Summary
Receptors carrying immunoreceptor tyrosine-based inhibition motifs (ITIM) in their cytoplasmic tail control a vast array of cellular responses, ranging from autoimmunity, allergy, phagocytosis of red blood cells, graft versus host disease, to even neuronal plasticity in the brain. The inhibitory function of many receptors has been deduced on the basis of cytoplasmic ITIM sequences. Tight regulation of natural killer (NK) cell cytotoxicity and cytokine production by inhibitory receptors specific for MHC class I molecules has served as a model system to study the negative signalling pathway triggered by an ITIM-containing receptor in the physiological context of NK–target cell interactions. Advances in our understanding of the molecular details of inhibitory signalling in NK cells have provided a conceptual framework to address how ITIM-mediated regulation controls cellular reactivity in diverse cell types.
Keywords: inhibition, innate immunity, natural killer cell, signalling, tyrosine phosphatase
Introduction
This review covers the regulation of cellular activation by cell surface receptors that carry immunoreceptor tyrosine-based inhibition motifs (ITIM) in their cytoplasmic tail, with an emphasis on the mechanism of inhibition. The sequence of the ITIM was defined as V/I/LxYxxL/V by sequence alignment, phosphorylated peptide binding specificity, and functional tests (1–4). Since then, ITIM-containing receptors (ITIM-R) have grown into a large family with expression in many different cell types, and inhibitory functions for the control of various cell activation pathways. This type of negative regulation may be an ancestral system, as many ITIM-Rs are expressed in bony fish (5). In general, inhibition exerted by ITIM-Rs is only local and transient. It does not induce a cell-wide or sustained non-responsiveness, but abrogates activation signals when and where they occur. NK cells are well suited as a model system for biochemical and functional analysis of the inhibitory signals delivered by ITIM-R, which allow NK cells to kill infected cells but spare healthy ones. NK cells express several ITIM-Rs, for which the ligands on target cells are well defined. NK cells selectively kill target cells that have lost expression of MHC class I. Early observation of this phenomenon led to the “missing self” hypothesis (6). Target cell killing by NK cells is not occurring by default in the absence of inhibition, but requires signalling by activation receptors upon recognition of their specific ligands on target cells. An inhibitory receptor specific for MHC class I was first identified in mice, as Ly49A (7). Similar negative regulation operates in human NK cells, which express several inhibitory receptors specific for different HLA molecules. These include NKG2/CD94 with specificity for HLA-E and killer-cell immunoglobulin-like receptors (KIR) that are specific for allotypes of HLA-C and HLA-B (8, 9).
The primary questions about the role of ITIM-Rs were: What are the ligands of ITIM-Rs? How is the inhibitory signal transmitted? What specific functions are regulated in vivo, and how? While genetic approaches using mouse knockouts have been very informative in this respect, for many of the receptors within the ITIM family the negative signalling pathways that control specific cellular functions have not been delineated. Progress in unravelling the inhibitory mechanism that operates in NK cells has provided some insight into how ITIM-Rs function.
ITIM: Early history
The ITIM was initially defined as a tyrosine-containing sequence in the cytoplasmic tail of FcγRIIb, which caused inhibition of B-cell activation and influx of extracellular Ca2+ when FcγRIIb is co-engaged with the BCR (10, 11). The sequence was further mapped to within 13 amino acids that include tyrosine 309 (12). It was identified as ITIM, by analogy with the immunoreceptor tyrosine-based activation motif (ITAM), which is found in several signalling adaptor proteins associated with receptors such as the TCR, BCR, and Fc receptors. The tyrosine-phosphorylated cytoplasmic tail of FcγRIIb binds to protein tyrosine phosphatase SHP-1 (13) and to the inositol phosphatase SHIP (14). Although phosphorylated FcγRIIb binds to both phosphatases, only SHIP is required for inhibition. SHIP dephosphorylates PI-3,4,5-P3 into PI-4,5-P2, thereby preventing membrane-association of Tec-family tyrosine kinases and sustained activation of PLC-γ (15, 16).
Functional characterization of an inhibitory KIR revealed binding of SHP-1 to cytoplasmic phospho-tyrosines and led to the definition of the ITIM sequence as V/I/LxYxxL/V (1). The requirement for a specific amino acid at position −2, relative to the tyrosine, was unusual. Sequences of peptides bound to the isolated SH2 domains of tyrosine phosphatases SHP-1 and SHP-2 from large peptide libraries were in agreement with the ITIM sequence (4, 17). SHP-1 and SHP-2 bind to the ITIMs of both KIR and mouse Ly49 inhibitory receptors (18). Selective binding of the SHIP SH2 domain to the FcγRIIb ITIM, and of SHP-1/2 to the KIR ITIM, is determined by a leucine at position +2 in FcγRIIb, and a leucine/isoleucine at position −2 in the KIR ITIM (19, 20). Whereas FcγRIIb requires SHIP but not SHP-1 for inhibition, KIR inhibition is mediated by SHP-1 and not SHIP. Co-ligation of BCR and FcγRIIb in the chicken DT40 B cell line resulted in inhibition in the absence of SHP-1, but not SHIP (21). Conversely, co-ligation of BCR and a chimeric FcγRIIb carrying a KIR cytoplasmic tail resulted in inhibition in the absence of SHIP but not SHP-1. Furthermore, the reverse chimeras were expressed in NK cells and inhibition of lysis by the HLA class I ligand of KIR on target cells was tested (22). Inhibition by the KIR cytoplasmic tail was prevented by a dominant-negative form of SHP-1, but not SHIP. Conversely, a dominant-negative form of SHIP, but not SHP-1, prevented inhibition by the KIR/FcγRIIb chimeric receptor expressing the FcγRIIb cytoplasmic tail. Therefore, SHIP is dispensable for inhibition by KIR, while SHP-1 is dispensable for inhibition by FcγRIIb.
Amino acid substitutions in the two ITIMs present in the cytoplasmic tail of KIR provided functional evidence for the role of both ITIMs in transmitting inhibitory signals (3, 23). The first, N-terminal and membrane-proximal, ITIM is not only essential, but also sufficient for inhibition. Phosphorylation of the first ITIM alone results in the preferential binding to SHP-2 (24). The first ITIM of receptors NKG2A (25) and Siglec-7/9 (26) are also dominant in their inhibitory function. However, due to the opposite transmembrane orientation of lectin-like type II proteins, the first, N-terminal, ITIM in NKG2A is membrane-distal. The lectin-like inhibitory Ly49 receptors carry a single ITIM. It is likely that assembly of Ly49 into a homodimer provides two ITIMs for optimal inhibitory function.
Recruitment of the tyrosine phosphatases SHP-1 and SHP-2 through binding of their tandem SH2 domains releases the catalytic domain from an inhibitory, intramolecular interaction with the N-terminal SH2 domain (27). Thus, recruitment and activation of tyrosine phosphatases by phosphorylated ITIMs provided a potentially simple explanation for inhibition. By dephosphorylation of tyrosines in activation receptors (such as ITAM-associated receptors, DAP10-associated receptor NKG2D, and the tyrosine-bearing receptor 2B4) and various tyrosine–dependent signalling components of the activation pathway, SHP would simply shut down any phospho-tyrosine-dependent signal (Fig. 1). However, this prevailing model for inhibition by KIR via SHP dephosphorylation of phosphotyrosine-dependent activation signals demanded experimental validation. Is ITIM-bound SHP well suited for rapid dephosphorylation of a large number of phosphotyrosines, which are distributed among multiple receptors and other proteins? Perhaps inhibition relies on a more sophisticated and reliable mechanism, which is designed to bring together active SHP and specific proteins that control activation signals.
Fig. 1. Early model for inhibitory signalling by KIR in NK cells.
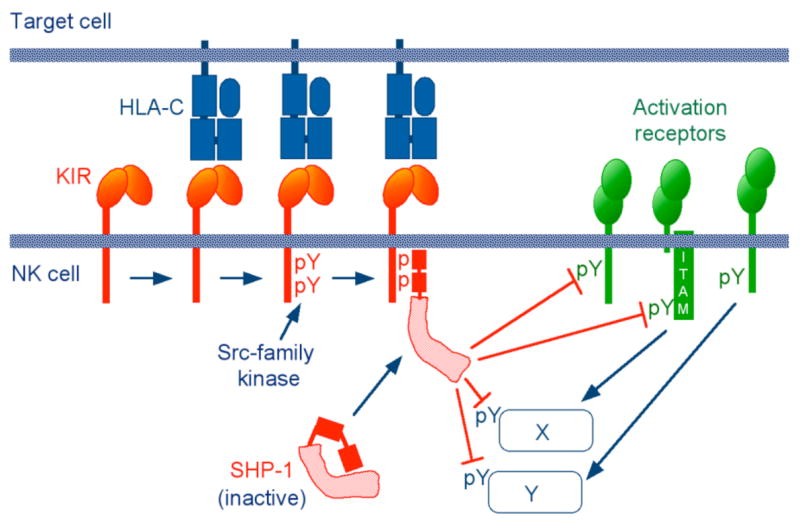
Sequential steps in the inhibition of NK cells by KIR: Binding of inhibitory KIR to HLA-C on target cells; KIR clustering; phosphorylation of two tyrosines within cytoplasmic ITIM sequences; recruitment and activation of the tyrosine phosphatase SHP-1 to the tyrosine-phosphorylated ITIMs; dephosphorylation of multiple substrates, such as activation receptors and signalling molecules (X, Y) by catalytically active SHP-1. The Src-family kinase that phosphorylates the ITIMs may be provided in trans by activation receptors. Inhibitory molecules are indicated in red, activation receptors in green.
The negative signalling pathway in NK cells
Early studies with NK cells, which were mixed with target cells that were either sensitive or resistant to lysis, showed that inhibition occurs at a very early signalling step, even prior to release of intracellular Ca2+ (28). The formation of PLC-γ–LAT complexes is also inhibited by the binding of inhibitory KIR to HLA class I in NK cells (29). The coalescence of lipid rafts at the interface with target cells is also inhibited by KIR engagement (30). Studies of the inhibitory mechanism have also been conducted with T cells. Engagement of inhibitory receptor CD94/NKG2A on T cells by MHC class I on tumor cells inhibited cytotoxicity, TNF-α release, and tyrosine phosphorylation of Lck and ZAP70, but not the TCR ζ chain (31). A KIR2DL1+ tumor-specific T cell clone in contact with tumor cells that expressed an HLA-C ligand failed to phosphorylate ZAP70, LAT, and Vav, did not polarize lipid rafts, and failed to accumulate F-actin towards the tumor cell (32). Consistent with a block that is upstream of actin cytoskeleton rearrangements, engagement of an inhibitory KIR by MHC class I ligand on target cells greatly reduced the recruitment of actin and myosin IIA to the WIP–WASp complex (33). Myosin II is required for the degranulation but not granule polarization towards target cells (34). HLA-E attached to beads was sufficient to induce CD94/NKG2A accumulation, tyrosine phosphorylation, and SHP-1 recruitment to the zone of NK cell–bead contact, while causing exclusion of F-actin and ERM proteins (35). Several questions arise: How do ITIM-Rs accumulate at the inhibitory synapse and how is their phosphorylation achieved? How does inhibitory KIR prevent proximal activation signals, before Ca2+ flux and actin cytoskeleton–dependent processes? Could it be due to dephosphorylation of a specific substrate by ITIM–bound SHP?
The rapid accumulation of KIR at inhibitory immune synapses, through binding to HLA class I on target cells, has unusual properties. Accumulation occurs even in the absence of actin polymerization and ATP production (36). On the other hand, KIR accumulation is enhanced by Zn2+ (36), which is also required for optimal inhibitory function (37). Binding to HLA class I ligands alone is sufficient to induce receptor clustering, as KIR2DL1 accumulated at the site of contact with insect cells expressing HLA-Cw4 (38), and CD94/NKG2A clustered towards HLA-E coupled to beads (35). Accumulated KIR at inhibitory NK cell immune synapses segregates from LFA-1 into distinct regions (36, 39). The distribution pattern varies according to the density of the HLA-C ligand on target cells: Segregation of HLA-C from ICAM-1 into a central cluster on target cells occurred only at high HLA-C density (40).
