Abstract
The nature of cross-priming immunogens for CD8+ T cell responses is highly controversial. Using a panel of T cell receptor-like antibodies specific for viral peptides bound to mouse Db major histocompatibility complex class I molecules, we show that an exceptional peptide (PA224-233) expressed as a viral minigene product formed a sizeable cytosolic pool continuously presented for hours after protein synthesis was inhibited. PA224-233 pool formation required active cytosolic heat shock protein 90 but not ER g96, and uniquely enabled cross-priming by this peptide. These findings demonstrate that exceptional class I binding oligopeptides that escape proteolytic degradation are potent cross-priming agents. Thus, the feeble immunogenicity of natural proteasome products in cross-priming can be attributed to their evanescence in donor cells, and not an absolute inability of cytosolic oligopeptides to be transferred to and presented by professional antigen presenting cells..
Direct antigen presentation is based on proteasome initiated degradation of defective ribosomal products (DRiPs) and other rapidly degraded endogenous translation products (Eisenlohr et al., 2007; Yewdell, 2007). Typically, peptides of 8 or more residues are transported by the transporter associated with antigen processing (TAP) into the endoplasmic reticulum (ER) where final trimming of COOH-terminal residues is mediated by ER associated aminopeptidase (ERAAP). Peptide binding releases nascent class I molecules from the loading complex, initiating their transport to the plasma membrane for recognition by CD8+ T cells.
In viral infections, direct presentation enables CD8+ T cell activation by virus infected cells, including dendritic cells (DCs), which can initiate naïve CD8+ T cell response in lymph nodes and spleen. But what if viruses cannot (or will not) express their proteins in DCs? In this case, an alterative pathway exists, termed cross-priming, where DCs process antigens acquired from infected cells and present them to naïve CD8+ T cells (Shen and Rock, 2006). Controversy swirls around virtually every aspect of cross-priming.
A substantial literature has grown around the concept that cross-priming is mediated by DC acquisition of glucose regulated protein 94 (GRP94) (also known as glycoprotein 96 (gp96)) and other chaperones bearing oligopeptides derived from proteasomal antigen degradation (Srivastava, 2002). It has proven difficult, however, for many laboratories to recover peptides bound to gp96 purified from antigen presenting cells, or even to demonstrate that gp96 binds to oligopeptides with an affinity consistent with its proposed role in cross-priming (Demine and Walden, 2005; Javid et al., 2007; Nicchitta et al., 2004; Wallin et al., 2002; Ying and Flatmark, 2006). Purified gp96 and a variety of other chaperones demonstrate antigen-specific cross-priming activity, but this activity is weak, and the adjuvant role of contaminants (e.g. lectins, bacterial lipopolysaccharide) in the phenomenon is a concern. Heat shock protein 90 (HSP90) has been recovered from cells bound to truncated forms of a model antigen (Kunisawa and Shastri, 2006), but the cross-priming activity of such complexes has not been demonstrated in the context of intact cells as immunogens. Moreover, seven laboratories recently published studies pointing to proteasome substrates rather than proteasome products as the source of cross-priming antigens (Basta et al., 2005; Donohue et al., 2006; Freigang et al., 2007; Gasteiger et al., 2007; Norbury et al., 2004; Shen and Rock, 2004; Wolkers et al., 2004).This conclusion jibes with the original observation by Rammensee and colleagues that antigenic oligopeptides are only recovered from cells expressing class I molecules capable of presenting the peptides (Falk et al., 1990). Further the finding that cytosolic oligopeptides are rapidly destroyed by endopeptidases and aminopeptidases (half-life on the order of 10 seconds) (Reits et al., 2004), is inconsistent with the hypothesis that oligopeptides exist in stable complexes with molecular chaperones.
A key advance in the antigen presentation field was the introduction of antibodies with TCR like specificities, generated by standard hybridoma technology or by screening of antibody phage display libraries (Andersen et al., 1996; Denkberg and Reiter, 2006; Porgador et al., 1997). Such reagents enable measurement of cell surface major histocompatibility complex (MHC) class I -peptide complexes with great precision by flow cytometry. Here we describe the generation and characterization of a panel of TCR-like phage displayed antibodies specific for well defined influenza A virus (IAV) peptides bound to H-2Db molecules. We then use these phage antibodies to study the cell biology and immunology of oligopeptides introduced into the cytosol and ER of antigen presenting cell and cross-priming donor cells as biosynthesized minigene products.
