genes. However, further studies are needed to confirm this hypothesis. In particular, atpF is part of an ATP synthase transcriptional unit consisting of atpI/H/F/A and full polycistronic and partial length transcripts due to post-transcriptional processing have been detected (Stahl et al. 1993; Miyagi et al. 1998; Knauf and Hachtel 2002). Editing also occurs in atpA (Tillich et al. 2006) and two sites (codons 264 and 383) do not appear to be edited in cassava. A third site, codon 265, is ambiguous and would need experimental verification as although editing is possible (i.e., a cytosine is present in cassava), it appears dispensible and the third-codon position change is synonymous (Corneille et al. 2000). If a transcriptional unit was involved in recombination, then such additional editing changes beyond atpF might be expected. Other possible mechanisms involved in the loss of introns include maturases that have reverse transcriptase and endonuclease activity for intron homing, and retro-transposition. However, the only maturase (matK) identified in land plant chloroplast genomes lacks reverse transcriptase activity (Barkan 2004) and therefore may not play any significant role in the loss of introns.
Phylogenetic distribution of atpF intron losses
To determine the distribution and phylogenetic utility of the atpF intron loss, we screened a broad diversity of Euphor-biaceae and other Malpighiales DNAs mostly used in prior phylogenetic studies (i.e., Davis and Wurdack 2004; Wurdack et al. 2004, 2005; Berry et al. 2005; Davis et al. 2005). Our results, when considered in the context of these studies, suggest that the intron has been lost at least seven times in Malpighiales (losses each from Elatinaceae, Euphorbiaceae, Picrodendraceae, Lophopyxidaceae, and Passifloraceae s.l., and twice from Phyllanthaceae). The minimum number of losses estimated here can be established with confidence even though there is uncertainty in the relationships of some groups (i.e., between groups of Malpighiales families and at the subfamily level in Euphorbiaceae), as prior studies indicate the seven clades with losses all have strongly supported sister-groups that possess the intron. Optimizations with fewer losses would imply the unlikely secondary gain of an intron at this site.
The intron loss in Euphorbiaceae is confined to the articulated crotonoids (sensu Wurdack et al. 2005), a clade of the subfamily Crotonoideae with many members possessing articulated laticifers that are the source of rubber production. This strongly supported (100% bootstrap; Wurdack et al. 2005; Tokuoka 2007) group is nested within the family and comprises approximately eight genera and ca 175 species (six genera sampled here, Cnidoscolus, Elateriospermum, Glycydendron, Hevea, Manihot, and Micrandra; not sampled, Micrandropsis and Cunuria). The affinities of Elateriospermum (traditionally treated as a monotypic tribe), the first-branching lineage of the articulated crotonoids, had been disputed prior to molecular phylogenetic analyses (Wurdack et al. 2005). The absence of the intron in Elateriospermum confirms its affinities. Although our sampling in this clade is incomplete, we believe this represents a single loss before the divergence of Elateriospermum and all nested members are expected to lack the intron. The intron absences in Phyllanthaceae are confined to two nested clades: (1) all Phyllantheae and (2) part of Poranthereae (clades F1 and F3, respectively; sensu Wurdack et al. 2004; Kathriarachchi et al. 2005; Hoffmann et al. 2006) and probably represent two separate losses. Additional sampling is needed for Poranthereae to see if the loss is confined to Leptopus or extends to other genera. Monophyly of this clade is strongly supported and although internal intergeneric relationships are poorly resolved (Wurdack et al. 2004; Kathriarachchi et al. 2005), the presence of both intron and intronless taxa confirms this loss is separate from that in Phyllantheae. The loss in Picrodendraceae is confined to nested closely related Australasian members of the family and probably represents a single loss. Podocalyx, sister to the rest of the family (Davis and Wurdack 2004; Davis et al. 2005), possesses the intron. Malesherbiaceae, Passifloraceae, and Turneraceae are closely related and have been proposed to be united in a single family (i.e., Passifloraceae sensu lato; APG 2003). The absence of the intron may reflect a single loss and represent an additional synapomorphy uniting the smaller segregate families. These families are nested within a large clade of Malpighiales with mostly parietal placentation whose other sampled members (including Populus) contain the intron. Passifloraceae are known to have other chloroplast gene and intron losses (Downie et al. 1994a, b; Hansen et al. 2006; Jansen et al. 2007). The loss of the intron in Lophopyxidaceae appears isolated as the family is monotypic, and because its strongly supported sister group (Davis et al. 2005), Putranjivaceae, possesses the intron. Only one member of digeneric Elatinaceae was sampled and it is unclear if this is an isolated loss or characterizes the entire small family (samples of Bergia, the second genus in the family failed to amplify); members of the strongly supported sister family, Malpighiaceae (Davis and Chase 2004), possess the intron. Given the isolated taxonomic positions of these losses and our uneven sampling outside of Euphorbiaceae, additional intron losses are likely not only in Malpighiales but also throughout angiosperms. We expect densely sampled future studies among other groups of angiosperms to uncover similar patterns of sporadic loss.
Phylogenetic analyses of the atpF coding sequence recovered trees that are largely congruent with established angiosperm relationships (Fig. 4) and with surprisingly high levels of support. This indicates that the molecular evolution of intronless alleles has not dramatically changed with the loss of the intron. Such changes might occur if some of the data represented paralogs in a new genetic environment (i.e., transferred to the nucleus or mitochnodrion) and can also negatively impact phylogenetic reconstruction (Bowe and dePamphilis 1996), which does not appear to be the case here. Higher-level relationships among families of Malpighiales are poorly resolved even with four or more genes (e.g., Davis et al. 2005) and supported relationships would not be expected with a single short gene such as the coding region of atpF. The unsupported paraphyletic relationship of Phyllanthaceae and with its sister group Picrodendraceae is probably an artifact of short branches and sparse taxon sampling.
In conclusion, this is the first published chloroplast genome sequence of a member of the family Euphorbiaceae. In order to achieve efficient homologous recombination in chloroplast transformation it is important for vectors to have 100% sequence identity of flanking sequences. In addition, having endogenous regulatory elements in the vector is highly desirable to hyper-express foreign genes in the chloroplast. The chloroplast genome sequence provides the necessary information for the design and construction of an efficient chloroplast vector for this important crop species. The loss of atpF intron among members of Euphorbiaceae and other Malpighales provides a new structural change to define clades in this large angiosperm group. Furthermore, a strong association between C-to-T changes at nucleotide position 92 and the loss of the intron suggests that recombination between an edited mRNA and the atpF gene may be a possible mechanism for the intron loss.