Rapid phosphorylation of ligand-bound ITIM-Rs is an obvious prerequisite for inhibitory function. Lck is a candidate Src-family kinase for the phosphorylation of ITIMs (41–44). However, it is still unclear how phosphorylation is achieved. Two, non-exclusive, hypotheses have been proposed. One is that phosphorylation of ITIMs by a Src-family kinase occurs in trans during co-engagement of inhibitory and activating receptors at the inhibitory NK cell immune synapse. However, the highly efficient inhibition of proximal signals by KIR engagement is not easily reconciled with an ITIM phosphorylation that is itself dependent on the very signals ITIMs have to stop. The other hypothesis is that clustering of inhibitory receptors is sufficient to recruit a Src-family tyrosine kinase, which phosphorylates the ITIMs. However, association of a kinase with ITIMs has not been detected, except for the binding of Lck to the mouse inhibitory NKR-P1B receptor through a CxCP motif in the cytoplasmic tail (44). ITIM phosphorylation without detectable association with a kinase is obviously possible, and would explain how CD94/NKG2A recruits SHP-1 upon binding to HLA-E on beads (35). The strong tyrosine phosphorylation of ITIM-Rs obtained by the treatment of cells with the tyrosine phosphatase inhibitor pervanadate implies that ITIMs are constitutively phosphorylated, and that their basal unphosphorylated state is maintained by tyrosine phosphatase activity.
High-resolution imaging of KIR by detection of fluorescence resonance energy transfer (FRET) between KIR and an anti-phosphotyrosine antibody revealed that phosphorylated KIR forms a few small clusters within the NK–target cell contact area, whereas total KIR is distributed over a larger area (45). This result raises interesting questions. Are clusters of phospho-KIR–SHP-1 complexes scanning the contact area in search of activation signals, or do they co-localize with clusters of activation receptor signalling? As TCR signalling occurs in small clusters within immune synapses, which move from a peripheral region towards the center of the synapse (46), it is possible that activation signals in NK cell immune synapses are also delivered by discrete signalling complexes, and that phosphorylated KIR is specifically targeted to such signalling receptor clusters.
Identification of substrates targeted for dephosphorylation by ITIM-bound SHP-1 could provide new information about critical signals for NK cell activation, and about the mechanism of inhibition. As SHP-1 is active only when tethered through its SH2 domains to phospho-ITIMs, a generalized, global dephosphorylation of many different substrates (Fig. 1) seemed unlikely. A substrate selection restricted in space and time would more likely focus on a few proteins. Identification of a substrate during inhibition of NK cells by KIR binding to an HLA class I ligand on target cells was performed using a “functional substrate trapping” approach. A KIR2DL1–SHP-1 chimera was generated, which had a full-length SHP-1 inserted in place of the cytoplasmic ITIMs (Fig. 2). An Asp to Ala substitution (DA) in the catalytic domain of SHP-1 results in a catalytically inactive phosphatase, which can still bind phosphotyrosine substrates. The only detectable substrate trapped during KIR2DL1–SHP-1(DA) binding to cognate HLA-C was Vav1 (47). The essential role of Vav1 in T cell immunological synapse formation and receptor clustering (48), and the tyrosine phosphorylation–dependent Rac1 activation by the guanine exchange factor domain of Vav1, suggests that Vav1 dephosphorylation by SHP-1 may be sufficient to block NK cell cytotoxicity.
Fig. 2. Identification of Vav1 as the predominant substrate during inhibition of NK cells by KIR.
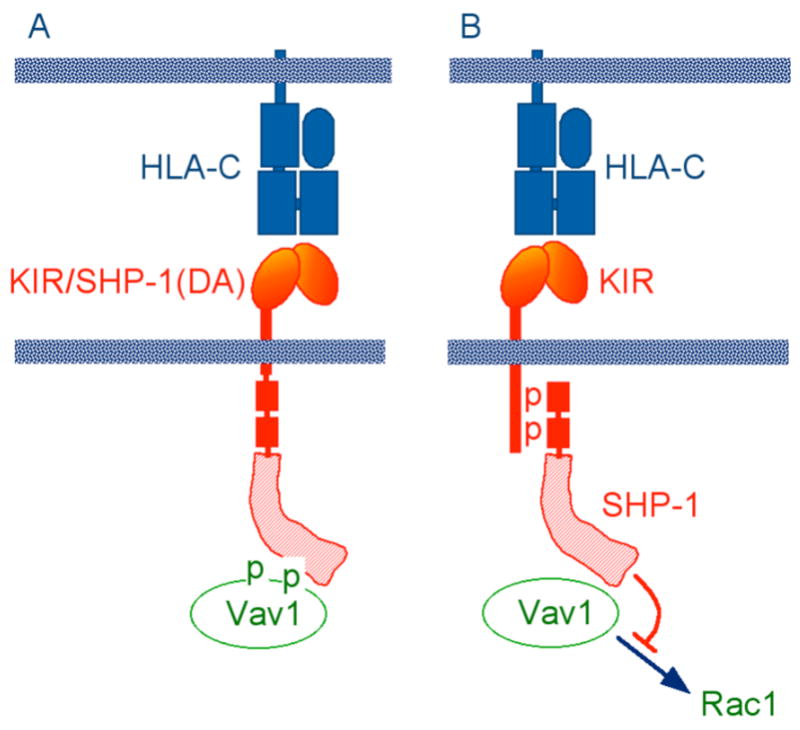
(A) Tyrosine-phosphorylated Vav1 was “trapped” by a chimeric KIR/SHP-1 receptor during inhibition of YTS NK cells by target cells expressing an HLA-C ligand of KIR. The trap was generated by an Asp to Ala mutation (DA) in the SHP-1 catalytic site, and by the fusion of SHP-1(DA) to the KIR cytoplasmic tail. (B) Vav1 trapping, as shown in panel A, implies that catalytically active SHP-1 recruited by KIR during inhibition blocks NK cell activation through dephosphorylation of Vav1, which prevents the guanine exchange factor activity of Vav1 towards the GTPase Rac1.
Phosphorylation of Vav1 would have to occur independently of actin polymerization, if Vav1 were to be a key target for dephosphorylation by SHP-1 during ITIM–dependent inhibition. Consistent with this prediction, trapping of Vav1 occurred in the presence of cytochalasin D (47). Furthermore, actin-independent phosphorylation of Vav1 is induced by binding of the β2 integrin LFA-1 to insect cells transfected with the ligand ICAM-1 alone (49). Inhibition of LFA-1–dependent lysis of insect cells expressing ICAM-1 and HLA-C by binding of inhibitory KIR to peptide-loaded HLA-C was reported, implying that LFA-1 signalling is sensitive to ITIM–dependent signalling (50). Furthermore, KIR engagement by HLA-C on target cells inhibits LFA-1–dependent adhesion (51). Phosphorylation of co-activation receptors 2B4 and NKG2D, and their recruitment to lipid rafts, are blocked by KIR binding to HLA-C on target cells (52, 53). Altogether, these results have led to a new model for activation and inhibition of NK cell responses to target cells (Fig. 3). By blocking actin-dependent processes, such as receptor clustering and coalescence of lipid rafts into larger signalling platforms, inhibitory KIR achieves inhibition at a very early step. This model can account for the ability of KIR to prevent NK cell activation by a large variety of receptors and signalling pathways. Placing ITIM-bound SHP-1 upstream of the full-strength signalling by clustered activating receptors is a better strategy for inhibition than to have SHP-1 chase after phosphorylated receptors and signalling components.
Fig. 3. Revised model for inhibitory signalling by KIR in NK cells.
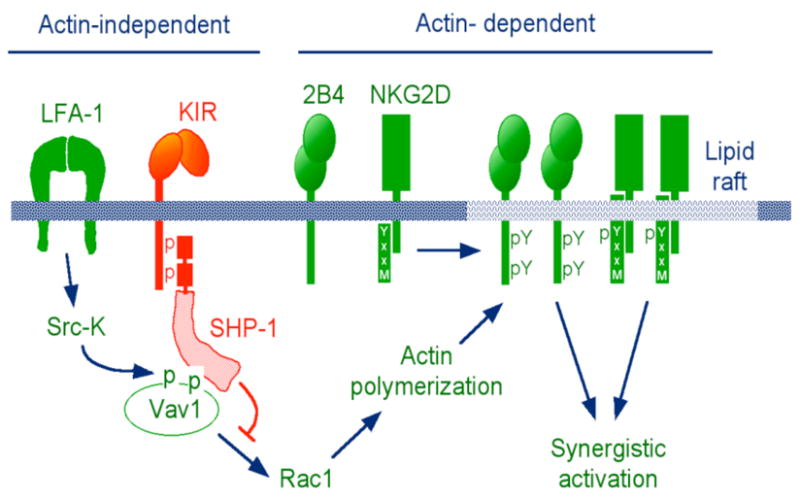
Early, actin-independent signalling by LFA-1 phosphorylates and activates Vav1. Actin-independent dephosphorylation of Vav1 by ITIM–bound SHP-1 prevents actin-dependent processes, such as recruitment of natural cytotoxicity receptors (e.g. NKG2D and 2B4) to lipid rafts, receptor tyrosine phosphorylation, and synergistic signalling by co-activation receptors.
The signalling complex, or “signalosome”, to which Vav1 is associated at the time of its dephosphorylation by SHP-1 remains to be identified. As both Vav1 and its GTPase effector Rac1 are multifunctional proteins, many potential interactions with signalling molecules exist. As inhibition of NK cells by engagement of KIR blocks activation upstream of Ca2+ flux, one can expect that Ca2+–dependent degranulation would be inhibited. It will be interesting to test whether ITIM–mediated inhibition can also prevent granule polarization, given that polarization and degranulation are controlled by separate signals (54).
The extended ITIM family
Receptors and cells
Since the original description of the ITIM sequence and, in part due to the value of the ITIM as predictor of inhibitory function, many ITIM-Rs have been discovered, some of which belong to their own subfamily within the larger ITIM-R superfamily. A stringent search for potential ITIM-Rs in the human proteome has identified 109 candidates, of which only 36 have been described previously as inhibitory receptors (55). ITIM-Rs exert their control in a very wide range of cell types and regulate diverse functions (Table 1). Specific recruitment of SHP-1 and SHP-2 to phosphorylated ITIM has been confirmed repeatedly. However, evidence that SHP plays a role in ITIM-R function is still missing in most cases. Binding of SHP to an ITIM does not imply that SHP is necessary for function, as shown with FcγRIIb (14). While selective binding of either SHP-1 or SHP-2 is often observed with ITIM-Rs, the functional significance of this selectivity is unclear.
Table 1.