Results
Isolation of human recombinant antibodies with TCR like specificity to influenza virus derived peptide-MHC complexes
To generate a panel of TCR-like antibodies specific for IAV-Db restricted determinants, we selected a large non-immune library consisting of 3.7 × 1010 unique human recombinant phage Fab antibody fragments (de Haard et al., 1999) for binding to purified H-2Db complexed with PA224-233, NP366-374 or PB1-F2-62-70. These determinants top the defined anti-IAV immunodominance hierarchy in B6 mice. Out of numerous candidates identified, we chose the Fabs that demonstrated the best signal to noise ratio and sensitivity. As seen in Supplementary Fig 1, the three Fabs chosen selectively recognized both purified Db and cell surface Db complexed with their cognate determinants. The threshold for peptide recognition on peptide pulsed EL4 cells with low temperature induced peptide receptive Db molecules occured at ~ 10−9 M. This is similar to the sensitivity of the 25-D1.16 mAb which is specific for Kb-SIINFEKL complexes. This reagent represents “gold standard” for highly sensitive and specific TCR like antibody recognition (Porgador et al., 1997)
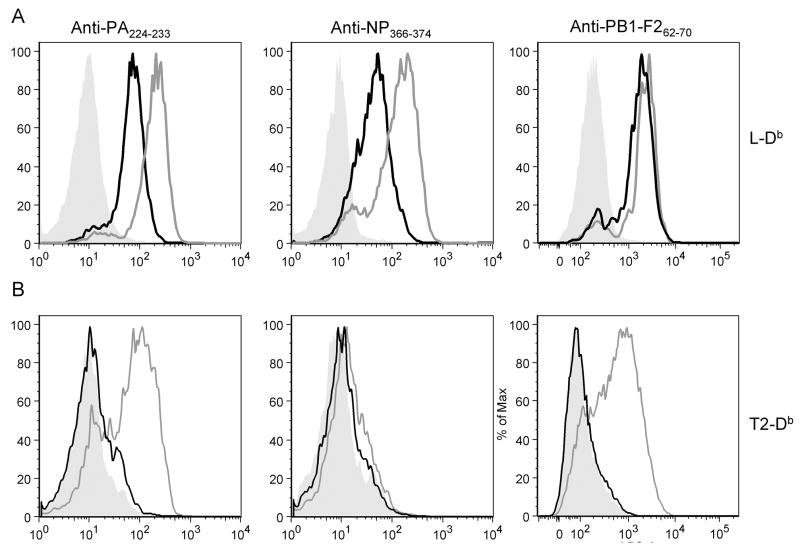
We next tested the ability of the TCR-like Fabs (FabTCRs) to bind to endogenous complexes generated from cytosolic or ER targeted minigene products expressed by recombinant vaccinia viruses (rVVs) (See Schematic of rVVs in supplementary figure 2). Minigene products are typically expressed at very high copy numbers (tens of thousands per cell) relative to peptides generated from full length proteins (10 to a few thousand copies per cell)(Princiotta et al., 2003). Each of the 6 minigene products tested was easily detected by its cognate FabTCR (Figure 1). Addition of a signal sequence increased the numbers of cell surface complexes, very slightly for PB1-F262-70, and ~5-fold for NP366-374 and PA224-233.
Figure 1.
Detection of Db -peptide complexes on cells infected with rVVs expressing cytosolic or ER targeted peptides.
(A) L-Db cells were infected with rVVs encoding for Mg-peptides (black line), ES-peptide (gray line) or uninfected control (solid gray). (B) T2-Db cells were infected with rVVs encoding for Mg-peptides (black line), ES-peptide (gray line) or uninfected control (solid gray). Each graph represents infections with rVVs encoding the peptide that the indicated antibody is directed to. Binding of Fabs to live cells was determined by flow cytometry. L-Db infections were repeated more than six times. T2-Db infections were repeated 2 times.