Acknowledgments
Investigations reported in this article were supported in part by grants from the USDA (3611-21000-017-00D) and NIH (R01 GM 63879) to Henry Daniell. Research by Kenneth J. Wurdack and Robert K. Jansen was supported, in part, by NSF AToL grants EF 0431242 and DEB 0120709, respectively. The authors thank Kenneth Olsen, the Royal Botanic Gardens, Kew, and the herbaria cited for tissue or DNA samples.
Appendix
Taxa and voucher information for 250 accessions (251 taxa) of Malpighiales and related families examined for atpF intron status. Taxa sequenced for atpF are followed by the GenBank accession number. Herbarium acronyms follow Index Herbariorum (http://sciweb.nybg.org/science2/IndexHerbariorum.asp).
Intron present
(i) Malpighiales
Achariaceae: Acharia tragodes Thunb., Cloete s.n. (BOL); Erythrospermum phytolaccoides Gardn., Chase 1277 (K); Hydnocarpus sp., Chase 1301 (K); Kiggelaria africana L., Chase 5607 (K). Balanopaceae: Balanops vieillardii Baill., Chase 1816 (K). Bonnetiaceae: Bonnetia sessilis Benth., Berry s.n. 25.7.98 (MO). Caryocaraceae: Anthodiscus amazonicus Gleason & A. C. Sm., van der Werff 13889 (MO); Caryocar glabrum Pers., Mori 22997 (NY). Clusiaceae s.l.: Mammea siamensis T. Anders., Chase 1216 (K); “Mesua” sp. (probably Kayea sp.), Coode 7884 (K); Montrouziera cauliflora Planch. & Triana, Lowry 5601 (MO); Pentaphalangium latissimum Lauterb., Chase 2100 (K); Rheedia macrophylla Planch. & Triana, Chase 1219 (K). Ctenolophonaceae: Ctenolophon englerianus Mildbr., McPherson 16911 (MO). Dichapetalaceae: Dichapetalum sp., Chase 624 (K?). Euphorbiaceae s.s.: Acalypha californica Benth., Levin 2192 (SD) GenBank EU293932; Adriana tomentosa (Thunb.) Gaudich. var. tomentosa, Cameron s.n. (US); Aleurites moluccana (L.) Willd., Wurdack s.n. (US); Amperea xiphoclada (Sieber ex Spreng.) Druce, Chase 1936 (K); Anthostema madagascariense Baill., Hoffmann 291 (K); Aparisthmium cordatum (A. Juss.) Baill., Bell et al. 93–60 (US); Astraea lobatum Klotzsch, Steinmann 2024 (RSA); Baloghia inophylla (G. Forst.) P. S. Green, Chase 3062 (K); Bernardia myricifolia (Scheele) S. Watson, McGill & Sundell 5459 (K); Bertya rosmarinifolia Planch., Chase 1938 (K); Beyeria leschenaultii (DC.) Baill., Chase 1939 (K); Blumeodendron sp., Chase 1252 (K); Calycopeplus casuarinoides L.S. Sm., Steinmann 1407 (RSA); Caperonia palustris (L.) A. St.-Hil., Wurdack D073 (US); Cavacoa aurea (Cavaco) J. Léonard, Luke & Robertson 1922 (US); Cephalomappa malloticarpa J.J. Sm., M. Chase 1256 (K); Chaetocarpus africanus Pax, Gabon, White 1319 (MO); Cladogynos orientalis Zipp., Chamchanroon 2010 (L); Claoxylon australe Baill. ex Müll. Arg., Chase 1937 (K); Clutia pulchella L., Chase 5876 (K); Cnesmone javanica Blume, Huq 10189 (US); Codiaeum variegatum (L.) Blume, Wurdack D033 (US), GenBank EU293933; Colliguaja integerrima Gillies & Hook., Landrum 8234 (MO); Conceveiba martiana Baill., Bell 93–176 (US); Croton alabamensis E.A. Sm. ex Chapm. var. alabamensis, Wurdack D008 (US), GenBankEU293934 Croton alabamensis E.A. Smith ex Chapman var. texensis Ginzbarg, Nesom 7850 (NY); Croton betulinus Vahl, Steinmann 2026 (RSA); Croton chilensis Müll. Arg., Wurdack D646 (US); Croton conduplicatus Kunth, Berry 5542 (MO); Croton cuneatus Klotzsch, Berry 7589 (PORT); Croton elliottii Chapm., Wurdack D455 (US); Croton ffavens L., Steinmann 2015 (RSA); Croton guianensis Aubl., Berry 5535 (MO); Croton martinianus V.W.Steinm., Steinmann 606 (RSA); Croton nanus Gagnep., Pooma 4256 (BKF); Croton repens Schltdl., Steinmann 1062 (RSA); Croton setigerus Hook., Hughey s.n. (US); Croton socotranus Balf. f., Wurdack D644 (US); Croton speciosus Müll. Arg., Berry 7590 (MO); Croton suaveolens Torr., Devender 96–257 (RSA); Croton trinitatis Millsp., Berry 7586 (MO); Crotonogynopsis usambarica Pax, Tanzania, Polhill et al. 5129 (K); Dalechampia spathulata (Scheidw.) Baill., Wurdack D010 (US); Dichostemma glaucescens Pierre, Reitsma 1285 (NY); Ditaxis argothamnoides (Bertol. ex Spreng.) Radcl.-Sm. & Govaerts, Wurdack D105 (US); Ditrysinia fruticosa (Bartram) Govaerts & Frodin, Wurdack D149 (US); Ditta myricoides Griseb., Cedeño s.n. (US); Dodecastigma amazonicum Ducke, Mori 22811 (US); Endospermum moluccanum (Teijsm. & Binn.) Kurz, Chase 1258 (K); Enriquebeltrania crenatifolia (Miranda) Rzed., Cabrera 10768 (NY); Erythrococca sp., Cameroon, Cheek s.n. (K); Euphorbia epithymoides L., Chase 102 (NCU); Euphorbia mesembryanthemifolia Jacq., Wurdack D102 (US); Euphorbia pulcherrima Willd. ex Klotzsch cv. ‘Brilliant Diamonds’, Wurdack D084 (US); Excoecaria agallocha L., Motley & Cameron 2053 (NY); Excoecaria cochinchinensis Lour., Chase 1260 (K); Garcia nutans Vahl ex Rohr, Wurdack D051 (US); Gitara venezolana Pax & K. Hoffm., Bell et al. 94–312 (US); Gymnanthes lucida Sw., Wurdack D055 (US); Hippomane mancinella L., Wurdack D053 (US); Homalanthus populneus (Geiseler) Pax, Chase 1266 (K); Hura crepitans L., Wurdack D089 (US) GenBankEU293935; Jatropha gossypifolia L., Tayor 11763 (MO); Jatropha integerrima Jacq., Wurdack D047 (US); Joannesia princeps Vell., Chase 1262 (K); Klaineanthus gaboniae Pierre, White (ser. 2) 334 (MO); Koilodepas bantamense