Expression, ligands, signaling properties, and function of selected ITEM-containing receptors (ITIM-R)
| Ligands
|
Selected Examples
|
|||||
|---|---|---|---|---|---|---|
| Receptor | Expression | Extracellular | Intracellular | SHP substrate | Inhibition (activation)* of: | Refs |
| FcγRIIb | B, mast, myeloid | Multivalent IgG | SHIP, SHP† | Sustained Ca2+ flux, FcR signaling | (108, 109) | |
| KIR‡ (human) (CD158) | NK,T§ | MHC class I (MHC-I) | SHP | Vavl | Early Ca2+ flux, Tyr-phosphorylation, cytotoxicity, cytokine secretion
β2 integrin-mediated adhesion |
(28,47,110)
(111) (112) (51) |
| Ly49‡ (mouse) | NK,T | MHC-I | SHP | NK cell activation | (7, 18) | |
| CD94/NKG2A‡ | NK,T | HLA-E (human), Qal (mouse) | SHP | NK cell activation
Lysis of CD4+ activated T cells |
(113,114)
(98) |
|
| NKR-P1 (CD161) | NK,T | LLT1 (human) Ocil/Clrb (mouse) | SHP | NK cell activation | (115,116)
(44,117,118) |
|
| LILR‡ (human) (ILT, LIR, CD85) | Broad | MHC-I | SHP, Csk | NK and T cell activation, T cell stimulation by DC | (42,119)
(120) |
|
| PIRB‡ (mouse) | Broad, including brain | MHC-I | SHP | BCR and FcεRI signaling, DC maturation, integrin signaling, GvHD, neuronal plasticity | (62, 121)
(122) (63,64) |
|
| Kirl2 (mouse) | Regions of neuro-genesis in brain | (123) | ||||
| SIRPα‡ (CD172a) | Myeloid | CD47, SP-A/D | SHP, Csk, Pyk2, Grb2, etc. | Cell growth, phagocytosis, adhesion | (65, 124–126) | |
| PD-1 | T | PD-1L | SHP | Autoimmunity, anti-viral
CDS T cell responses |
(56)
(58) |
|
| BTLA | B, T | SHP | Autoimmunity, proliferation | (60,61) | ||
| CD22 (Siglec-2) | B | Sialic acids | SHP, Grb2, SLP76 | SLP-76 | BCR basal signals, (Ca2+ efflux) | (66, 76, 127)
(67) |
| Siglecs‡ (CD33 and CD33-related) | Broad | Sialic acids | SHP, SOCS3 | Proliferation, adhesion | (128, 129) | |
| CD72 | B | CDS | SHP, Grb2 | CD72 | Complex regulation of BCR signaling | (73) |
| LAIR-1 | Broad | Collagen | SHP, Csk | (77, 130) | ||
| KLRG1 | NK, T, mast | Cadherins | SHP, SHIP | NK cell anti-viral response, TCR signaling | (131-134) | |
| gp49B (mouse) | Mast, NK, T neutrophils, eosinophils | αvβ3 integrin | SHP | Allergy, T cell activation, intravascular neutrophil adhesion, eosinophilia | (135, 136)
(137) (138) |
|
| PECAM-1 (CD31) | Platelets, T, endothelium, others | PECAM-1 | SHP | TCR signaling, (NK cell transmigration, endothelial cell motility, integrin-mediated adhesion) | (139-142)
(143) (144) (145) |
|
| CEACAM-1 (CD66a) | Broad | CEACAM-1/5/6/8, CD62E, galectin-3 | SHP | NK and T cell activation, tumor cell growth | (146, 147)
(148) |
|
| TLT-1 | Myeloid, platelets | SHP | (Ca2+ flux) | (149) | ||
| CD300a/f (IRp60, IREM-1/2) | Myeloid, NK | SHP, PI3K | NK cell activation, eosinophil activation | (150-152) | ||
| CLM-1 (mouse) (CMRF-35-like) | Myeloid | SHP | Osteoclastogenesis | (153) | ||
| PILRᇠ| Myeloid | CD99 | SHP | (154, 155) | ||
Activating signals of ITIM-Rs are given in parentheses.
Includes SHP-1 and SHP-2. Preferential association with SHP-1 or SHP-2 occurs with some ITIMs.
Receptor family, which includes activating and inhibitory receptors.
Some T cells, primarily those in the memory subset and IL-15-activated intra-epithelial lymphocytes, express NK cell receptors.
In vivo analysis of ITIM-R function through genetic ablation of genes encoding ITIM-Rs has provided some striking examples of the importance of this negative regulatory system (Table 1). Ablation of the programmed cell death 1 (PD-1) gene results in spontaneous autoimmunity (56). PD-1 binding to its ligands (PD-L) on antigen presenting cells induces peripheral tolerance, whereas binding to PD-L on parenchymal cells prevents tissue destruction (56). CD8+ T cell tolerance is controlled by the combination of CTLA-4 and PD-1 binding to their ligands on dendritic cells (57). Negative signalling by PD-1 can also be detrimental, as it causes exhaustion of CD8+ T cells during chronic virus infection (58). This exhaustion can be reversed by blockade of the PD-1–PD-1L interactions. Yet another role of PD-1 is to promote feto-maternal tolerance by restraining alloreactive T cells through binding to PD-L1 expressed in decidua basalis (maternal tissue in contact with the fetal trophoblast) (59). The B and T lymphocyte attenuator (BTLA) is another ITIM-R, expressed in B and T cells, which collaborates with PD-1 to achieve tolerance (60, 61). BTLA-deficient lymphocytes are hyper-responsive to antigen-receptor signals.
PIRB is an ITIM-R in mice, which is expressed in B and myeloid cells, and is considered an ortholog of human LILR. PIRB and LILR bind MHC class I molecules. PIRB−/− mice have augmented Th2 responses (62), and accelerated graft versus host disease (GvHD) due to higher DC activation upon transfer of allogeneic splenocytes (63). Strikingly, mutant mice lacking functional PIRB exhibit more robust cortical ocular-dominance plasticity, demonstrating an important function of an ITIM-R in the central nervous system (64). Expression of PIRB in several other regions of the brain suggests that it may function broadly to stabilize neural circuits. A specific function of the ITIM-R SIRPα was revealed in SIRPα−/− mice, in which red blood cells were rapidly cleared from the blood. Red blood cells are normally protected from phagocytosis by macrophages through expression of CD47, a ligand of SIRPα (65).
Regulation by ITIM-Rs can be complex. For example, CD22, which is a member of the Siglec receptor family, is an ITIM-R associated with membrane immunoglobulin on B cells. Upon BCR ligation and signalling, CD22 becomes phosphorylated and recruits SHP-1, which dampens the BCR signals. However, independent ligation of CD22 releases the BCR signals from inhibition by CD22-associated SHP-1, thereby lowering the activation threshold for B cell activation (66). Therefore, signalling by an ITIM-R does not necessarily result in inhibition of cellular activation. Another complex mode of BCR regulation by CD22 has been revealed: Tyrosine phosphorylated CD22 attenuates Ca2+ responses by promoting plasma membrane calcium-ATPase (PMCA) activity (67). PMCA extrudes cytoplasmic Ca2+ through the plasma membrane, thereby blocking BCR activation signals. The SHP-1–dependent activation of PMCA, and the association of PMCA with CD22, suggest that PMCA may be activated through dephosphorylation by SHP-1 (67). Additional examples of the diverse ITIM-R functions are listed in Table 1.
ITIM-Rs often belong to small families of closely related members with similar extracellular domains but shorter cytoplasmic tails. Some of the receptors lack ITIM sequences in their tails and transmit positive signals through association with ITAM-containing subunits. The diversification of inhibitory receptor families to include allelic polymorphism and activating isoforms may have been driven by selection for resistance to pathogens. Indeed, activating members in both KIR and Ly49 receptor families have evolved more recently from ancestral inhibitory KIR and Ly49 receptors (68). Activating KIRs contribute to disease susceptibility and to resistance to pathogens, as shown by studies of genetic linkage between KIR and HLA haplotypes (69). Another example of the potential advantage of receptor diversity, including inhibitory and activating forms, is illustrated by genetic resistance to the mouse Herpes virus CMV (MCMV). Among its many strategies to evade immune responses, MCMV expresses the MHC class I-like molecule m157, which binds to the inhibitory receptor Ly49I (70). Expression of m157 compensates for the inhibition of host MHC class I expression by other MCMV proteins. However, C57BL/6 mice are resistant to MCMV because m157 is recognized by the activating isoform Ly49H (70, 71).
Although the ITAM-associated members of ITIM-R families must have important functions, as suggested by their evolution under strong selective forces and by genetic studies of their disease associations, some common misconceptions about their role should be addressed here. The ITIM-containing and ITAM-associated members of these receptor families are sometimes referred to as “paired” receptors. However, it is clear that there is no obligate pairing, either in their expression or function. Healthy individuals with normal NK cell cytotoxic function but with a complete lack of activating KIRs are fairly common. Activating isoforms of ITIM-R families in NK cells (e.g. KIR2DS, Ly49H, CD94/NKG2C) are clearly dispensable in the activation of NK cell cytotoxicity. Furthermore, ITIM-containing receptors are not limited to blocking activation signals delivered by ITAM-associated receptors only.
An outstanding question about most of the ITIM-Rs is how they signal, and whether the ITIM-bound SHP-1/2 is always important for receptor function. Is there a common inhibitory mechanism used by all or most ITIM-Rs? For many of the ITIM-Rs described so far, there is only limited understanding of the negative signalling pathway involved.
Signalling by ITIM-Rs
The integration of signals transmitted during contact between cells through the engagement of several activating and inhibitory receptors results in a finely tuned cellular response. Therefore, when evaluating receptor function, it is critical to use experimental approaches that preserve the physiological situation under which those receptors are engaged (72). Receptor crosslinking with antibodies eliminates the unique environment of cell contacts and the formation of immune synapses. The use of antibodies in solution, on solid supports, or in redirected assays with FcR+ cells, creates artificial conditions in which receptors are bound with an affinity that is several orders of magnitude greater than the binding affinity of natural ligands.
There are cases where ITIM-R crosslinking with antibodies has led to erroneous conclusions about their function. For instance, antibody co-crosslinking of CD22 with BCR inhibits BCR-induced signals. However, CD22 engagement by its natural ligand releases the inhibitory effect of CD22 on BCR signals by separating CD22 from BCR clusters (66). Redirected assays using soluble antibodies to engage an ITIM-R, and FcR+ cells to obtain a multivalent interaction, can be used to evaluate the inhibitory potential of a given ITIM-R, but are not suited to draw conclusions about ITIM-R function. Therefore, studies of signalling properties and receptors function should be carried out in the natural situation of receptor–ligand interactions during contact between cells.
In NK cells, SHP is required for inhibition by an ITIM–containing KIR. Vav1 is the likely substrate of SHP-1 during the inhibition exerted through KIR binding to HLA class I on target cells. It is not known whether this mechanism for inhibition applies to other ITIM-Rs. However, the general concept of a single, or a few molecules that are selectively targeted for dephosphorylation by SHP may apply to inhibition by other ITIM-Rs on NK cells or other cells. Substrate selection during inhibition is most likely dictated, not by sequence preference of the SHP catalytic site, but by substrate accessibility in space and time.
An experimental approach to identify SHP substrates is to monitor protection of tyrosine-phosphorylated proteins through expression of catalytically inactive mutants of SHP. Using this approach in cells that were stimulated by engagement of activation receptors, but without engagement of inhibitory receptors, CD72, actin, and myosin were identified as SHP substrates in B cells (73–75). As phosphorylated CD72 promotes growth arrest and apoptosis, dephosphorylation by SHP blocks this negative regulation. By the same approach, IgM crosslinking led to the association of SLP76 with CD22, and protection of SLP76 phosphotyrosines by a catalytically inactive SHP (76). CD22 may therefore recruit substrates for dephosphorylation by CD22–associated SHP.
Inhibition through an ITIM-R can also be independent of SHP-1 and SHP-2. Phosphorylated LAIR-1 binds the tyrosine kinase Csk, which is a negative regulator of Src-family kinases. Furthermore, co-crosslinking of LAIR-1 and BCR in DT40 cells inhibited BCR-induced activation, even in the absence of SHP-1 and SHP-2. In those cells, LAIR-1 bound Csk but not SHIP, implying a role of Csk in the inhibitory signal (77). The ITIM-Rs ILT2 and SIRPα also bind Csk (Table 1).