To determine the TAP-dependence of complex formation, we infected human T2-Db cells with the rVVs (T2 cells are EBV-transformed lymphoblasts that lack both TAP1 and TAP2 genes and consequently, cytosolic peptides have limited access to the ER). This revealed that, as expected, each of the cytosolic minigene products demonstrated only slight TAP-independent presentation, while ER-targeting bypassed the requirement for TAP for PB1-F262-70 and PA224-233. Surprisingly, presentation of ER-targeted NP366-374 remained largely TAP dependent. A possible explanation is the presence of Ala as the NH2-terminal residue in NP366-374, because Ala is a preferred residue at the P1 site of signal peptidase cleavage, which may therefore destroy the peptide when it is translocated via the translocon. If this case, the enhanced ability of ES-NP366-374 to access Db in L-Db cells would presumably be due to the ability of the leader sequence to enhance TAP mediated antigen presentation. This could be due to resistance to degradation by cytosolic or ER proteases or enhanced TAP-mediated transport into the ER of the full length ES-peptide, or more likely, a NH2-terminally trimmed version.
These findings demonstrate that FabTCRs can be used as highly specific reagents to precisely quantitate cell surface Db-peptide complexes generated from VV-encoded minigenes.
Kinetics of cell surface complex presentation using TCR like Fabs
We next examined the kinetics of cell surface complex expression at 30 min intervals following infection of L-Db cells with rVVs expressing the three determinants. In each case, cognate peptide-MHC complexes were first detected by the FabTCRs 60–90 min post-infection and quickly reached Vmax of complex generation (i.e. the maximal increase in the rate of complex accumulation). This behavior is similar to expression of Kb-Ova257-264 complexes (Princiotta et al., 2003), and is exactly what is expected for peptides that rapidly achieve saturating amounts of expression for feeding the class I pathway and require little or no processing for binding to class I molecules.
Addition of BFA, which blocks MHC egress from the secretory pathway by inducing Golgi complex disassembly and redistribution of Golgi complex proteins into the ER (Yewdell and Bennink, 1989), resulted in an immediate block of cell surface complex accretion. In the case of ES-PB1-F262-70, BFA caused a steep and steady decline in surface complexes, indicative of a relatively low complex stability (t1/2 of ~ 120 min). Under the same conditions, both ER-targeted and cytosolic NP366-374 demonstrated intermediate complex stability while both cytosolic and ER-targeted PA224-233 generated highly stable complexes. This hierarchy of complex stability agrees with the stability of complexes generated with synthetic peptides that we previously reported (Figure 3 in Chen et al. (Chen et al., 2004), findings that we repeated using the FabTCRs (data not shown).
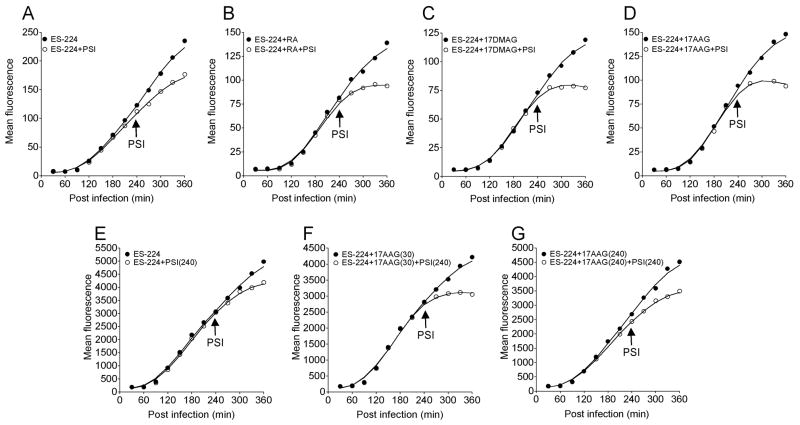
Figure 3.
Effect of HSP90 inhibitors on PA224-233 pool generation.
Cell surface Db-PA224-233 complexes were determined by flow cytometry at indicated times after infection with VV-ES-224. Cells were treated with PSIs 240 min post infection (open circles), or left untreated (solid circles) in the presence or absence of HSP90 inhibitors added 30 min or 240 min post infection as indicated. (A) No inhibitor, (B) treated with 2uM 17AAG, (C) treated with 2μM 17DMAG, (D) treated with 2μM 17AAG, (E) no inhibitor, (F) 17AAG added 30 min post infection and (G) 17AAG added 240 min post infection. Panels A–D are from the same experiment, panels E–F are from another experiment. Each experiment was repeated at least 3 times.