Hassk., Chase 1263 (K); Lasiocroton bahamensis Pax & K. Hoffm., Wurdack D058 (US); Leucocroton microphyllus (A. Rich.) Pax & K. Hoffm., HAJB 81915 (HAJB); Mabea sp., Bell et al. 94–30 (US); Macaranga grandifolia (Blanco) Merr., Wurdack D046 (US); Mallotus japonicus (L. f.) Müll. Arg., Wurdack D004 (US); Mallotus philippensis (Lam.) Müll. Arg., Wurdack D822 (US); Manniophyton africanum Müll. Arg., White 3366 (MO); Maprounea guianensis Aubl., Wurdack D332 (US); Melanolepis multiglandulosa Reichb. & Zoll., Motley 2493 (NY); Micrococca capensis (Baill.) Prain, Kurzweil 463/89 (K). Monadenium guentheri Pax, Chase 1007 (K); Monotaxis grandiflora Endl., Horn 2386 (DUKE); Moultonianthus leembruggianus (Boerl. & Koord.) Steenis, Challen et al. 3 (K); Nealchornea yapurensis Huber, Fine s.n. (US); Neoboutonia mannii Benth. & Hook. f., Fay 6701 (MO); Neoscortechinia kingii (Hook. f.) Pax & K. Hoffm., Chase 1265 (K) GenBank EU293936; Neoshirakia japonica (Siebold & Zucc.) Esser, Wurdack D005 (US); Omphalea diandra L., Chase 570 (K); Ostodes paniculata Blume, Chase 1267 (K); Pachystroma longifolium (Nees) I. M. Johnst., Prince s.n. (US); Pausandra martinii Baill., Berry 7466 (MO); Pedilanthus tithymaloides (L.) Poit., Wurdack D034 (US); Pera bicolor (Klotzsch) Müll. Arg., Gillespie 4300 (US), GenBank EU293937 Pimelodendron zoanthogyne J.J. Sm., Chase 1268 (K); Plukenetia volubilis L., Armbruster 38 (?); Pogonophora schomburgkiana Miers ex Benth., Larpin 1022 (US); Pseudosenefeldera inclinata (Müll. Arg.) Esser, Wurdack D385 (US); Ricinocarpos tuberculatus Müll. Arg., Chase 2164 (K); Ricinodendron heudelotii (Baill.) Heckel, Chase 1269 (K); Ricinus communis L., Wurdack D009 (US); Seidelia triandra (E. Mey.) Pax, Giess 13381 (MO); Senefelderopsis croizatii Steyerm., Berry s.n. (MO); Spathiostemon javensis Blume, Chase 1261 (K); Stillingia sylvatica L. subsp. tenuis (Small) D. J. Rogers, Wurdack D117 (US); Strophioblachia fimbricalyx Boerl., Chase 1270 (K); Sumbaviopsis albicans (Blume) J.J. Sm., Chase 1271 (K); Suregada glomerulata (Blume) Baill., Chase 1272 (K); Synadenium grantii Hook. f., Wurdack D259 (US); Syndyophyllum occidentale (Airy Shaw) Welzen, Aik et al. SAN-111913 (L); Tannodia cordifolia (Baill.) Baill., Ralimanana 293 (K); Tetrorchidium cf. macrophyllum Müll. Arg., Bell et al. 93–204 (US), GenBank EU293938; Tragia fallax Müll. Arg., Bell et al. 94–378 (US); Tragia urticifolia Michx., Wurdack D074 (US); Tragiella anomala (Prain) Pax & K. Hoffm., Mwasumbi et al. 16202 (MO); Trewia nudiflora L., Chase 1273 (K); Triadica sebifera (L.) Small, Wurdack D059 (US); Trigonostemon verrucosus J. J. Sm., Chase 1274 (K); Vernicia montana Lour., Chase 2105 (K). Euphroniaceae: Euphronia guianensis (R. H. Schomb.) H. Hallier, Mori 23699 (NY). Goupiaceae: Goupia glabra Aubl., Prevost 3031 (CAY). Humiriaceae: Humiria wurdackii Cuatrec., Wurdack s.n. (US); Sacoglottis gabonensis Urb., Stone 3283 (MO); Vantanea guianensis Aubl., Pennington 13855 (K). Hypericaceae: Cratoxylum formosum (Jack) Benth & Hook. f. ex Dyer, Chase 1218 (K). Irvingiaceae: Irvingia malayana Oliv., Simpson 2638 (K?); Klainedoxa gabonensis Pierre, Bradley et al. 1092 (MO). Ixonanthaceae: Cyrillopsis paraensis Kuhlm., Hentrich 68 (NY); Ochthocosmus longipedicellatus Steyerm. & Luteyn, Berry 6561 (MO). Lacistemataceae: Lacistema aggregatum Rusby, Pennington et al. 583 (K); Lozania pittieri (Blake) L. B. Sm., Pennington et al. 584 (K). Linaceae: Durandea pentagyna K. Schum., Takeuchi 7103 (MO); Linum perenne L., Chase 111 (NCU); Reinwardtia indica Dumort., Chase 230 (NCU). Malpighiaceae: Dicella nucifera Chodat, Anderson 13607 (MICH); Galphimia gracilis Bartl., Adelson s.n. (MICH), GenBank EU293939. Medusagynaceae: Medusagyne oppositifolia Baker, Fay s.n. (K) [Kew 1981–2059]. Ochnaceae s.s.: Elvasia calophyllea DC., Amaral s.n. (K?); Lophira lanceolata Tiegh., Schmidt 1902 (MO); Ochna multiflora DC., Chase 229 (NCU). Pandaceae: Galearia filiformis (Blume) Boerl., Chase 1334 (K); Microdesmis pierlotiana J. Léonard, Gereau et al. 5654 (MO); Panda oleosa Pierre., Schmidt et al. 2048 (MO). Phyllanthaceae: Amanoa strobilacea Müll. Arg., McPherson 16826 (MO); Apodiscus chevalieri Hutch., Schmidt et al. 2094 (MO); Aporusa frutescens Blume, Chase 1251 (K); Astrocasia neurocarpa (Müll. Arg.) I. M. Johnst. ex Standl., Wurdack D743 (US); Bischofia javanica Blume, Levin 2200 (SD), GenBank EU293940; Cleistanthus oblongifolius (Roxb.) Müll. Arg., Chase 1257 (K); Croizatia brevipetiolata (Secco) Dorr, Dorr et al. 8555 (US), GenBank EU293941; Didymocistus chrysadenius Kuhlm., Gillespie et al. 4805 (US); Discocarpus essequeboensis Klotzsch, Hoffman 996 (US); Gonatogyne brasiliensis (Baill.) Müll. Arg., Cordeiro & Esteves 1384 (K); Heywoodia lucens Sim, Saufferer & Muchai SS-1544 (US); Hieronyma oblonga (Tul.) Müll. Arg., Bell 94–252 (US); Hymenocardia acida Tul., Walters et al. 897 (MO); Lachnostylis bilocularis R. A. Dyer, Kurzweil 83/88 (K); Leptonema glabrum (Leandri) Leandri, McPherson & Rabenantoandro 18389 (MO); Maesobotrya vermeulenii (de Wild.) J. Léonard, Bradley et al. 1032 (MO); Poranthera huegelii Klotzsch, Spjut 7369 (US); Richeria grandis