The phosphorylation status of different ITIM-Rs varies. Some ITIM-Rs, such as PIRB, are constitutively phosphorylated (78). The kinases responsible for PIRB phosphorylation in DC and neutrophils are the Src-family kinases Hck and Fgr (79). SHP-1 is constitutively associated with phosphorylated LAIR-1 in T cells (80). In addition, tyrosine residues outside of ITIM sequences can modulate ITIM phosphorylation, as shown in ILT2 (81). Although SHP-1 and SHP-2 are interchangeable for inhibition by the PIRB cytoplasmic tail in DT40 B cells (82), these two phosphatases have interesting differences in their properties. Considering that preferential association with either SHP-1 or SHP-2 occurs with a number of ITIM-Rs, differences in negative signalling between SHP-1 and SHP-2 could be relevant for ITIM-R function. SHP-2 contributes to several activation pathways, independently of its ability to bind phosphorylated ITIMs (83, 84). Unique structural features suggest that recruitment of SHP-1 may have a different outcome on signalling than recruitment of SHP-2. Binding of SHP-1 to ITIM-Rs requires phosphorylation of two ITIM sequences. Upon recruitment, SHP-1 becomes phosphorylated at two C-terminal tyrosines. Each one of these tyrosines has the potential to form an intramolecular interaction with an SH2 domain (Fig. 4) (85). The spacing between the C-terminal tyrosines in SHP-1 is too short to allow simultaneous binding to the two SH2 domains (85). Therefore, SHP-1 should be most active when bound to the ITIM-R, as dissociation results in two, mutually exclusive, monovalent SH2 domain–phosphotyrosine interactions, one of which is catalytically inactive.
Fig. 4. Distinct structural properties of SHP-1 and SHP-2 suggest different inhibitory potential.
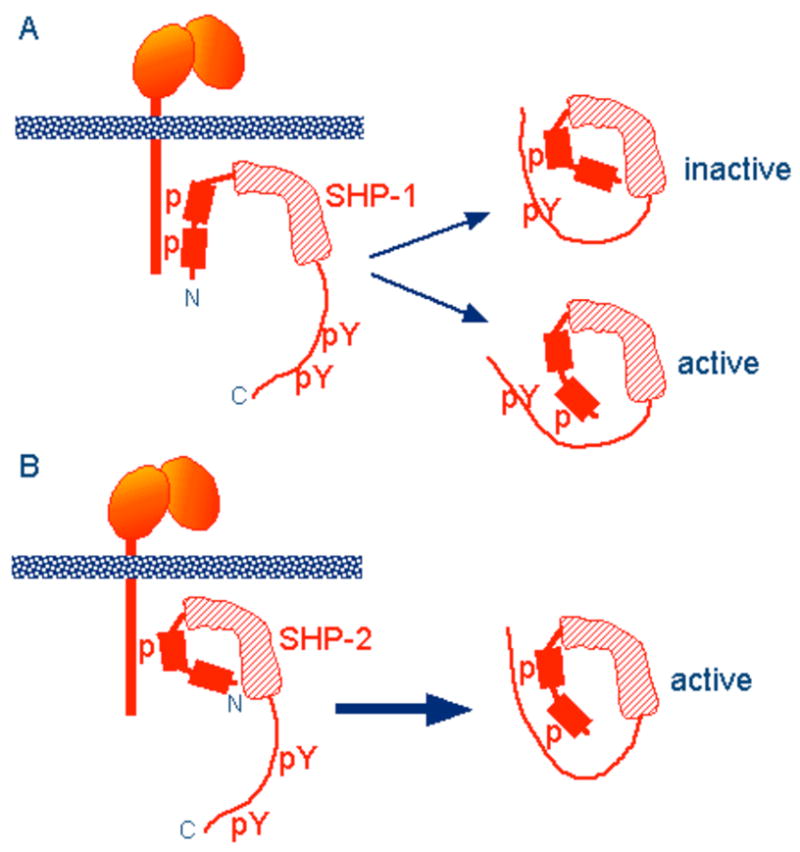
These diagrams are adapted from (86) and (85). (A) The preferential binding of phosphorylated ITIM to the second SH2 domain of SHP-1 (1, 4), and the crystal structure of SHP-1 (107) suggest that the first ITIM of inhibitory KIR binds to the second SH2 domain of SHP-1. The two C-terminal tyrosines of SHP-1 can engage in intramolecular interactions with the SH2 domains, when phosphorylated. However, the short spacing (28 amino acids) between the tyrosines preclude intramolecular binding to both SH2 domains simultaneously. (B) SHP-2 phosphorylated at both C-terminal tyrosines (38 amino acids apart) can form a divalent, intramolecular complex with its own SH2 domains, which retains catalytic activity. Unlike SHP-1, which requires phosphorylation of both ITIMs for binding, SHP-2 binds to either both phosphorylated ITIMs or to the first phosphorylated ITIM only, as indicated.
In contrast, the longer spacing between the C-terminal tyrosines of SHP-2 is compatible with intramolecular engagement of both tyrosines with the tandem SH2 domains (86). Therefore, SHP-2 has the potential to be released from the ITIM-R in a catalytically active conformation. Furthermore, as SHP-2 can bind to inhibitory KIR that is phosphorylated at the first ITIM only, release of SHP-2 in its divalent intra-molecular form may be favoured over the monovalent, intermolecular association with the ITIM-R (Fig. 4). Even a transient association of SHP-2 with a phosphorylated ITIM, together with phosphorylation of both C-terminal tyrosines in SHP-2, may result in release of SHP-2 with sustained catalytic activity. Therefore, SHP-2 has the potential to dephosphorylate molecules while it is no longer bound to the ITIM-R. The functional consequences of these different properties of SHP-1 and SHP-2 have yet to be tested.
Inhibition by “pseudo”-ITIM receptors
Among the many receptors with inhibitory function, some recruit SHP-1 through phosphorylated tyrosines that are not within typical ITIM sequences (Table 2). It is worth describing what is known about their inhibitory mechanism, as it may shed light on how SHP recruited by bona-fide ITIM-Rs may inhibit activation signals. Early on, it was shown that signalling by the erythropoietin receptor (EPO-R) was terminated by recruitment of SHP-1 to the phosphorylated tyrosine at position 429, and dephosphorylation of the EPO-R–associated kinase JAK2 (87). This type of inhibition, namely the termination of a signal delivered by the receptor to which SHP-1 is recruited, is quite different from the function of ITIM-Rs, which prevent other activation receptors from signalling at all. Inhibition of signals from the EPO-R and the prolactin receptor through SHP-1 can also occur through recruitment of a complex of the suppressor of cytokine signalling-1 (SOCS-1) and Grb2 by SHP-1 (88). In this case, inhibition is independent of the SHP-1 catalytic activity.
Table 2.
Inhibition by non-ITEM and pseudo-ITEM containing receptors
| Ligands
|
||||||
|---|---|---|---|---|---|---|
| Receptor | Expression | Extracellular | Intracellular | SHP substrate | Inhibition (activation)* of | Refs |
| CTLA-4 | T | CD80, CD86 | T cell activation | (156) | ||
| CDS | B,T | CD72, other | Ca2+ flux, Erk2, cytokine production | (91, 157) | ||
| GF-R† | Broad | Growth factors | SHP‡ | JAK | JAK-Stat signaling | (87, 88, 158) |
| IL-4Rα | Broad | IL-4 | SHP, SHIP | Stat6, IL-4 induced proliferation | (159) | |
| TNF-R family | Broad | TNF-R ligands | SHP, SHIP | Lyn | (Apoptosis), anti-apoptotic signals | (89) |
| CD47 | Hemopoietic | SIRPα, thrombospondin | IL-12R, Thl response (Actin, PKCθ) | (160)
(161) |
||
| CD200R | Myeloid, mast | CD200 | Dokl/2 | MAPK, autoimmunity, degranulation | (162–164) | |
| FcαRI | Myeloid | Soluble IgA | SHP | Syk, LAT, Erk, phagocytosis, allergy | (92) | |
| DAP 1 2-associated receptors | Broad | Multiple | TLR signals, macrophage activation, antimicrobial defense | (94–96) | ||
Activating functions are given in parentheses.
Growth factor receptors (e.g. EPO-R, PDGF-R).
Includes SHP-1 and SHP-2. Preferential association with SHP-1 or SHP-2 occurs with some ITIMs.
The death receptors Fas, TNF-R, and TRAIL bind SHP-1, SHP-2, and SHIP through a YxxL motif in their cytoplasmic tail (89). Signaling by these death receptors prevents GM-CSF–induced activation of the Src-kinase Lyn. Presumably, dephosphorylation of Lyn by SHP-1 blocks the anti-apoptotic signalling pathway induced by GM-CSF. Additional examples of inhibition by non-ITIM receptors are listed in Table 2.
Two receptors involved in inhibition of T cell activation, namely CTLA-4 and CD5, had been proposed to inhibit by a mechanism similar to that of ITIM-Rs. However, CTLA-4 inhibitory function is tyrosine-independent (90), and the inhibition of Ca2+ flux, Erk activation, and cytokine production by CD5 depends on a cytoplasmic tyrosine that does not bind SHP and SHIP (91).
Surprisingly, there are examples of signalling by ITAM-containing receptors that result in inhibition. The high affinity IgA receptor FcαRI, which is associated with the ITAM-containing FcR γ chain, inhibits signalling by FcγR or FcεRI when bound to soluble IgA (92). The interaction of FcαRI with low avidity monomeric IgA results in recruitment of SHP-1, and inactivation of the early signalling components, Syk, LAT, and Erk (92). This inhibitory property of ITAM could explain how TLR signalling is enhanced in the absence of ITAM-based signals in macrophages (93). Inhibition of TLR responses occurs through DAP12–associated receptors, such as TREM2 (94, 95). These findings provide a new framework for understanding how upregulation of DAP12–associated LIR7 in lesions of lepromatous patients had a suppressive effect towards TLR-mediated antimicrobial activity (96).
Regulation of NK cells by a complex repertoire of inhibitory receptors
The repertoire of inhibitory receptors on NK cells is complex at several levels. First, three families of MHC class I-specific inhibitory receptors are expressed in human (KIR, CD94/NKG2, LILR) and mouse (Ly49, CD94/NKG2, PIR) NK cells. Second, expression of receptors in the KIR, Ly49, and CD94 families is turned on in a stochastic manner, such that each NK cell expresses its own repertoire of receptors. Third, there is very high genetic diversity in the KIR and Ly49 receptor families. KIR genes vary in number, genomic organization, and allelic polymorphism among individual haplotypes. This combination generates diversity such that most individuals have a unique KIR repertoire (97). And finally, NK cells express other inhibitory receptors that bind non- MHC class I ligands such as NKR-P1, LAIR-1, Siglec-7/9, and CEACAM1 (Table 1). It is not known yet if this multiplicity of inhibitory receptor–ligand systems serves to protect different types of cells, which may express only a subset of inhibitory receptor ligands. Alternatively, the additive effects of several inhibitory receptor–ligand interactions may be required to achieve full inhibition. Another hypothesis is that different inhibitory receptors may specialize in the type of signals and cellular function they control.
MHC class I recognition by inhibitory receptors is crucial for protection of healthy cells from NK cells. For example, CD94/NKG2A binding to Qa1 on activated CD4+ T cells is essential for their protection from NK cell-mediated lysis (98). However, NK cell tolerance develops in the complete absence of MHC class I (99, 100). This tolerance towards MHC class I-negative cells is actively maintained, and is quickly lost when tolerant NK cells are mixed with MHC class I-positive cells (101, 102). Blocking of the inhibitory Ly49–MHC class I interaction in vivo with antibodies interfered with the development of a functional NK cell population (103). NK cells that lack inhibitory receptors for self MHC class I exist but are hypo-responsive (104). The intrinsic responsiveness of NK cells is calibrated by an ITIM-dependent process (105). NK cells that receive stronger inhibitory signals through MHC class I recognition acquire a stronger capacity to respond to activation signals (105, 106). An interesting implication is that genetic associations between combinations of KIR and HLA haplotypes with disease could be accounted for, not only on the basis of inhibition by ITIM-Rs, but also by the calibration of NK cell responsiveness acquired through engagement of ITIM-Rs. It should be noted that these two properties of ITIM-Rs have opposite effects. For example, the combination of an inhibitory KIR and its cognate HLA-C can, on the one hand, provide inhibition during contact with target cells, but, on the other hand, confer greater intrinsic ability to respond to activation signals. Perhaps such opposite effects will explain the complex patterns of disease associations with ITIM-Rs that have emerged in genetic studies (69).