As cellular oligopeptides are believed to be nearly completely degraded in a few seconds (Reits et al. 2004), the kinetic difference between blocking protein synthesis and blocking surface delivery with BFA should be minimal, and difficult to detect when sampling cells at 30 min intervals. This was indeed the case for NP366-374-and PB1-F262-70- Db complexes, (Figure 2B, 2C, 2E, 2F). PA224-233-Db complexes behaved in a completely different and highly surprising manner.
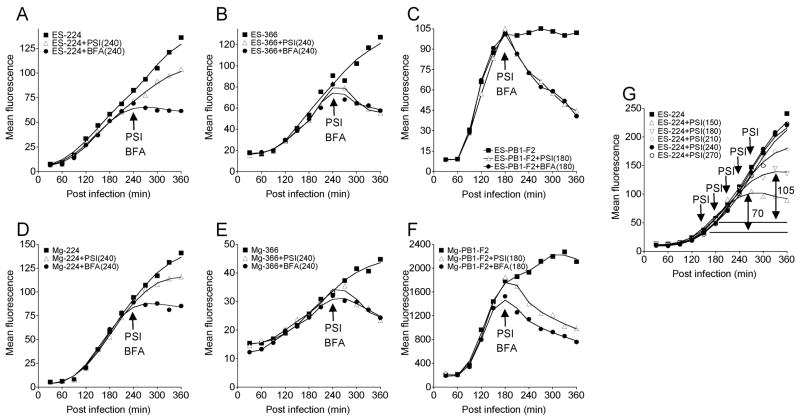
Figure 2.
Kinetics of Db-peptide presentation on L-Db cells following rVV infections.
Cell surface Db-peptide amounts were measured on L-Db cells infected with rVVs expressing ES-224 (A), ES-366 (B), ES-PB1-F2 (C), Mg-224 (D), Mg-366 (E) or Mg-PB1-F2 (F). Infected cells were treated with protein synthesis inhibitors (PSI) or BFA at times indicated by arrows, (G) Surface Db-PA224 amounts on cells infected with ES-224 followed by addition of protein synthesis inhibitors at various times post infection. Addition of PSI at 150 min and 180 min reveals PA224-233 pools of 70 and 105 fluorescence units, respectively. Each experiment was repeated at least 3 times.
The delivery of PA224-233 complexes to the cell surface continued at nearly the same rate up to two hours following addition of protein synthesis inhibitors (PSI) (Figure 2A, 2D).
A trivial explanation for this finding is that the translation of PA224-233 is resistant to the protein synthesis inhibitors we utilized. Based on the mode of action of protein synthesis inhibitors, we considered this very unlikely (particularly because a number of potent protein synthesis inhibitors [anisomycin, cycloheximide, emetine, pactamycin] used individually gave similar results). We eliminated this possibility by treating cells with protein synthesis inhibitors at the start of infection and showing that Db-PA224-233 complex formation was completely inhibited (not shown). Rather, these findings demonstrate the formation of a pool of PA224-233 peptide or PA224-233-Db complexes that feeds the class I pathway for hours following protein synthesis shutdown.
The kinetics of pool formation is most consistent with the former possibility. When we blocked protein synthesis at increasing times following infection, the size of the pool increased with time of expression. In the interval between 150 and 180 min the pool increased from 70 flow units to 105 units (Figure 2G). Notably, this is twice the amount of complexes delivered to the cell surface over the identical interval. Because the Vmax of cell surface complex delivery has been reached by 150 min, the large increase in the size of the pool over the next 30 min is difficult to reconcile with a pool of Db-PA224-233 complexes in the secretory pathway if the size of this pool is directly related to the rate of ER export and cell surface complex delivery.
These experiments demonstrate that PB1-F262-70 and NP366-374 demonstrate kinetic behavior completely consistent rapid degradation unless bound to MHC class I molecules, while PA224-233 demonstrates kinetics not previously observed (for either proteins or minigene products) that are consistent with it forming a substantial pool that forms a steady source of peptides for hours after shutting off its biosynthetic source.