Vahl, Merello et al. 1714 (MO); Sauropus racemosus Beille, Soejarto et al. 10648 (NY); Spondianthus preussii Engl., Merello et al. 1661 (MO); Zimmermania ovata E.A. Bruce, Faden s.n. (US). Picrodendraceae: Androstachys johnsonii Prain, Chase 1904 (K); Hyaenanche globosa (Gaertn.) Lamb. & Vahl, Chase 1445 (K); Picrodendron baccatum Krug & Urb. ex Urban, Wurdack D050; Podocalyx loranthoides Klotzsch, Berry & Aymard 7226 (MO), GenBank EU293955 (partial sequence not used in phylogenetic analyses); Tetracoccus dioicus Parry, Levin 2202 (DUKE), GenBank EU293942. Podostemaceae: Marathrum c.f. oxycarpum Tul., Gabriel da Cachoeirc 9/1996 (AM); Podostemum ceratophyllum Michx., Horn & Wurdack s.n. (DUKE). Putranjivaceae: Drypetes capillipes Pax & K. Hoffm., Harris 4884 (K); Drypetes fallax Pax & K. Hoffm., Harris 4892 (K); Drypetes macrostigma J.J. Sm., Chase 1259 (K); Putranjiva roxburghii Wall., Wurdack D057 (US), GenBank EU293956 (partial sequence not used in phylogenetic analyses). Quiinaceae: Quiina pteridophylla (Radlk.) Pires, Pires s.n. (CPATU) Touroulia guianensis Aubl., Pires (CPATU). Rhizophoraceae s.s.: Bruguiera gymnorhiza Lam., Chase 12838 (K); Carallia brachiata (Lour.) Merr., Chase 2151 (K); Crossostylis multiflora Brongn. & Gris ex Pancher & Sebert, Lowry 5686 (MO); Paradrypetes subintegrifolia G. A. Levin, Acevedo-Rdgz. & Cedeño 7560 (US); Cassipourea lanceolata Tul., Schatz 3689 (MO). Salicaceae s.l.: Abatia parviflora Ruiz & Pav., Pennington 676 (K); Casearia nitida Jacq., FTG 72496; Flacourtia jangomas Steud., Chase 2150 (K); Idesia polycarpa Maxim., Wurdack D22 (US); Poliothyrsis sinensis Oliver, Wurdack D029 (US); Scyphostegia borneensis Stapf, Beaman 911 (BH). Trigoniaceae: Trigoniastrum hypoleucum Miq., Og B128 (L). Violaceae: Hybanthus concolor Spreng., Wurdack D148 (US); Hymenanthera alpina Oliv., Chase 501 (K); Rinorea bengalensis Kuntze, Chase 2148 (K).
(ii) Other related rosids (Celastrales, Oxalidales + Huaceae)
Brunelliaceae: Brunellia acutangula Humb. & Bonpl., Stergios 20646 (US), GenBank EU293943. Celastraceae: Brexia madagascariensis Thouars, Wurdack D764 (US); Maytenus senegalensis (Lam.) Exell, Collenette 4/93 (K); Plagiopteron suaveolens Griff., Chase 1335 (K); Siphonodon celastrineus Griff., Chase 2097 (K); Stackhousia minima Hook. f., Molloy s.n. (CHR); Tripterygium regelii Sprague & Takeda, Chase 1003 (K). Cephalotaceae: Cephalotus folicularis Labill., Chase 147 (NCU). Connaraceae: Rourea minor Leenh., Chase 1221 (K). Cunoniaceae: Eucryphia milliganii Hook. f., Chase 2528 (K). Elaeocarpaceae: Crinodendron hookerianum Gay, Chase 909 (K); Sloanea sp., Chase 343 (NCU). Huaceae: Afrostyrax sp., Cheek 5007 (K). Lepidobotryaceae: Ruptiliocarpon caracolito Hammel & Zamora, Hammel 19102 (MO). Oxalidaceae: Averrhoa carambola L., Chase 214 (NCU). Parnassiaceae: Parnassia grandifolia DC., Wurdack D795 (US), GenBank EU293944.
Intron absent
(i) Malpighiales
Elatinaceae: Elatine hexandra DC., Chase 2978 (K), GenBank EU293945. Euphorbiaceae s.s.: Cnidoscolus urens (L.) Arthur var. stimulosus (Michx.) Govaerts, Wurdack D002 (US); Elateriospermum tapos Blume, Soepadmo & Suhaimi s193 (NY), GenBank EU293946; Glycydendron amazonicum Ducke, Gillespie et al. 4546 (US); Glycydendron cf. amazonicum Ducke, Mori et al. 23265 (US); Hevea sp. [cf. pauciflora (Spruce ex Benth.) Müll. Arg.], Gillespie 4272 (US), GenBankEU293947; Manihot esculenta subsp. esculenta Crantz, EU117376; anihot foetida Pohl, Olsen s.n.; Manihot grahamii Hook., Wurdack s.n. (US); Manihot pauciflora Brandegee, Villasenor 1248 (NY); Manihot walkerae Croizat, Olsen s.n.; Micrandra inundata P. E. Berry & A. Wiedenhoeft, Berry 6350 (MO). Lophopyxidaceae: Lophopyxis maingayi Hook. f., Adelbai P-10203 (US), GenBank EU293948. Malesherbiaceae: Malesherbia weberbaueri Gilg, Weigend 2000/365 (NY), GenBank EU293949. Passifloraceae s.s.: Paropsia madagascariensis (Baill.) H. Perrier, Zyhra 949 (WIS); Passiflora coccinea Aubl., Chase 2475 (K), GenBank EU293950. Phyllanthaceae: Breynia disticha J. R. Forst. & G. Forst. cv. ‘Rosea Picta’, Wurdack D012 (US); Flueggea virosa (Roxb. ex. Willd.) Voigt subsp. virosa, Wurdack D101 (US); Glochidion puberum (L.) Hutch., Wurdack D003 (US); Leptopus colchicus (Fisch. & C. A. Mey. ex Boiss.) Pojark., Wurdack D778 (US), GenBank EU293951; Leptopus esquirolii (H. Lév.) P. T. Li, 1980 Sino-American Bot. Exped. 1678 (US); Margaritaria tetracocca (Baill.) G. L. Webster, Wurdack D044 (US); Phyllanthus epiphyllanthus L., Wurdack D056 (US), GenBank EU293952; Phyllanthus fluitans Benth. ex Müll. Arg., Wurdack D761 (US); Phyllanthus juglandifolius Willd., Wurdack D759 (US); Phyllanthus nutans Sw., Wurdack D762 (US); Reverchonia arenaria A. Gray, Atwood 17245 (NY); Savia bahamensis Britton, Wurdack D048 (US). Picrodendraceae: Austrobuxus megacarpus P. I. Forster, Forster 21239 (BRI), GenBank EU293953; Choriceras majus Airy Shaw, Forster 21230 (BRI); Dissiliaria muelleri Baill., Forster 14629 (BRI); Whyanbeelia terraereginae Airy Shaw & B. Hyland, Forster 21231 (BRI). Turneraceae: Tricliceras longipedunculatum (Mast.) R. Fern., Kanji et al. 304 (NY); Turnera ulmifolia L., Wurdack s.n. (US), GenBank EU293954.