Concluding remarks
The prominent role of inhibitory receptors in the maintenance of a balanced immune system has been illustrated vividly with mice that are deficient in selected receptors of the ITIM-R superfamily. Identification of a vast array of specific inhibitory receptor–ligand combinations, and expression of those ligands in diverse cell types, has revealed a sophisticated system for the regulation of cellular responses in various environments. An important concept to emerge from studies of inhibitory signalling during NK cell contact with resistant target cells, is that a single predominant component of the activation pathway is targeted for dephosphorylation by SHP-1. That inhibition is not mediated simply by dephosphorylation of many signalling components in the activation pathway, but by the selection of a specific substrate for SHP, suggests that ITIM-Rs may be capable of a finely tuned regulation of activation signals. However, the orchestration of inhibitory signals and their intersection with specific activation signals are still unknown for most of the ITIM-Rs. A key challenge that lies ahead is to determine in molecular detail the initiation and propagation of inhibitory signals delivered by different ITIM-Rs in their natural physiological contexts.
Acknowledgments
I thank Sumati Rajagopalan for comments on the manuscript. Supported by the Intramural Research Program of the NIH, NIAID.
References
- 1.Burshtyn DN, et al. Recruitment of tyrosine phosphatase HCP by the killer cell inhibitor receptor. Immunity. 1996;4:77–85. doi: 10.1016/s1074-7613(00)80300-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Burshtyn DN, Yang WT, Yi TL, Long EO. A novel phosphotyrosine motif with a critical amino acid at position-2 for the SH2 domain-mediated activation of the tyrosine phosphatase SHP-1. J Biol Chem. 1997;272:13066–13072. doi: 10.1074/jbc.272.20.13066. [DOI] [PubMed] [Google Scholar]
- 3.Burshtyn DN, Lam AS, Weston M, Gupta N, Warmerdam PAM, Long EO. Conserved residues amino-terminal of cytoplasmic tyrosines contribute to the SHP-1-mediated inhibitory function of killer cell Ig-like receptors. J Immunol. 1999;162:897–902. [PubMed] [Google Scholar]
- 4.Beebe KD, Wang P, Arabaci G, Pei DH. Determination of the binding specificity of the SH2 domains of protein tyrosine phosphatase SHP-1 through the screening of a combinatorial phosphotyrosyl peptide library. Biochemistry. 2000;39:13251–13260. doi: 10.1021/bi0014397. [DOI] [PubMed] [Google Scholar]
- 5.Hawke NA, et al. Extraordinary variation in a diversified family of immune-type receptor genes. Proc Natl Acad Sci U S A. 2001;98:13832–13837. doi: 10.1073/pnas.231418598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ljunggren HG, Kärre K. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol Today. 1990;11:237–244. doi: 10.1016/0167-5699(90)90097-s. [DOI] [PubMed] [Google Scholar]
- 7.Karlhofer FM, Ribaudo RK, Yokoyama WM. MHC class I alloantigen specificity of Ly-49+ IL-2-activated natural killer cells. Nature. 1992;358:66–70. doi: 10.1038/358066a0. [DOI] [PubMed] [Google Scholar]
- 8.Moretta L, Ciccone E, Mingari MC, Biassoni R, Moretta A. Human Natural Killer Cells; Origin, Clonality, Specificity, and Receptors. Adv Immunol. 1994;55:341–380. doi: 10.1016/s0065-2776(08)60513-1. [DOI] [PubMed] [Google Scholar]
- 9.Wagtmann N, Rajagopalan S, Winter CC, Peruzzi M, Long EO. Killer cell inhibitory receptors specific for HLA-C and HLA-B identified by direct binding and by functional transfer. Immunity. 1995;3:801–809. doi: 10.1016/1074-7613(95)90069-1. [DOI] [PubMed] [Google Scholar]
- 10.Amigorena S, et al. Cytoplasmic domain heterogeneity and functions of IgG Fc receptors in B lymphocytes. Science. 1992;256:1808–1812. doi: 10.1126/science.1535455. [DOI] [PubMed] [Google Scholar]
- 11.Choquet D, Partiseti M, Amigorena S, Bonnerot C, Fridman WH, Korn H. Cross-linking of IgG receptors inhibits membrane immunoglobulin-stimulated calcium influx in B lymphocytes. J Cell Biol. 1993;121:355–363. doi: 10.1083/jcb.121.2.355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muta T, Kurosaki T, Misulovin Z, Sanchez M, Nussenzweig MC, Ravetch JV. A 13-amino-acid motif in the cytoplasmic domain of Fc gamma RIIB modulates B-cell receptor signalling. Nature. 1994;368:70–73. doi: 10.1038/368070a0. [DOI] [PubMed] [Google Scholar]
- 13.D’Ambrosio D, et al. Recruitment and activation of PTP1C in negative regulation of antigen receptor signaling by FcgammaRIIB1. Science. 1995;268:293–297. doi: 10.1126/science.7716523. [DOI] [PubMed] [Google Scholar]
- 14.Ono M, Bolland S, Tempst P, Ravetch JV. Role of the inositol phosphatase SHIP in negative regulation of the immune system by the receptor FcgammaRIIB. Nature. 1996;383:263–266. doi: 10.1038/383263a0. [DOI] [PubMed] [Google Scholar]
- 15.Bolland S, Pearse RN, Kurosaki T, Ravetch JV. SHIP modulates immune receptor responses by regulating membrane association of Btk. Immunity. 1998;8:509–516. doi: 10.1016/s1074-7613(00)80555-5. [DOI] [PubMed] [Google Scholar]
- 16.Scharenberg AM, et al. Phosphatidylinositol-3,4,5-trisphosphate (PtdIns-3,4,5-P3) Tec kinase-dependent calcium signaling pathway: a target for SHIP-mediated inhibitory signals. EMBO J. 1998;17:1961–1972. doi: 10.1093/emboj/17.7.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huyer G, Li ZM, Adam M, Huckle WR, Ramachandran C. Direct determination of the sequence recognition requirements of the SH2 domains of SH-PTP2. Biochemistry. 1995;34:1040–1049. doi: 10.1021/bi00003a039. [DOI] [PubMed] [Google Scholar]
- 18.Olcese L, et al. Human and mouse killer-cell inhibitory receptors recruit PTP1C and PTPlD protein tyrosine phosphatases. J Immunol. 1996;156:4531–4534. [PubMed] [Google Scholar]
- 19.Vély F, et al. Differential association of phosphatases with hematopoietic co-receptors bearing immunoreceptor tyrosine-based inhibition motifs. Eur J Immunol. 1997;27:1994–2000. doi: 10.1002/eji.1830270825. [DOI] [PubMed] [Google Scholar]
- 20.Bruhns P, Vély F, Malbec O, Fridman WH, Vivier E, Daëron M. Molecular basis of the recruitment of the SH2 domain-containing inositol 5-phosphatases SHIP1 and SHIP2 by FcgammaRIIB. J Biol Chem. 2000;275:37357–37364. doi: 10.1074/jbc.M003518200. [DOI] [PubMed] [Google Scholar]
- 21.Ono M, Okada H, Bolland S, Yanagi S, Kurosaki T, Ravetch JV. Deletion of SHIP or SHP-1 reveals two distinct pathways for inhibitory signaling. Cell. 1997;90:293–301. doi: 10.1016/s0092-8674(00)80337-2. [DOI] [PubMed] [Google Scholar]
- 22.Gupta N, et al. Negative signaling pathways of the killer cell inhibitory receptor and FcgammaRIIb1 require distinct phosphatases. J Exp Med. 1997;186:473–478. doi: 10.1084/jem.186.3.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fry AM, Lanier LL, Weiss A. Phosphotyrosines in the killer cell inhibitory receptor motif of NKB1 are required for negative signaling and for association with protein tyrosine phosphatase 1C. J Exp Med. 1996;184:295–300. doi: 10.1084/jem.184.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bruhns P, Marchetti P, Fridman WH, Vivier E, Daëron M. Differential roles of N- and C-terminal immunoreceptor tyrosine-based inhibition motifs during inhibition of cell activation by killer cell inhibitory receptors. J Immunol. 1999;162:3168–3175. [PubMed] [Google Scholar]
- 25.Kabat J, Borrego F, Brooks A, Coligan JE. Role that each NKG2A immunoreceptor tyrosine-based inhibitory motif plays in mediating the human CD94/NKG2A inhibitory signal. J Immunol. 2002;169:1948–1958. doi: 10.4049/jimmunol.169.4.1948. [DOI] [PubMed] [Google Scholar]
- 26.Avril T, Floyd H, Lopez F, Vivier E, Crocker PR. The membrane-proximal immunoreceptor tyrosine-based inhibitory motif is critical for the inhibitory signaling mediated by siglecs-7 and-9, CD33-related Siglecs expressed on human monocytes and NK cells. J Immunol. 2004;173:6841–6849. doi: 10.4049/jimmunol.173.11.6841. [DOI] [PubMed] [Google Scholar]
- 27.Hof P, Pluskey S, Dhe-Paganon S, Eck MJ, Shoelson SE. Crystal structure of the tyrosine phosphatase SHP-2. Cell. 1998;92:441–450. doi: 10.1016/s0092-8674(00)80938-1. [DOI] [PubMed] [Google Scholar]
- 28.Kaufman DS, Schoon RA, Robertson MJ, Leibson PJ. Inhibition of selective signaling events in natural killer cells recognizing major histocompatibility complex class I. Proc Natl Acad Sci USA. 1995;92:6484–6488. doi: 10.1073/pnas.92.14.6484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Valiante NM, Phillips JH, Lanier LL, Parham P. Killer cell inhibitory receptor recognition of human leukocyte antigen (HLA) class I blocks formation of a pp36/PLC-gamma signaling complex in human natural killer (NK) cells. J Exp Med. 1996;184:2243–2250. doi: 10.1084/jem.184.6.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fassett MS, Davis DM, Valter MM, Cohen GB, Strominger JL. Signaling at the inhibitory natural killer cell immune synapse regulates lipid raft polarization but not class I MHC clustering. Proc Natl Acad Sci U S A. 2001;98:14547–14552. doi: 10.1073/pnas.211563598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carena I, Shamshiev A, Donda A, Colonna M, Libero GD. Major histocompatibility complex class I molecules modulate activation threshold and early signaling of T cell antigen receptor-gamma/delta stimulated by nonpeptidic ligands. J Exp Med. 1997;186:1769–1774. doi: 10.1084/jem.186.10.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guerra N, et al. Engagement of the inhibitory receptor CD158a interrupts TCR signaling, preventing dynamic membrane reorganization in CTL/tumor cell interaction. Blood. 2002;100:2874–2881. doi: 10.1182/blood-2002-02-0643. [DOI] [PubMed] [Google Scholar]
- 33.Krzewski K, Chen X, Orange JS, Strominger JL. Formation of a WIP-, WASp-, actin-, and myosin IIA-containing multiprotein complex in activated NK cells and its alteration by KIR inhibitory signaling. J Cell Biol. 2006;173:121–132. doi: 10.1083/jcb.200509076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Andzelm MM, Chen X, Krzewski K, Orange JS, Strominger JL. Myosin IIA is required for cytolytic granule exocytosis in human NK cells. J Exp Med. 2007;204:2285–2291. doi: 10.1084/jem.20071143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masilamani M, Nguyen C, Kabat J, Borrego F, Coligan JE. CD94/NKG2A inhibits NK cell activation by disrupting the actin network at the immunological synapse. J Immunol. 2006;177:3590–3596. doi: 10.4049/jimmunol.177.6.3590. [DOI] [PubMed] [Google Scholar]