PA224-233 peptide cellular pool is dependent on cellular HSP90 activity
The recent report from Kunisawa and Shastri (2003) demonstrating that HSP90α associates with proteolytic fragments of KOVAK, a misfolded cytosolic model antigen, prompted us to examine the effect of cell permeant small molecule inhibitors of HSP90. These inhibitors are among the most selective inhibitors available to probe intracellular pathways. All of the inhibitors used target the NH2- terminal ATP binding domain of HSP90 family members. Two of the inhibitors used, 17AAG and 17DMAG, are 2nd generation geldanamycin analogs with higher potency than the parent, while the third, radicicol is chemically distinct from geldanamycin, and so would be expected to have a distinct off-target profile. Cells were treated with the given HSP90 inhibitor at 30 min post infection to allow viral penetration to occur and minimize effects on viral gene expression (Figure 3). Treatment with each of the inhibitors inhibited Db-PA224-233 cell surface complex expression by approximately 20–30%. This is not specific for PA224-233 as NP366-374 and PB1-F262-70 decreased similarly (data not shown), and probably reflects a requirement for active HSP90 for maximal VV gene expression.
To assess the effect of HSP90 inhibitors on PA224-233 pool formation in this experiment, we incubated cell with protein synthesis inhibitors at 240 min. This revealed that each of the HSP90 inhibitors tested eliminated the PA224-233 peptide pool. Importantly, addition of HSP90 inhibitors simultaneously with PSI, failed to diminish complex expression (panels E-G), indicating that maintenance of the peptide pool does not require active HSP90.
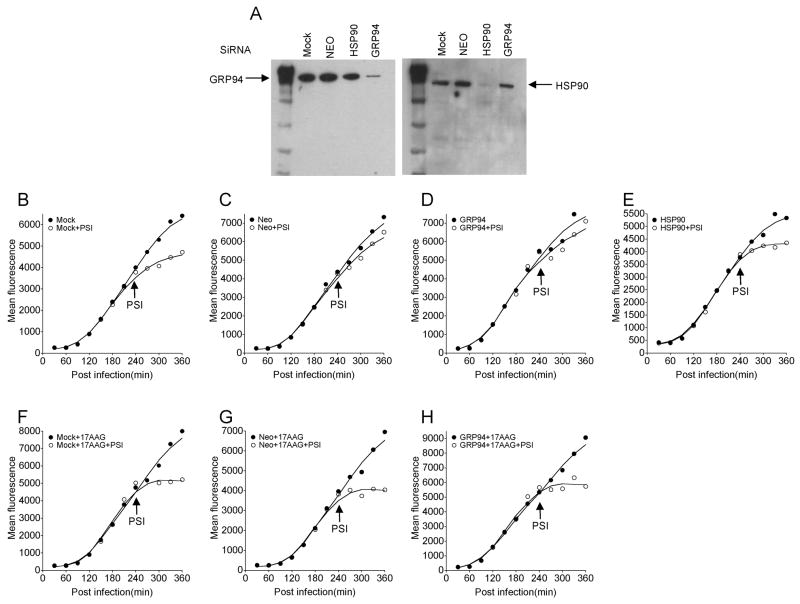
The Hsp90 family consists of cytosolic Hsp90α Hsp90β, the ER paralog GRP94, and mitochondrial Trap-1, all of which are sensitive to the panel of HSP90 inhibitors. To identify the relevant targets involved in PA224-233 peptide pool formation we transfected L-Db cells with siRNAs specific for HSP90 (α&β) (the siRNA sequences do not distinguish between these highly similar genes), GRP94 or Neomycin (negative control). Cells were re-transfected 4 days after the initial transfection and used 4 days later. This resulted in greater than a 10-fold specific reduction in HSP90 and GRP94 as determined by immunoblotting of total cell lysates (Figure 4A). These cells were infected with the rVV expressing ER targeted PA224-233 and tested for PA224-233 pool formation as described above by the effect of protein synthesis inhibition on continued complex expression (Figure 4B, 4C). This revealed that mock transfected cells or cells transfected with control siRNA or with GRP94 siRNA exhibited a large PA224-233 pool that remained completely sensitive to HSP90 inhibition by 17AAG. By contrast, the pool was nearly completely depleted by HSP90 (α&β) knockdown.