(ii) Other related rosids (Celastrales, Oxalidales + Huaceae): none noted.
References
- APG . An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Bot J Linn Soc. 2003;141:399–436. [Google Scholar]
- Asakura Y, Barkan A. Arabidopsis orthologs of maize chloroplast splicing factors promote splicing of orthologous and species-specific group II introns. Plant Physiol. 2006;142:1656–1663. doi: 10.1104/pp.106.088096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. Intron splicing in plant organelles. In: Daniell H, Chase CD, editors. Molecular biology and biotechnology of plant organelles: chloroplast and mitochondria. Springer; Netherlands: 2004. pp. 295–322. [Google Scholar]
- Bausher MG, Singh ND, Lee SB, Jansen RK, Daniell H. The complete chloroplast genome sequence of Citrus sinensis (L.) Osbeck var ‘Ridge Pineapple’: organization and phylogenetic relationships to other angiosperms. BMC Plant Biol. 2006;6:21. doi: 10.1186/1471-2229-6-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry PE, Hipp AL, Wurdack KJ, van Ee B, Riina R. Molecular phylogenetics of the giant genus Croton and tribe Crotoneae (Euphorbiaceae sensu stricto) using ITS and trnL-trnF DNA sequence data. Am J Bot. 2005;92:1520–1534. doi: 10.3732/ajb.92.9.1520. [DOI] [PubMed] [Google Scholar]
- Bonen L, Vogel J. The ins and outs of group II introns. Trends Genet. 2001;17:322–331. doi: 10.1016/s0168-9525(01)02324-1. [DOI] [PubMed] [Google Scholar]
- Bowe LM, dePamphilis CW. Effects of RNA editing and gene processing on phylogenetic reconstruction. Mol Biol Evol. 1996;13:1159–1166. doi: 10.1093/oxfordjournals.molbev.a025680. [DOI] [PubMed] [Google Scholar]
- Corneille S, Lutz K, Maliga P. Conservation of RNA editing between rice and maize plastids: are most editing events dispensable? Mol Gen Genet. 2000;264:419–424. doi: 10.1007/s004380000295. [DOI] [PubMed] [Google Scholar]
- Daniell H. Molecular strategies for gene containment in transgenic crops. Nat Biotechnol. 2002;20:581–586. doi: 10.1038/nbt0602-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H. Production of biopharmaceuticals and vaccines in plants via the chloroplast genome. Biotechnol J. 2006;1:1071–1079. doi: 10.1002/biot.200600145. [DOI] [PubMed] [Google Scholar]
- Daniell H. Transgene containment by maternal inheritance: effective or elusive? Proc Natl Acad Sci USA. 2007;104:6879–6880. doi: 10.1073/pnas.0702219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Datta R, Varma S, Gray S, Lee S-B. Containment of herbicide resistance through genetic engineering of the chloroplast genome. Nat Biotechnol. 1998;16:345–348. doi: 10.1038/nbt0498-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Kumar S, Dufourmantel N. Breakthrough in chloroplast genetic engineering of agronomically important crops. Trends Biotechnol. 2005;5:238–245. doi: 10.1016/j.tibtech.2005.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Lee S-B, Grevich J, Saski C, Quesada-Vargas T, Guda C, Tomkins J, Jansen RK. Complete chloroplast genome sequences of Solanum bulbocastanum, Solanum lycopersicum and comparative analyses with other Solanaceae genomes. Theor Appl Genet. 2006;112:1503–1518. doi: 10.1007/s00122-006-0254-x. [DOI] [PubMed] [Google Scholar]
- Davis CC, Chase MW. Elatinaceae are sister to Malpighiaceae; Peridiscaceae belong to Saxifragales. Am J Bot. 2004;91:262–273. doi: 10.3732/ajb.91.2.262. [DOI] [PubMed] [Google Scholar]
- Davis CC, Webb CO, Wurdack KJ, Jaramillo CA, Donoghue MJ. Explosive radiation of Malpighiales supports a mid-cretaceous origin of modern tropical rain forests. Am Nat. 2005;165:E36–E65. doi: 10.1086/428296. [DOI] [PubMed] [Google Scholar]
- Davis CC, Wurdack KJ. Host-to-parasite gene transfer in flowering plants: phylogenetic evidence from Malpighiales. Science. 2004;305:676–678. doi: 10.1126/science.1100671. [DOI] [PubMed] [Google Scholar]
- DeCosa B, Moar W, Lee S-B, Miller M, Daniell H. Overexpression of the Bt cry2Aa2 operon in chloroplasts leads to formation of insecticidal crystals. Nat Biotechnol. 2001;9:71–74. doi: 10.1038/83559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGray G, Rajasekaran K, Smith F, Sanford J, Daniell H. Expression of an antimicrobial peptide via the chloroplast genome to control phytopathogenic bacteria and fungi. Plant Physiol. 2001;127:852–862. [PMC free article] [PubMed] [Google Scholar]
- Deshpande NN, Hollingsworth M, Herrin DL. The atpF group-II intron-containing gene from spinach chloroplasts is not spliced in transgenic Chlamydomonas chloroplasts. Curr Genet. 1995;28:122–127. doi: 10.1007/BF00315777. [DOI] [PubMed] [Google Scholar]
- Dickau R, Ranere AJ, Cooke RG. Starch grain evidence for the preceramic dispersals of maize and root crops into tropical dry and humid forests of Panama. Proc Natl Acad Sci USA. 2007;104:3651–3656. doi: 10.1073/pnas.0611605104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie SR, Katz-Downie DS, Wolfe KH, Calie PJ, Palmer JD. Structure and evolution of the largest chloroplast gene (ORF2280): internal plasticity and multiple gene loss during angiosperm evolution. Curr Genet. 1994a;25:367–378. doi: 10.1007/BF00351492. [DOI] [PubMed] [Google Scholar]
- Downie SR, Llanas E, Katz-Downie DS. Multiple independent losses of the rpoC1 intron in angiosperm chloroplast DNAs. Syst Bot. 1994b;21:135–151. [Google Scholar]
- Downie SR, Olmstead RG, Zurawski G, Soltis DE, Soltis PS, Watson JC, Palmer JD. Six independent losses of the chloroplast DNA rpl2 intron in dicotyledons: molecular and phylogenetic implications. Evolution. 1991;45:1245–1259. doi: 10.1111/j.1558-5646.1991.tb04390.x. [DOI] [PubMed] [Google Scholar]