- 36.Davis DM, Chiu I, Fassett M, Cohen GB, Mandelboim O, Strominger JL. The human natural killer cell immune synapse. Proc Natl Acad Sci USA. 1999;96:15062–15067. doi: 10.1073/pnas.96.26.15062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rajagopalan S, Winter CC, Wagtmann N, Long EO. Cutting Edge: The Ig-related killer cell inhibitory receptor binds zinc and requires zinc for recognition of HLA-C on target cells. J Immunol. 1995;155:4143–4146. [PubMed] [Google Scholar]
- 38.Faure M, Barber DF, Takahashi SM, Jin T, Long EO. Spontaneous clustering and tyrosine phosphorylation of NK cell inhibitory receptor induced by ligand binding. J Immunol. 2003;170:6107–6114. doi: 10.4049/jimmunol.170.12.6107. [DOI] [PubMed] [Google Scholar]
- 39.Vyas YM, et al. Spatial organization of signal transduction molecules in the NK cell immune synapses during MHC class I-regulated noncytolytic and cytolytic interactions. J Immunol. 2001;167:4358–4367. doi: 10.4049/jimmunol.167.8.4358. [DOI] [PubMed] [Google Scholar]
- 40.Almeida CR, Davis DM. Segregation of HLA-C from ICAM-1 at NK cell immune synapses is controlled by its cell surface density. J Immunol. 2006;177:6904–6910. doi: 10.4049/jimmunol.177.10.6904. [DOI] [PubMed] [Google Scholar]
- 41.Binstadt BA, et al. Sequential involvement of Lck and SHP-1 with MHC-recognizing receptors on NK cells inhibits FcR-initiated tyrosine kinase activation. Immunity. 1996;5:629–638. doi: 10.1016/s1074-7613(00)80276-9. [DOI] [PubMed] [Google Scholar]
- 42.Dietrich J, Cella M, Colonna M. Ig-like transcript 2 (ILT2)/leukocyte Ig-like receptor 1 (LIR1) inhibits TCR signaling and actin cytoskeleton reorganization. J Immunol. 2001;166:2514–2521. doi: 10.4049/jimmunol.166.4.2514. [DOI] [PubMed] [Google Scholar]
- 43.Henshall TL, Jones KL, Wilkinson R, Jackson DE. Src homology 2 domain-containing protein-tyrosine phosphatases, SHP-1 and SHP-2, are required for platelet endothelial cell adhesion molecule-1/CD31-mediated inhibitory signaling. J Immunol. 2001;166:3098–3106. doi: 10.4049/jimmunol.166.5.3098. [DOI] [PubMed] [Google Scholar]
- 44.Ljutic B, Carlyle JR, Filipp D, Nakagawa R, Julius M, Zúñiga-Pflücker JC. Functional requirements for signaling through the stimulatory and inhibitory mouse NKR-P1 (CD161) NK cell receptors. J Immunol. 2005;174:4789–4796. doi: 10.4049/jimmunol.174.8.4789. [DOI] [PubMed] [Google Scholar]
- 45.Treanor B, et al. Microclusters of inhibitory killer immunoglobulin-like receptor signaling at natural killer cell immunological synapses. J Cell Biol. 2006;174:153–161. doi: 10.1083/jcb.200601108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yokosuka T, et al. Newly generated T cell receptor microclusters initiate and sustain T cell activation by recruitment of Zap70 and SLP-76. Nat Immunol. 2005;6:1253–1262. doi: 10.1038/ni1272. [DOI] [PubMed] [Google Scholar]
- 47.Stebbins CC, Watzl C, Billadeau DD, Leibson PJ, Burshtyn DN, Long EO. Vav1 dephosphorylation by the tyrosine phosphatase SHP-1 as a mechanism for inhibition of cellular cytotoxicity. Mol Cell Biol. 2003;23:6291–6299. doi: 10.1128/MCB.23.17.6291-6299.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tybulewicz VL. Vav-family proteins in T-cell signalling. Curr Opin Immunol. 2005;17:267–274. doi: 10.1016/j.coi.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 49.Riteau B, Barber DF, Long EO. Vav1 phosphorylation is induced by beta2 integrin engagement on natural killer cells upstream of actin cytoskeleton and lipid raft reorganization. J Exp Med. 2003;198:469–474. doi: 10.1084/jem.20021995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barber DF, Faure M, Long EO. LFA-1 contributes an early signal for NK cell cytotoxicity. J Immunol. 2004;173:3653–3659. doi: 10.4049/jimmunol.173.6.3653. [DOI] [PubMed] [Google Scholar]
- 51.Burshtyn DN, Shin J, Stebbins C, Long EO. Adhesion to target cells is disrupted by the killer cell inhibitory receptor. Curr Biol. 2000;10:777–780. doi: 10.1016/s0960-9822(00)00568-6. [DOI] [PubMed] [Google Scholar]
- 52.Watzl C, Long EO. Natural killer cell inhibitory receptors block actin cytoskeleton-dependent recruitment of 2B4 (CD244) to lipid rafts. J Exp Med. 2003;197:77–85. doi: 10.1084/jem.20020427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Endt J, et al. Inhibitory receptor signals suppress ligation-induced recruitment of NKG2D to GM1-rich membrane domains at the human NK cell immune synapse. J Immunol. 2007;178:5606–5611. doi: 10.4049/jimmunol.178.9.5606. [DOI] [PubMed] [Google Scholar]
- 54.Bryceson YT, March ME, Barber DF, Ljunggren HG, Long EO. Cytolytic granule polarization and degranulation controlled by different receptors in resting NK cells. J Exp Med. 2005;202:1001–1012. doi: 10.1084/jem.20051143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Staub E, Rosenthal A, Hinzmann B. Systematic identification of immunoreceptor tyrosine-based inhibitory motifs in the human proteome. Cell Signal. 2004;16:435–456. doi: 10.1016/j.cellsig.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 56.Okazaki T, Honjo T. The PD-1-PD-L pathway in immunological tolerance. Trends Immunol. 2006;27:195–201. doi: 10.1016/j.it.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 57.Probst HC, McCoy K, Okazaki T, Honjo T, van den Broek M. Resting dendritic cells induce peripheral CD8+ T cell tolerance through PD-1 and CTLA-4. Nat Immunol. 2005;6:280–286. doi: 10.1038/ni1165. [DOI] [PubMed] [Google Scholar]
- 58.Barber DL, et al. Restoring function in exhausted CD8 T cells during chronic viral infection. Nature. 2006;439:682–687. doi: 10.1038/nature04444. [DOI] [PubMed] [Google Scholar]
- 59.Guleria I, et al. A critical role for the programmed death ligand 1 in fetomaternal tolerance. J Exp Med. 2005;202:231–237. doi: 10.1084/jem.20050019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Watanabe N, et al. BTLA is a lymphocyte inhibitory receptor with similarities to CTLA-4 and PD-1. Nat Immunol. 2003;4:670–679. doi: 10.1038/ni944. [DOI] [PubMed] [Google Scholar]
- 61.Han P, Goularte OD, Rufner K, Wilkinson B, Kaye J. An inhibitory Ig superfamily protein expressed by lymphocytes and APCs is also an early marker of thymocyte positive selection. J Immunol. 2004;172:5931–5939. doi: 10.4049/jimmunol.172.10.5931. [DOI] [PubMed] [Google Scholar]
- 62.Ujike A, Takeda K, Nakamura A, Ebihara S, Akiyama K, Takai T. Impaired dendritic cell maturation and increased T(H)2 responses in PIR-B(−/−) mice. Nat Immunol. 2002;3:542–548. doi: 10.1038/ni801. [DOI] [PubMed] [Google Scholar]
- 63.Nakamura A, Kobayashi E, Takai T. Exacerbated graft-versus-host disease in Pirb−/− mice. Nat Immunol. 2004;5:623–629. doi: 10.1038/ni1074. [DOI] [PubMed] [Google Scholar]
- 64.Syken J, Grandpre T, Kanold PO, Shatz CJ. PirB restricts ocular-dominance plasticity in visual cortex. Science. 2006;313:1795–1800. doi: 10.1126/science.1128232. [DOI] [PubMed] [Google Scholar]
- 65.Oldenborg PA, Zheleznyak A, Fang YF, Lagenaur CF, Gresham HD, Lindberg FP. Role of CD47 as a marker of self on red blood cells. Science. 2000;288:2051–2054. doi: 10.1126/science.288.5473.2051. [DOI] [PubMed] [Google Scholar]
- 66.Doody GM, et al. A role in B cell activation for CD22 and the protein tyrosine phosphatase SHP. Science. 1995;269:242–244. doi: 10.1126/science.7618087. [DOI] [PubMed] [Google Scholar]
- 67.Chen J, McLean PA, Neel BG, Okunade G, Shull GE, Wortis HH. CD22 attenuates calcium signaling by potentiating plasma membrane calcium-ATPase activity. Nat Immunol. 2004;5:651–657. doi: 10.1038/ni1072. [DOI] [PubMed] [Google Scholar]
- 68.Abi-Rached L, Parham P. Natural selection drives recurrent formation of activating killer cell immunoglobulin-like receptor and Ly49 from inhibitory homologues. J Exp Med. 2005;201:1319–1332. doi: 10.1084/jem.20042558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Khakoo SI, Carrington M. KIR and disease: a model system or system of models? Immunol Rev. 2006;214:186–201. doi: 10.1111/j.1600-065X.2006.00459.x. [DOI] [PubMed] [Google Scholar]
- 70.Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 2002;296:1323–1326. doi: 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- 71.Voigt V, et al. Murine cytomegalovirus m157 mutation and variation leads to immune evasion of natural killer cells. Proc Natl Acad Sci USA. 2003;100:13483–13488. doi: 10.1073/pnas.2233572100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Long EO, et al. Inhibition of natural killer cell activation signals by killer cell immunoglobulin-like receptors (CD158) Immunol Rev. 2001;181:223–233. doi: 10.1034/j.1600-065x.2001.1810119.x. [DOI] [PubMed] [Google Scholar]
- 73.Wu YJ, et al. The B-cell transmembrane protein CD72 binds to and is an in vivo substrate of the protein tyrosine phosphatase SHP-1. Curr Biol. 1998;8:1009–1017. doi: 10.1016/s0960-9822(07)00421-6. [DOI] [PubMed] [Google Scholar]
- 74.Baba T, Fusaki N, Shinya N, Iwamatsu A, Hozumi N. Actin tyrosine dephosphorylation by the Src homology 1-containing protein tyrosine phosphatase is essential for actin depolymerization after membrane IgM cross-linking. J Immunol. 2003;170:3762–3768. doi: 10.4049/jimmunol.170.7.3762. [DOI] [PubMed] [Google Scholar]
- 75.Baba T, Fusaki N, Shinya N, Iwamatsu A, Hozumi N. Myosin is an in vivo substrate of the protein tyrosine phosphatase (SHP-1) after mIgM cross-linking. Biochem Biophys Res Commun. 2003;304:67–72. doi: 10.1016/s0006-291x(03)00542-4. [DOI] [PubMed] [Google Scholar]
- 76.Mizuno K, Tagawa Y, Watanabe N, Ogimoto M, Yakura H. SLP-76 is recruited to CD22 and dephosphorylated by SHP-1, thereby regulating B cell receptor-induced c-Jun N-terminal kinase activation. Eur J Immunol. 2005;35:644–654. doi: 10.1002/eji.200425465. [DOI] [PubMed] [Google Scholar]
- 77.Verbrugge A, Rijkers ESK, De Ruiter T, Meyaard L. Leukocyte-associated Ig-like receptor-1 has SH2 domain-containing phosphatase-independent function and recruits C-terminal Src kinase. Eur J Immunol. 2006;36:190–198. doi: 10.1002/eji.200535226. [DOI] [PubMed] [Google Scholar]