Figure 4.
PA224-233 Pool formation is dependent on HSP90 expression
Cell surface Db-PA224-233 complexes were determined by flow cytometry at indicated times after infection with VV-ES-224. Cells were transfected 8 d and repeated transfection 4d earlier with siRNA to knock down HSP90 or GRP94. (A) Knock down of HSP90 and GRP94 in L-Db cells following siRNA transfection was confirmed by immunoblotting. B–H. Kinetic flow cytometry of infected cells transfected with siRNA indicated (with Neo siRNA as a control siRNA transfection) or non-transfected. In F–H cells were treated with 17AAG at 30 min post-infection to demonstrate that pool remained sensitive to HSP90 inhibition. SiRNA knock down followed by kinetics experiment was repeated 2 times.
These data demonstrate that the PA224-233 pool is formed independently of GRP94 but requires functional cytosolic HSP90.
PA224-233 physically associates with a HSP90 inhibitor sensitive receptor in living cells
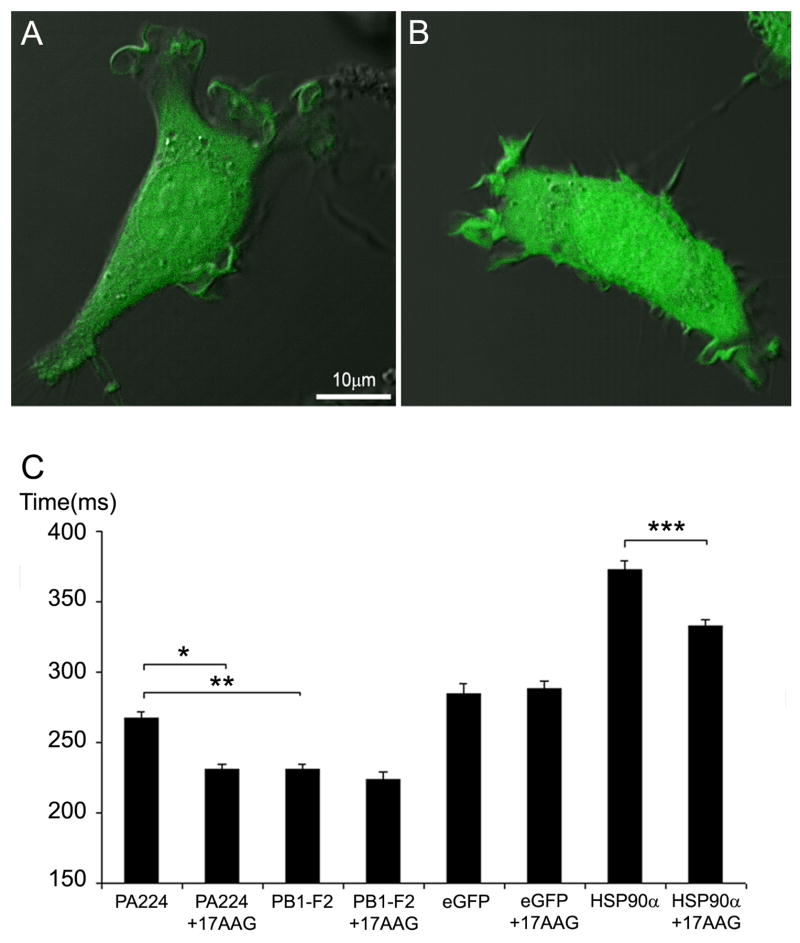
These findings were consistent with the idea that PA224-233 physically associates with HSP90 or a HSP90 sensitive client. To investigate the possible interaction of PA224-233 with a cytosolic receptor we microinjected L-Db cells with synthetic PA224-233 or PB1-F262-70 (as a control) with a COOH-terminal Lys residue conjugated with fluorescein at its ε-NH2-group (PA224-233KFl, PB1-F262-70KFl). Each peptide distributed evenly between cytosolic and nuclear compartments (Figure 5A). This contrasts with the findings of Reits et al. (2003) that microinjected peptides typically concentrate in the nucleus due to their protection from proteolysis by chromatin. In other experiments (unpublished observations), we observed that another microinjected peptide with Lys-fluorescein (SIINFEKFLL) demonstrates nuclear localization. It is likely that these differences reflect chemical differences in the peptides used in these disparate studies that govern their interaction with cellular constituents.