- Downie SR, Palmer JD. Use of chloroplast DNA rearrangements in reconstructing plant phylogeny. In: Soltis PS, Soltis DE, Doyle JJ, editors. Molecular systematics of plants. Chapman and Hall; New York: 1992. pp. 14–35. [Google Scholar]
- Dufourmantel N, Dubald M, Matringe M, Canard H, Garcon F, Job C, Kay E, Wisniewski JP, Ferullo JM, Pelissier B, Sailland A, Tissot G. Generation and characterization of soybean and marker-free tobacco plastid transformants overexpressing a bacterial 4-hydroxyphenylpyruvate dioxygenase which provides strong herbicide tolerance. Plant Biotechnol J. 2007;5:118–133. doi: 10.1111/j.1467-7652.2006.00226.x. [DOI] [PubMed] [Google Scholar]
- El-Sharkawy MA. Cassava biology and physiology. Plant Mol Biol. 2004;56:481–501. doi: 10.1007/s11103-005-2270-7. [DOI] [PubMed] [Google Scholar]
- Ewing B, Green P. Base-calling of automated sequencer traces using phred. II. Error probabilities. Gen Res. 1998;8:186–194. [PubMed] [Google Scholar]
- Fregene M. Marking progress: collaboration to improve cassava. In: Kinley D, editor. Synergies in science. Consultative Group on International Agricultural Research; Washington: 2000. pp. 6–7. [Google Scholar]
- Fregene M, Angel F, Gómez R, Rodríguez F, Chavarriaga P, Roca W, Tohme J, Bonierbale M. A molecular genetic map of cassava (Manihot esculenta Crantz) Theor Appl Genet. 1997;95:431–441. [Google Scholar]
- Grevich JJ, Daniell H. Chloroplast genetic engineering: recent advances and future perspectives. Crit Rev Plant Sci. 2005;24:83–108. [Google Scholar]
- Hagemann R. The Sexual inheritance of plant organelles. In: Daniell H, Chase C, editors. Molecular biology and biotechnology of plant organelles: chloroplasts and mitochondria. Springer; Netherlands: 2004. pp. 93–113. [Google Scholar]
- Hansen AK, Gilbert LE, Simpson BB, Downie SR, Cervi AC, Jansen RK. Phylogenetic relationships and chromosome number evolution in Passiflora. Syst Bot. 2006;31:138–150. [Google Scholar]
- Hillocks RJ. Cassava in Africa. In: Hillocks RJ, Thresh JM, Bellotti AC, editors. Cassava: biology, production and utilization. CABI Publishing; New York: 2002. pp. 41–54. [Google Scholar]
- Hoffmann P, Kathriarachchi H, Wurdack KJ. A phylogenetic classification of Phyllanthaceae (Malpighiales; Euphorbiaceae sensu lato) Kew Bull. 2006;61:37–53. [Google Scholar]
- Huelsenbeck JP, Ronquist FR. MrBayes: Bayesian inference of phylogenetic trees. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [MrBayes available at http://mrbayes.scs.fsu.edu/] [DOI] [PubMed]
- Itchoda N, Nishizawa S, Nagano H, Kubo T, Mikami T. The sugar beet mitochondrial nad4 gene: an intron loss and its phylogenetic implication in the Caryophyllales. Theor Appl Genet. 2002;104:209–213. doi: 10.1007/s001220100744. [DOI] [PubMed] [Google Scholar]
- Jansen RK, Cai Z, Raubeson LA, Daniell H, dePamphilis CW, Leebens-Mack J, Müller KF, Guisinger-Bellian M, Haberle RC, Hansen AK, Chumley TW, Lee S-B, Peery R, McNeal J, Kuehl JV, Boore JL. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA. 2007;104:19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins BD, Kulhanek DJ, Barkan A. Nuclear mutations that block group II RNA splicing in maize chloroplasts reveal several intron classes with distinct requirements for splicing factors. Plant Cell. 1997;9:283–296. doi: 10.1105/tpc.9.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahlau S, Aspinall S, Gray JC, Bock R. Sequence of the tomato chloroplast DNA and evolutionary comparison of solanaceous plastid genomes. J Mol Evol. 2006;63:194–207. doi: 10.1007/s00239-005-0254-5. [DOI] [PubMed] [Google Scholar]
- Kanamoto H, Yamashita A, Asao H, Okumura S, Takase H, Hattori M, Yokota A, Tomizawa K. Efficient and stable transformation of Lactuca sativa L. cv. Cisco (lettuce) plastids. Transgenic Res. 2006;15:205–217. doi: 10.1007/s11248-005-3997-2. [DOI] [PubMed] [Google Scholar]
- Kathriarachchi H, Hoffmann P, Samuel R, Wurdack KJ, Chase MW. Molecular phylogenetics of Phyllanthaceae inferred from five genes (plastid atpB, matK, 3′ ndhF, and nuclear PHYC) Mol Phylog Evol. 2005;36:112–134. doi: 10.1016/j.ympev.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Knauf U, Hachtel W. The genes encoding subunits of ATP synthase are conserved in the reduced plastid genome of the heterotrophic alga Prototheca wickerhamii. Mol Genet Genomics. 2002;267:492–497. doi: 10.1007/s00438-002-0681-6. [DOI] [PubMed] [Google Scholar]
- Kota M, Daniell H, Varma S, Garczynski SF, Gould F, Moar WJ. Overexpression of the Bacillus thuringiensis (Bt) Cry2Aa2 protein in chloroplasts confers resistance to plants against susceptible and Bt-resistant insects. Proc Natl Acad Sci USA. 1999;96:1840–1845. doi: 10.1073/pnas.96.5.1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Dhingra A, Daniell H. Plastid expressed betaine aldehyde dehydrogenase gene in carrot cultured cells, roots, and leaves confer enhanced salt tolerance. Plant Physiol. 2004a;136:2843–2854. doi: 10.1104/pp.104.045187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Dhingra A, Daniell H. Stable transformation of the cotton plastid genome and maternal inheritance of transgenes. Plant Mol Biol. 2004b;56:203–216. doi: 10.1007/s11103-004-2907-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SB, Kaittanis C, Jansen RK, Hostetler JB, Tallon LJ, Town CD, Daniell H. The complete chloroplast genome sequence of Gossypium hirsutum: organization and phylogenetic relationships to other angiosperms. BMC Genomics. 2006;7:61. doi: 10.1186/1471-2164-7-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S-B, Kwon HB, Kwon SJ, Park SC, Jeong MJ, Han SE, Byun MO, Daniell H. Accumulation of trehalose within transgenic chloroplasts confers drought tolerance. Mol Breed. 2003;11:1–13. [Google Scholar]