- 78.Ho LH, Uehara T, Chen CC, Kubagawa H, Cooper MD. Constitutive tyrosine phosphorylation of the inhibitory paired Ig-like receptor PIR-B. Proc Natl Acad Sci U S A. 1999;96:15086–15090. doi: 10.1073/pnas.96.26.15086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang H, Meng FY, Chu CL, Takai T, Lowell CA. The Src family kinases Hck and Fgr negatively regulate neutrophil and dendritic cell chemokine signaling via PIR-B. Immunity. 2005;22:235–246. doi: 10.1016/j.immuni.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 80.Sathish JG, et al. Constitutive association of SHP-1 with leukocyte-associated Ig-like receptor-1 in human T cells. J Immunol. 2001;166:1763–1770. doi: 10.4049/jimmunol.166.3.1763. [DOI] [PubMed] [Google Scholar]
- 81.Bellon T, Kitzig F, Sayos J, López-Botet M. Mutational analysis of immunoreceptor tyrosine-based inhibition motifs of the Ig-like transcript 2 (CD85j) leukocyte receptor. J Immunol. 2002;168:3351–3359. doi: 10.4049/jimmunol.168.7.3351. [DOI] [PubMed] [Google Scholar]
- 82.Maeda A, Kurosaki M, Ono M, Takai T, Kurosaki T. Requirement of SH2-containing protein tyrosine phosphatases SHP-1 and SHP-2 for paired immunoglobulin-like receptor B (PIR-B)-mediated inhibitory signal. J Exp Med. 1998;187:1355–1360. doi: 10.1084/jem.187.8.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim SO, Jiang J, Yi WS, Feng GS, Frank SJ. Involvement of the Src homology 2-containing tyrosine phosphatase SHP-2 in growth hormone signaling. J Biol Chem. 1998;273:2344–2354. doi: 10.1074/jbc.273.4.2344. [DOI] [PubMed] [Google Scholar]
- 84.Salmond RJ, Alexander DR. SHP2 forecast for the immune system: fog gradually clearing. Trends Immunol. 2006;27:154–160. doi: 10.1016/j.it.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 85.Zhang ZS, Shen K, Lu W, Cole PA. The role of C-terminal tyrosine phosphorylation in the regulation of SHP-1 explored via expressed protein ligation. J Biol Chem. 2003;278:4668–4674. doi: 10.1074/jbc.M210028200. [DOI] [PubMed] [Google Scholar]
- 86.Lu W, Gong DQ, Bar-Sagi D, Cole PA. Site-specific incorporation of a phosphotyrosine mimetic reveals a role for tyrosine phosphorylation of SHP-2 in cell signaling. Mol Cell. 2001;8:759–769. doi: 10.1016/s1097-2765(01)00369-0. [DOI] [PubMed] [Google Scholar]
- 87.Klingmüller U, Lorenz U, Cantley LC, Neel BG, Lodish HF. Specific recruitment of SH-PTP1 to the erythropoietin receptor causes inactivation of JAK2 and termination of proliferative signals. Cell. 1995;80:729–738. doi: 10.1016/0092-8674(95)90351-8. [DOI] [PubMed] [Google Scholar]
- 88.Minoo P, Zadeh MM, Rottapel R, Lebrun JJ, Ali S. A novel SHP-1/Grb2-dependent mechanism of negative regulation of cytokine-receptor signaling: contribution of SHP-1C-terminal tyrosines in cytokine signaling. Blood. 2004;103:1398–1407. doi: 10.1182/blood-2003-07-2617. [DOI] [PubMed] [Google Scholar]
- 89.Daigle I, Yousefi S, Colonna M, Green DR, Simon HU. Death receptors bind SHP-1 and block cytokine-induced anti-apoptotic signaling in neutrophils. Nat Med. 2002;8:61–67. doi: 10.1038/nm0102-61. [DOI] [PubMed] [Google Scholar]
- 90.Baroja ML, et al. The inhibitory function of CTLA-4 does not require its tyrosine phosphorylation. J Immunol. 2000;164:49–55. doi: 10.4049/jimmunol.164.1.49. [DOI] [PubMed] [Google Scholar]
- 91.Gary-Gouy H, Bruhns P, Schmitt C, Dalloul A, Daëron M, Bismuth G. The pseudo-immunoreceptor tyrosine-based activation motif of CD5 mediates its inhibitory action on B-cell receptor signaling. J Biol Chem. 2000;275:548–556. doi: 10.1074/jbc.275.1.548. [DOI] [PubMed] [Google Scholar]
- 92.Pasquier B, et al. Identification of FcalphaRI as an inhibitory receptor that controls inflammation: Dual role of FcRgamma ITAM. Immunity. 2005;22:31–42. doi: 10.1016/j.immuni.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 93.Hamerman JA, Tchao NK, Lowell CA, Lanier LL. Enhanced Toll-like receptor responses in the absence of signaling adaptor DAP12. Nat Immunol. 2005;6:579–586. doi: 10.1038/ni1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Hamerman JA, Jarjoura JR, Humphrey MB, Nakamura MC, Seaman WE, Lanier LL. Cutting edge: inhibition of TLR and FcR responses in macrophages by triggering receptor expressed on myeloid cells (TREM)-2 and DAP12. J Immunol. 2006;177:2051–2055. doi: 10.4049/jimmunol.177.4.2051. [DOI] [PubMed] [Google Scholar]
- 95.Turnbull IR, Colonna M. Activating and inhibitory functions of DAP12. Nat Rev Immunol. 2007;7:155–161. doi: 10.1038/nri2014. [DOI] [PubMed] [Google Scholar]
- 96.Bleharski JR, et al. Use of genetic profiling in leprosy to discriminate clinical forms of the disease. Science. 2003;301:1527–1530. doi: 10.1126/science.1087785. [DOI] [PubMed] [Google Scholar]
- 97.Parham P. MHC class I molecules and KIRs in human history, health and survival. Nature Rev Immunol. 2005;5:201–214. doi: 10.1038/nri1570. [DOI] [PubMed] [Google Scholar]
- 98.Lu L, Ikizawa K, Hu D, Werneck MB, Wucherpfennig KW, Cantor H. Regulation of activated CD4+ T cells by NK cells via the Qa-1-NKG2A inhibitory pathway. Immunity. 2007;26:593–604. doi: 10.1016/j.immuni.2007.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Liao NS, Bix M, Zijlstra M, Jaenisch R, Raulet D. MHC class I deficiency: susceptibility to natural killer (NK) cells and impaired NK activity. Science. 1991;253:199–202. doi: 10.1126/science.1853205. [DOI] [PubMed] [Google Scholar]
- 100.Hoglund P, et al. Recognition of beta 2-microglobulin-negative (beta 2m-) T-cell blasts by natural killer cells from normal but not from beta 2m- mice: nonresponsiveness controlled by beta 2m- bone marrow in chimeric mice. Proc Natl Acad Sci USA. 1991;88:10332–10336. doi: 10.1073/pnas.88.22.10332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Johansson MH, Bieberich C, Jay G, Kärre K, Höglund P. Natural killer cell tolerance in mice with mosaic expression of major histocompatibility complex class I transgene. J Exp Med. 1997;186:353–364. doi: 10.1084/jem.186.3.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Johansson S, et al. Natural killer cell education in mice with single or multiple major histocompatibility complex class I molecules. J Exp Med. 2005;201:1145–1155. doi: 10.1084/jem.20050167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Das A, Saxena RK. Role of interaction between Ly49 inhibitory receptors and cognate MHC I molecules in IL2-induced development of NK cells in murine bone marrow cell cultures. Immunol Lett. 2004;94:209–214. doi: 10.1016/j.imlet.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 104.Fernandez NC, Treiner E, Vance RE, Jamieson AM, Lemieux S, Raulet DH. A subset of natural killer cells achieves self-tolerance without expressing inhibitory receptors specific for self-MHC molecules. Blood. 2005;105:4416–4423. doi: 10.1182/blood-2004-08-3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kim S, et al. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436:709–713. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- 106.Anfossi N, et al. Human NK cell education by inhibitory receptors for MHC class I. Immunity. 2006;25:331–342. doi: 10.1016/j.immuni.2006.06.013. [DOI] [PubMed] [Google Scholar]
- 107.Yang J, et al. Crystal structure of human protein-tyrosine phosphatase SHP-1. J Biol Chem. 2003;278:6516–6520. doi: 10.1074/jbc.M210430200. [DOI] [PubMed] [Google Scholar]
- 108.Nimmerjahn F, Ravetch JV. Fcgamma receptors as regulators of immune responses. Nat Rev Immunol. 2008;8:34–47. doi: 10.1038/nri2206. [DOI] [PubMed] [Google Scholar]
- 109.Cambier JC. Immunol Rev. 2008 this issue. [Google Scholar]
- 110.Biassoni R, et al. Human natural killer cell receptors and co-receptors. Immunol Rev. 2001;181:203–214. doi: 10.1034/j.1600-065x.2001.1810117.x. [DOI] [PubMed] [Google Scholar]
- 111.Moretta A. Immunol Rev. 2008 this issue. [Google Scholar]
- 112.D’Andrea A, Chang C, Phillips JH, Lanier LL. Regulation of T cell lymphokine production by killer cell inhibitory receptor recognition of self HLA class I alleles. J Exp Med. 1996;184:789–794. doi: 10.1084/jem.184.2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Borrego F, Ulbrecht M, Weiss EH, Coligan JE, Brooks AG. Recognition of human histocompatibility leukocyte antigen (HLA)-E complexed with HLA class I signal sequence-derived peptides by CD94/NKG2 confers protection from natural killer cell-mediated lysis. J Exp Med. 1998;187:813–818. doi: 10.1084/jem.187.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gunturi A, Berg RE, Forman J. The role of CD94/NKG2 in innate and adaptive immunity. Immunol Res. 2004;30:29–34. doi: 10.1385/IR:30:1:029. [DOI] [PubMed] [Google Scholar]
- 115.Aldemir H, et al. Cutting edge: lectin-like transcript 1 is a ligand for the CD161 receptor. J Immunol. 2005;175:7791–7795. doi: 10.4049/jimmunol.175.12.7791. [DOI] [PubMed] [Google Scholar]
- 116.Rosen DB, Bettadapura Y, Alsharifi M, Mathew PA, Warren HS, Lanier LL. Cutting edge: Lectin-like transcript-1 is a ligand for the inhibitory human NKR-P1A receptor. J Immunol. 2005;175:7796–7799. doi: 10.4049/jimmunol.175.12.7796. [DOI] [PubMed] [Google Scholar]
- 117.Iizuka K, Naidenko OV, Plougastel BF, Fremont DH, Yokoyama WM. Genetically linked C-type lectin-related ligands for the NKRP1 family of natural killer cell receptors. Nat Immunol. 2003;4:801–807. doi: 10.1038/ni954. [DOI] [PubMed] [Google Scholar]
- 118.Carlyle JR, Jamieson AM, Gasser S, Clingan CS, Arase H, Raulet DH. Missing self-recognition of Ocil/Clr-b by inhibitory NKR-P1 natural killer cell receptors. Proc Natl Acad Sci U S A. 2004;101:3527–3532. doi: 10.1073/pnas.0308304101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Sayós J, Martínez-Barriocanal A, Kitzig F, Bellón T, López-Botet M. Recruitment of C-terminal Src kinase by the leukocyte inhibitory receptor CD85j. Biochem Biophys Res Commun. 2004;324:640–647. doi: 10.1016/j.bbrc.2004.09.097. [DOI] [PubMed] [Google Scholar]
- 120.Chang CC, et al. Tolerization of dendritic cells by T(S) cells: the crucial role of inhibitory receptors ILT3 and ILT4. Nat Immunol. 2002;3:237–243. doi: 10.1038/ni760. [DOI] [PubMed] [Google Scholar]
- 121.Kubagawa H, et al. Biochemical nature and cellular distribution of the paired immunoglobulin-like receptors, PIR-A and PIR-B. J Exp Med. 1999;189:309–318. doi: 10.1084/jem.189.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Pereira S, Zhang H, Takai T, Lowell CA. The inhibitory receptor PIR-B negatively regulates neutrophil and macrophage integrin signaling. J Immunol. 2004;173:5757–5765. doi: 10.4049/jimmunol.173.9.5757. [DOI] [PubMed] [Google Scholar]