Figure 5.
PA224-233 cytosolic diffusion is retarded in a HSP90-dependent manner.
Synthetic peptides with an additional COOH-terminal Lys-ε-FITC were microinjected into L-Db cells. (A) PA224-233 peptide cellular distribution. (B) PB1-F262-70 peptide cellular distribution. (C) Diffusion rates measured by FRAP for microinjected peptides compared to control GFP and mCherry-HSP90α Open bars represent cells treated with 17-AAG ~20 min prior to microinjection. FRAP analysis was performed in at least 50 different cells for each condition shown. Error bars represent standard error of the mean. Starred pairs represent significant differences between conditions (p values < .0001).
We measured the mobility of cytosolic PA224-233KFl by fluorescence recovery after photobleaching (FRAP) analysis and compared its properties to PB1-F262-70KFl as a control (Figure 5). PA224-233KFl demonstrated a substantially slower diffusion co-efficient than PB1-F262-70KFl. Critically, treating cells with 17AAG for 20 min prior to microinjection did not affect the diffusion coefficient of PB1-F262-70KFL but increased the diffusion co-efficient of PA224-233KFl to match that of PB1-F262-70KFL. It was of interest to compare the mobility peptides to full length proteins. Consistent with Reits et al. (2003), the mobility of PB1-F262-70KFl was ~20% greater than GFP, which in turn was substantially more mobile than a HSP90α-GFPcherry construct (Figure 5). Importantly while GFP mobility was unaffected by 17AAG, HSP90α-GFPcherry demonstrated a substantial increase in mobility (though less mobile than GFP, as expected form its larger radius of gyration), providing the initial FRAP evidence for the physical interaction of a HSP90 family member peptide binding site. This contrasts with HSP90β, whose mobility is reported to be insensitive to HSP-90 inhibitors (Picard et al., 2006).
These data demonstrate that 17AAG detectably affects HSP90 mobility in living cells and are consistent with the conclusion that PA224-233 associates with a cytosolic partner that requires HSP90 peptide binding activity: either HSP90 itself, or a client that is rather rapidly altered by HSP90 inhibition.
PA224-233 peptide minigene cross-primes CD8+ T cells in B6 mice
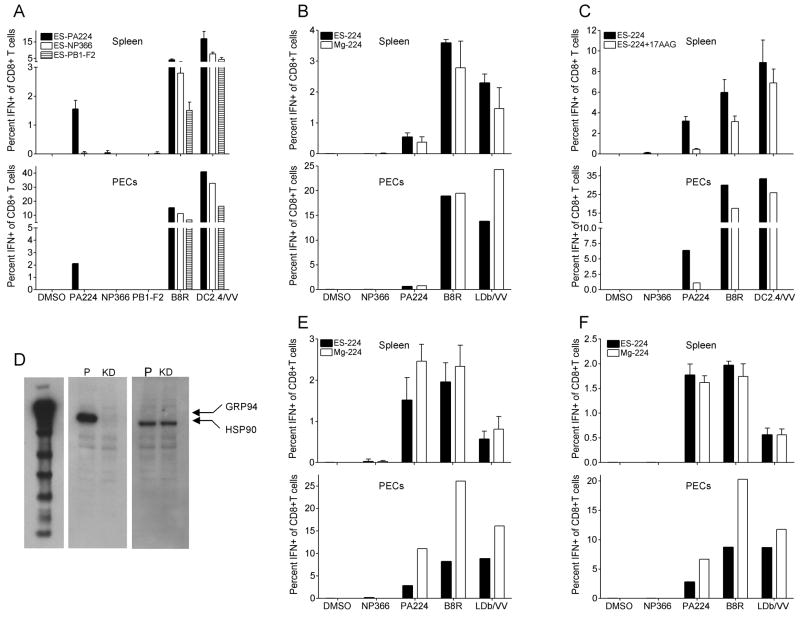
The observation that PA224-233 accumulates in a substantial stable pool in L-Db cells prompted the question of whether the pool can be tapped for cross-priming in vivo. To address this question, we immunized B6 mice via intraperitoneal (i.p.) injection with rVV infected L929 cells (i.e. the original non-transfected cells that do not express Db) that were irradiated with UV light to destroy residual viral infectivity. We determined the frequency of local (peritoneal exudate cells [PEC]) and splenic responding CD8+ T cells by intracellular staining for INF-γ following incubation of antigen presenting cells (APCs) with peptides corresponding to the cognate minigene product or to a peptide corresponding to residues B8R20-27, the immunodominant determinant in VV infections in B6 mice. For an estimate of overall anti-VV responses we also stimulated responding CD8+ T cells ex vivo with VV infected DC2.4 or L-Db cells.