- Maliga P. Plastid transformation in higher plants. Annu Rev Plant Biol. 2004;55:289–313. doi: 10.1146/annurev.arplant.55.031903.141633. [DOI] [PubMed] [Google Scholar]
- Mba REC, Stephenson P, Edwards K, Melzer S, Nkumbira J, Gullberg U, Apel K, Gale M, Tohme J, Fregene M. Simple sequence repeat (SSR) markers survey of the cassava (Manihot esculenta Crantz) genome: towards a SSR-based molecular genetic map of cassava. Theor Appl Genet. 2001;102:21–31. [Google Scholar]
- McPherson MA, Fay MF, Chase MW, Graham SW. Parallel loss of a slowly evolving intron from two closely related families in Asparagales. Syst Bot. 2004;29:296–307. [Google Scholar]
- Millen RS, Olmstead RG, Adams KL, Palmer JD, Lao NT, Heggie L, Kavanagh TA, Hibberd JM, Gray JC, Morden CW, Calie PJ, Jermiin LS, Wolfe KH. Many parallel losses of infA from chloroplast DNA during angiosperm evolution with multiple independent transfers to the nucleus. Plant Cell. 2001;13:645–658. doi: 10.1105/tpc.13.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagi T, Kapoor S, Sugita M, Sugiura M. Transcript analysis of the tobacco plastid operon rps2/atpI/H/F/A reveals the existence of a non-consensus type II (NCII) promoter upstream of the atpI coding sequence. Mol Gen Genet. 1998;257:299–307. doi: 10.1007/s004380050651. [DOI] [PubMed] [Google Scholar]
- Okezie BO, Kosikowski FV. Cassava as a food. Crit Rev Food Sci Nutr. 1982;17:259–275. doi: 10.1080/10408398209527349. [DOI] [PubMed] [Google Scholar]
- Okumura S, Sawada M, Park YW, Hayashi T, Shimamura M, Takase H, Tomizawa K. Transformation of poplar (Populus alba) plastids and expression of foreign proteins in tree chloroplasts. Transgenic Res. 2006;15:637–646. doi: 10.1007/s11248-006-9009-3. [DOI] [PubMed] [Google Scholar]
- Olsen KM, Schaal BA. Evidence on the origin of cassava: phylogeography of Manihot esculenta. Proc Natl Acad Sci USA. 1999;96:5586–5591. doi: 10.1073/pnas.96.10.5586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen KM, Schaal BA. Microsatellite variation in cassava (Manihot esculenta, Euphorbiaceae) and its wild relatives: further evidence for a southern Amazonian origin of domestication. Am J Bot. 2001;88:131–142. [PubMed] [Google Scholar]
- Ostersetzer O, Cooke AM, Watkins KP, Barkan A. CRS1, a chloroplast group II intron splicing factor, promotes intron folding through specific interactions with two intron domains. Plant Cell. 2005;17:241–255. doi: 10.1105/tpc.104.027516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagel M, Meade A. Bayesian analysis of correlated evolution of discrete characters by reversible-jump markov chain Monte Carlo. Am Nat. 2006;167:808–825. doi: 10.1086/503444. [BayesTraits available at http://www.evolution.reading.ac.uk/BayesTraits.html] [DOI] [PubMed]
- Plader W, Yukawa Y, Sugiura M, Malepszy S. The complete structure of the cucumber (Cucumis sativus L.) chloroplast genome: its composition and comparative analysis. Cell Mol Biol Lett. 2007;12:584–594. doi: 10.2478/s11658-007-0029-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posada D, Crandall KA. MODELTEST: testing the model of DNA substitution. Bioinformatics. 1998;14:817–818. doi: 10.1093/bioinformatics/14.9.817. [MODELTEST available at http://darwin.uvigo.es/software/modeltest.html] [DOI] [PubMed]
- Quesada-Vargas T, Ruiz ON, Daniell H. Characterization of heterologous multigene operons in transgenic chloroplasts: transcription, processing, and translation. Plant Physiol. 2005;138:1746–1762. doi: 10.1104/pp.105.063040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raubeson LA, Peery R, Chumley TW, Dziubek C, Fourcade HM, Boore JL, Jansen RK. Comparative chloroplast genomics: analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus. BMC Genomics. 2007;8:174. doi: 10.1186/1471-2164-8-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raven P, Fauquet C, Swaminathan MS, Borlaug N, Samper C. Where next for genome sequencing? Science. 2006;311:468. doi: 10.1126/science.311.5760.468b. [DOI] [PubMed] [Google Scholar]
- Ruf S, Hermann M, Berger I, Carrer H, Bock R. Stable genetic transformation of tomato plastids and expression of a foreign protein in fruit. Nat Biotechnol. 2001;19:870–875. doi: 10.1038/nbt0901-870. [DOI] [PubMed] [Google Scholar]
- Ruhlman T, Lee SB, Jansen RK, Hostetler JB, Tallon LJ, Town CD, Daniell H. Complete plastid genome sequence of Daucus carota: implications for biotechnology and phylogeny of angiosperms. BMC Genomics. 2006;7:222. doi: 10.1186/1471-2164-7-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhlman T, Ahangari R, Devine A, Samsam M, Daniell H. Expression of cholera toxin B-proinsulin fusion protein in lettuce and tobacco chloroplasts—oral administration protects against development of insulitis in non-obese diabetic mice. Plant Biotech J. 2007;5:495–510. doi: 10.1111/j.1467-7652.2007.00259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz ON, Daniell H. Engineering the cytoplasmic male sterility via the chloroplast genome. Plant Physiol. 2005;138:1232–1246. doi: 10.1104/pp.104.057729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samson N, Bausher MG, Lee S-B, Jansen RK, Daniell H. The complete nucleotide sequence of the coffee (Coffea arabica L.) chloroplast genome: organization and implications for biotechnology and phylogenetic relationships among angiosperms. Plant Biotech J. 2007;5:339–353. doi: 10.1111/j.1467-7652.2007.00245.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saski C, Lee S-B, Daniell H, Wood TC, Tomkins J, Kim HG, Jansen RK. Complete chloroplast genome sequence of Glycine max and comparative analyses with other legume genomes. Plant Mol Biol. 2005;59:309–322. doi: 10.1007/s11103-005-8882-0. [DOI] [PubMed] [Google Scholar]