- 123.Bryceson YT, Foster JA, Kuppusamy SP, Herkenham M, Long EO. Expression of a killer cell receptor-like gene in plastic regions of the central nervous system. J Neuroimmunol. 2005;161:177–182. doi: 10.1016/j.jneuroim.2004.11.018. [DOI] [PubMed] [Google Scholar]
- 124.Kharitonenkov A, Chen ZJ, Sures I, Wang HY, Schilling J, Ullrich A. A family of proteins that inhibit signalling through tyrosine kinase receptors. Nature. 1997;386:181–186. doi: 10.1038/386181a0. [DOI] [PubMed] [Google Scholar]
- 125.Timms JF, et al. SHPS-1 is a scaffold for assembling distinct adhesion-regulated multi-protein complexes in macrophages. Curr Biol. 1999;9:927–930. doi: 10.1016/s0960-9822(99)80401-1. [DOI] [PubMed] [Google Scholar]
- 126.Barclay AN, Brown MH. The SIRP family of receptors and immune regulation. Nat Rev Immunol. 2006;6:457–464. doi: 10.1038/nri1859. [DOI] [PubMed] [Google Scholar]
- 127.Otipoby KL, Draves KE, Clark EA. CD22 regulates B cell receptor-mediated signals via two domains that independently recruit Grb2 and SHP-1. J Biol Chem. 2001;276:44315–44322. doi: 10.1074/jbc.M105446200. [DOI] [PubMed] [Google Scholar]
- 128.Crocker PR, Paulson JC, Varki A. Siglecs and their roles in the immune system. Nat Rev Immunol. 2007;7:255–266. doi: 10.1038/nri2056. [DOI] [PubMed] [Google Scholar]
- 129.Orr SJ, et al. SOCS3 targets Siglec 7 for proteasomal degradation and blocks Siglec 7-mediated responses. J Biol Chem. 2007;282:3418–3422. doi: 10.1074/jbc.C600216200. [DOI] [PubMed] [Google Scholar]
- 130.Lebbink RJ, et al. Collagens are functional, high affinity ligands for the inhibitory immune receptor LAIR-1. J Exp Med. 2006;203:1419–1425. doi: 10.1084/jem.20052554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Robbins SH, Nguyen KB, Takahashi N, Mikayama T, Biron CA, Brossay L. Cutting edge: Inhibitory functions of the killer cell lectin-like receptor G1 molecule during the activation of mouse NK cells. J Immunol. 2002;168:2585–2589. doi: 10.4049/jimmunol.168.6.2585. [DOI] [PubMed] [Google Scholar]
- 132.Grundemann C, et al. Cutting edge: identification of E-cadherin as a ligand for the murine killer cell lectin-like receptor G1. J Immunol. 2006;176:1311–1315. doi: 10.4049/jimmunol.176.3.1311. [DOI] [PubMed] [Google Scholar]
- 133.Ito M, Maruyama T, Saito N, Koganei S, Yamamoto K, Matsumoto N. Killer cell lectin-like receptor G1 binds three members of the classical cadherin family to inhibit NK cell cytotoxicity. J Exp Med. 2006;203:289–295. doi: 10.1084/jem.20051986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tessmer MS, et al. KLRG1 binds cadherins and preferentially associates with SHIP-1. Int Immunol. 2007;19:391–400. doi: 10.1093/intimm/dxm004. [DOI] [PubMed] [Google Scholar]
- 135.Feldweg AM, et al. gp49B1 suppresses stem cell factor-induced mast cell activation-secretion and attendant inflammation in vivo. Eur J Immunol. 2003;33:2262–2268. doi: 10.1002/eji.200323978. [DOI] [PubMed] [Google Scholar]
- 136.Gu XG, et al. The gp49B1 inhibitory receptor regulates the IFN-gamma responses of T cells and NK cells. J Immunol. 2003;170:4095–4101. doi: 10.4049/jimmunol.170.8.4095. [DOI] [PubMed] [Google Scholar]
- 137.Zhou JS, et al. Prevention of lipopolysaccharide-induced microangiopathy by gp49B1: Evidence for an important role for gp49B1 expression on neutrophils. J Exp Med. 2003;198:1243–1251. doi: 10.1084/jem.20030906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Norris HH, et al. Inhibitory receptor gp49B regulates eosinophil infiltration during allergic inflammation. J Leukoc Biol. 2007;82:1531–1541. doi: 10.1189/jlb.1106667. [DOI] [PubMed] [Google Scholar]
- 139.Berman ME, Xie Y, Muller WA. Roles of platelet/endothelial cell adhesion molecule-1 (PECAM-1, CD31) in natural killer cell transendothelial migration and beta 2 integrin activation. J Immunol. 1996;156:1515–1524. [PubMed] [Google Scholar]
- 140.Newton-Nash DK, Newman PJ. A new role for platelet-endothelial cell adhesion molecule-1 (CD31): inhibition of TCR-mediated signal transduction. J Immunol. 1999;163:682–688. [PubMed] [Google Scholar]
- 141.Zhao T, Newman PJ. Integrin activation by regulated dimerization and oligomerization of platelet endothelial cell adhesion molecule (PECAM)-1 from within the cell. J Cell Biol. 2001;152:65–73. doi: 10.1083/jcb.152.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Nagaishi T, et al. SHP1 phosphatase-dependent T cell inhibition by CEACAM1 adhesion molecule isoforms. Immunity. 2006;25:769–781. doi: 10.1016/j.immuni.2006.08.026. [DOI] [PubMed] [Google Scholar]
- 143.O’Brien CD, Cao G, Makrigiannakis A, DeLisser HM. Role of immunoreceptor tyrosine-based inhibitory motifs of PECAM-1 in PECAM-1-dependent cell migration. Am J Physiol Cell Physiol. 2004;287:C1103–1113. doi: 10.1152/ajpcell.00573.2003. [DOI] [PubMed] [Google Scholar]
- 144.Wee JL, Jackson DE. The Ig-ITIM superfamily member PECAM-1 regulates the “outside-in” signaling properties of integrin alphaIIbbeta3 in platelets. Blood. 2005;106:3816–3823. doi: 10.1182/blood-2005-03-0911. [DOI] [PubMed] [Google Scholar]
- 145.Kasirer-Friede A, Kahn ML, Shattil SJ. Platelet integrins and immunoreceptors. Immunol Rev. 2007;218:247–264. doi: 10.1111/j.1600-065X.2007.00532.x. [DOI] [PubMed] [Google Scholar]
- 146.Markel G, et al. CD66a interactions between human melanoma and NK cells: A novel class I MHC-independent inhibitory mechanism of cytotoxicity. J Immunol. 2002;168:2803–2810. doi: 10.4049/jimmunol.168.6.2803. [DOI] [PubMed] [Google Scholar]
- 147.Gray-Owen SD, Blumberg RS. CEACAM1: contact-dependent control of immunity. Nat Rev Immunol. 2006;6:433–446. doi: 10.1038/nri1864. [DOI] [PubMed] [Google Scholar]
- 148.Beauchemin N, et al. Association of biliary glycoprotein with protein tyrosine phosphatase SHP-1 in malignant colon epithelial cells. Oncogene. 1997;14:783–790. doi: 10.1038/sj.onc.1200888. [DOI] [PubMed] [Google Scholar]
- 149.Barrow AD, et al. Cutting edge: TREM-like transcript-1, a platelet immunoreceptor tyrosine-based inhibition motif encoding costimulatory immunoreceptor that enhances, rather than inhibits, calcium signaling via SHP-2. J Immunol. 2004;172:5838–5842. doi: 10.4049/jimmunol.172.10.5838. [DOI] [PubMed] [Google Scholar]
- 150.Cantoni C, et al. Molecular and functional characterization of IRp60, a member of the immunoglobulin superfamily that functions as an inhibitory receptor in human NK cells. Eur J Immunol. 1999;29:3148–3159. doi: 10.1002/(SICI)1521-4141(199910)29:10<3148::AID-IMMU3148>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 151.Munitz A, Bachelet I, Eliashar R, Moretta A, Moretta L, Levi-Schaffer F. The inhibitory receptor IRp60 (CD300a) suppresses the effects of IL-5, GM-CSF, and eotaxin on human peripheral blood eosinophils. Blood. 2006;107:1996–2003. doi: 10.1182/blood-2005-07-2926. [DOI] [PubMed] [Google Scholar]
- 152.Alvarez-Errico D, Sayos J, Lopez-Botet M. The IREM-1 (CD300f) inhibitory receptor associates with the p85alpha subunit of phosphoinositide 3-kinase. J Immunol. 2007;178:808–816. doi: 10.4049/jimmunol.178.2.808. [DOI] [PubMed] [Google Scholar]
- 153.Chung DH, Humphrey MB, Nakamura MC, Ginzinger DG, Seaman WE, Daws MR. CMRF-35-like molecule-1, a novel mouse myeloid receptor, can inhibit osteoclast formation. J Immunol. 2003;171:6541–6548. doi: 10.4049/jimmunol.171.12.6541. [DOI] [PubMed] [Google Scholar]
- 154.Fournier N, et al. FDF03, a novel inhibitory receptor of the immunoglobulin superfamily, is expressed by human dendritic and myeloid cells. J Immunol. 2000;165:1197–1209. doi: 10.4049/jimmunol.165.3.1197. [DOI] [PubMed] [Google Scholar]
- 155.Wang J, Shiratori I, Satoh T, Lanier LL, Arase H. An essential role of sialylated O-linked sugar chains in the recognition of mouse CD99 by paired Ig-like type 2 receptor (PILR) J Immunol. 2008;180:1686–1693. doi: 10.4049/jimmunol.180.3.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bluestone JA. Immunol Rev. 2008 this issue. [Google Scholar]
- 157.Brossard C, Semichon M, Trautmann A, Bismuth G. CD5 inhibits signaling at the immunological synapse without impairing its formation. J Immunol. 2003;170:4623–629. doi: 10.4049/jimmunol.170.9.4623. [DOI] [PubMed] [Google Scholar]
- 158.Jiao HY, Berrada K, Yang WT, Tabrizi M, Platanias LC, Yi TL. Direct association with and dephosphorylation of Jak2 kinase by the SH2-domain-containing protein tyrosine phosphatase SHP-1. Mol Cell Biol. 1996;16:6985–992. doi: 10.1128/mcb.16.12.6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Kashiwada M, Giallourakis CC, Pan PY, Rothman PB. Immunoreceptor tyrosine-based inhibitory motif of the IL-4 receptor associates with SH2-containing phosphatases and regulates IL-4-induced proliferation. J Immunol. 2001;167:6382–387. doi: 10.4049/jimmunol.167.11.6382. [DOI] [PubMed] [Google Scholar]
- 160.Latour S, et al. Bidirectional negative regulation of human T and dendritic cells by CD47 and its cognate receptor signal-regulator protein-alpha: Down-regulation of IL-12 responsiveness and inhibition of dendritic cell activation. J Immunol. 2001;167:2547–554. doi: 10.4049/jimmunol.167.5.2547. [DOI] [PubMed] [Google Scholar]
- 161.Rebres RA, Green JM, Reinhold MI, Ticchioni M, Brown EJ. Membrane raft association of CD47 is necessary for actin polymerization and protein kinase C theta translocation in its synergistic activation of T cells. J Biol Chem. 2001;276:7672–680. doi: 10.1074/jbc.M008858200. [DOI] [PubMed] [Google Scholar]
- 162.Hoek RM, et al. Down-regulation of the macrophage lineage through interaction with OX2 (CD200) Science. 2000;290:1768–771. doi: 10.1126/science.290.5497.1768. [DOI] [PubMed] [Google Scholar]
- 163.Zhang S, Cherwinski H, Sedgwick JD, Phillips JH. Molecular mechanisms of CD200 inhibition of mast cell activation. J Immunol. 2004;173:6786–793. doi: 10.4049/jimmunol.173.11.6786. [DOI] [PubMed] [Google Scholar]
- 164.Zhang S, Phillips JH. Identification of tyrosine residues crucial for CD200R-mediated inhibition of mast cell activation. J Leukoc Biol. 2006;79:363–68. doi: 10.1189/jlb.0705398. [DOI] [PubMed] [Google Scholar]