- Sato S, Nakamura Y, Kaneko T, Asamizu E, Tabata S. Complete structure of the chloroplast genome of Arabidopsis thaliana. DNA Res. 1999;6:283–290. doi: 10.1093/dnares/6.5.283. [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Maier RM, Alcaraz JP, Cottet A, Herrmann RG, Mache R. The plastid chromosome of spinach (Spinacia oleracea) complete nucleotide sequence and gene organization. Plant Mol Biol. 2001;45:307–315. doi: 10.1023/a:1006478403810. [DOI] [PubMed] [Google Scholar]
- Schmitz-Linneweber C, Regel R, Du TG, Hupfer H, Herrmann RG, Maier RM. The plastid chromosome of Atropa belladonna and its comparison with that of Nicotiana tabacum: the role of RNA editing in generating divergence in the process of plant speciation. Mol Biol Evol. 2002;19:1602–1612. doi: 10.1093/oxfordjournals.molbev.a004222. [DOI] [PubMed] [Google Scholar]
- Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K, Ohto C, Torazawa K, Meng BY, Sugita M, Deno H, Kamogashira T, Yamada K, Kusuda J, Takaiwa F, Kato A, Tohdoh N, Shimada H, Sugiura M. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986;5:2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorov VA, Kasten D, Pang SZ, Hajdukiewicz PT, Staub JM, Nehra NS. Stable chloroplast transformation in potato: use of green fluorescent protein as a plastid marker. Plant J. 1999;19:209–216. doi: 10.1046/j.1365-313x.1999.00508.x. [DOI] [PubMed] [Google Scholar]
- Sikdar SR, Serino G, Chaudhuri S, Maliga P. Plastid transformation in Arabidopsis thaliana. Plant Cell Rep. 1998;18:245–247. [Google Scholar]
- Sikes DS, Lewis PO. PAUPRat: PAUP* implementation of the parsimony ratchet. 2001 [PAUPRat available at http://www.ucalgary.ca/~dsikes/software2.htm]
- Simon D, Fewer D, Friedl T, Bhattacharya D. Phylogeny and self-splicing ability of the plastid tRNA-Leu group I intron. J Mol Evol. 2003;57:710–720. doi: 10.1007/s00239-003-2533-3. [DOI] [PubMed] [Google Scholar]
- Stahl DJ, Rodermel SR, Bogorad L, Subramanian AR. Co-transcription pattern of an introgressed operon in the maize chloroplast genome comprising four ATP synthase subunit genes and the ribosomal rps2. Plant Mol Biol. 1993;21:1069–1076. doi: 10.1007/BF00023603. [DOI] [PubMed] [Google Scholar]
- Swofford DL. Phylogenetic analysis using parsimony (*and other methods), vers. 4. Sinauer Associates; Sunderland, MA: 2003. PAUP*. [Google Scholar]
- Tillich M, Lehwark P, Morton BR, Maier UG. The evolution of chloroplast RNA editing. Mol Biol Evol. 2006;23:1912–1921. doi: 10.1093/molbev/msl054. [DOI] [PubMed] [Google Scholar]
- Tokuoka T. Molecular phylogenetic analysis of Euphorbiaceae sensu stricto based on plastid and nuclear DNA sequences and ovule and seed character evolution. J Plant Res. 2007;120:511–522. doi: 10.1007/s10265-007-0090-3. [DOI] [PubMed] [Google Scholar]
- Turmel M, Lemieux C, Burger G, Lang BF, Otis C, Plante I, Gray MW. The complete mitochondrial DNA sequences of Nephroselmis olivacea and Pedinomonas minor: two radically different evolutionary patterns within green algae. Plant Cell. 1999;11:1717–1730. doi: 10.1105/tpc.11.9.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmel M, Otis C, Lemieux C. The complete chloroplast DNA sequences of the charophycean green algae Staurastrum and Zygnema reveal that the chloroplast genome underwent extensive changes during the evolution of the Zygnematales. BMC Biol. 2005;3:22. doi: 10.1186/1741-7007-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turmel M, Otis C, Lemieux C. The chloroplast genome sequence of Chara vulgaris sheds new light into the closest green algal relatives of land plants. Mol Biol Evol. 2006;23:1324–1338. doi: 10.1093/molbev/msk018. [DOI] [PubMed] [Google Scholar]
- Ugent D, Pozorski S, Pozorski T. Archaeological manioc (Manihot) from coastal Peru. Econ Bot. 1986;40:78–102. [Google Scholar]
- Vogel J, Börner T. Lariat formation and a hydrolytic pathway in plant chloroplast group II intron splicing. EMBO J. 2002;21:3794–3803. doi: 10.1093/emboj/cdf359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J, Börner T, Hess WR. Comparative analysis of splicing of the complete set of chloroplast group II introns in three higher plant mutants. Nucleic Acids Res. 1999;27:3866–3874. doi: 10.1093/nar/27.19.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurdack KJ, Hoffmann P, Chase MW. Molecular phylogenetic analysis of uniovulate Euphorbiaceae (Euphorbiaceae sensu stricto) using plastid rbcL and trnLF sequences. Am J Bot. 2005;92:1397–1420. doi: 10.3732/ajb.92.8.1397. [DOI] [PubMed] [Google Scholar]
- Wurdack KJ, Hoffmann P, Samuel R, De Bruijn A, Van der Bank M, Chase MW. Molecular phylogenetic analysis of Phyllanthaceae (Phyllanthoideae pro parte, Euphorbiaceae sensu lato) using plastid rbcL DNA sequences. Amer J Bot. 2004;91:1882–1900. doi: 10.3732/ajb.91.11.1882. [DOI] [PubMed] [Google Scholar]
- Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOGMA available at http://dogma.ccbb.utexas.edu/] [DOI] [PubMed]
- Xu MQ, Kaathe S, Goodrich BH, Nierwicki BS, Shub D. Bacterial origin of a chloroplast intron: conserved self splicing group I introns in cyanobacteria. Science. 1990;250:1566–1569. doi: 10.1126/science.2125747. [DOI] [PubMed] [Google Scholar]
- Zubkot MK, Zubkot EI, van Zuilen K, Meyer P, Day A. Stable transformation of petunia plastids. Transgenic Res. 2004;13:523–530. doi: 10.1007/s11248-004-2374-x. [DOI] [PubMed] [Google Scholar]
- Zwickl DJ. Genetic algorithm approaches for the phylogenetic analysis of large biological sequence datasets under the maximum likelihood criterion. Dissertation, the university of Texas at Austin. 2006 [GARLI available at http://www.bio.utexas.edu/faculty/antisense/garli/Garli.html]