The data in Figure 6A clearly show that L929 cells expressing NP366-374 and PB1-F262-70 as ER targeted minigene products fail to elicit CD8+ T cells responses that are substantially above background values obtained by stimulating with an irrelevant peptide. By contrast, cells expressing ER targeted PA224-233 elicited robust PA224-233 specific responses (Figure 6A, 6B). Note that cells expressing the non-immunogenic minigene products elicited vigorous B8R20-27 and total VV-specific CD8+ T cell responses, indicating that the non- immunogenicity of their minigene products cannot be attributed to diminished infectivity or other factors that limit the cross-priming capacity of these rVVs.
Figure 6.
PA224-233 minigene product induces robust cross-priming
Mice were immunized intraperitoneally (i.p.) with UV irradiated L929, T2 or HEK293 cells infected for 6h with rVV-expressing the minigene products indicated. Six d later, splenocytes and PECs were tested for IFN-γ accumulation following restimulation with indicated peptides or VV- infected DC2.4 or L-Db cells. (A) Cross priming using L929 cells infected with ES-224(black), ES-366(white) and ES-PB1-F2 (striped). (B) T2 cells infected with ES-224(black) or Mg-224(white). (C) ES-224 cross priming using L929 cells treated with 17AAG (white), or untreated (black). (D) HSP90 and GRP94 amounts in HEK293 cells were measured by western blot with specific antibodies as indicated. (E) Cross priming using HEK293 cells with GRP94 stably knocked down infected with ES-224(black) or Mg-224(white). (F) Cross priming using parental HEK293 cells stably transfected with scramble sequence infected with ES-224(black) or Mg-224(white). Values are subtracted from background obtained from wells receiving no peptides and are expressed as mean ± SE of three individual mice in each group for splenic responses. For peritoneal responses, PECs were pooled. Each experiment was repeated at least 2 times.
To determine the TAP dependence of cross-priming, we performed a similar experiment in which we replaced L929 cells with T2 cells (again lacking Db). Both cytosolic and ER targeted PA224-233 minigenes cross primed to a similar extent for PA224-233-specific responses (Figure 6B). (Note that it is likely that substantial amounts of ER targeted peptides persist in the cytosol, due either to inefficient targeting of such small proteins to the ER, or the recycling of excess peptides from the ER to the cytosol (Koopmann et al., 2000; Roelse et al., 1994)). The TAP-independence of cytosolic PA224-233 cross-priming is in agreement with the finding that cytosolic HSP90 but not GRP94 is involved in peptide pool formation (Figure 4). We more directly tested the involvement of functional HSP90 in PA224-233 cross-priming by treating donor cells with 17AAG during rVV infection prior to their introduction into B6 mice (Figure 6C). This reduced PEC and splenic anti-PA224-233 responses by ~85%, while reducing B8R20-27 or overall VV-specific response by 15 to 50%..
Next, we examined the requirement for GRP94 in cross-priming by using human HEK293 cells stably transfected with GRP94-targeting siRNA (control HEK293 cells were stably transfected with the same sequence but scrambled). Via immunoblotting, GRP94 is undetectable in knockdown cells while HSP90 amounts are similar between control and knockdown cells (Figure 6D). Despite the absence of GRP94, both rVVs expressing cytosolic and ER-targeted PA224-233 were effective at cross-priming for PA224-233-, B8R20-27-, and total VV-specific CD8+ T cell responses (Figure 6E, 6F).
Cross-Priming is based on the presence of a large Db –independent pool
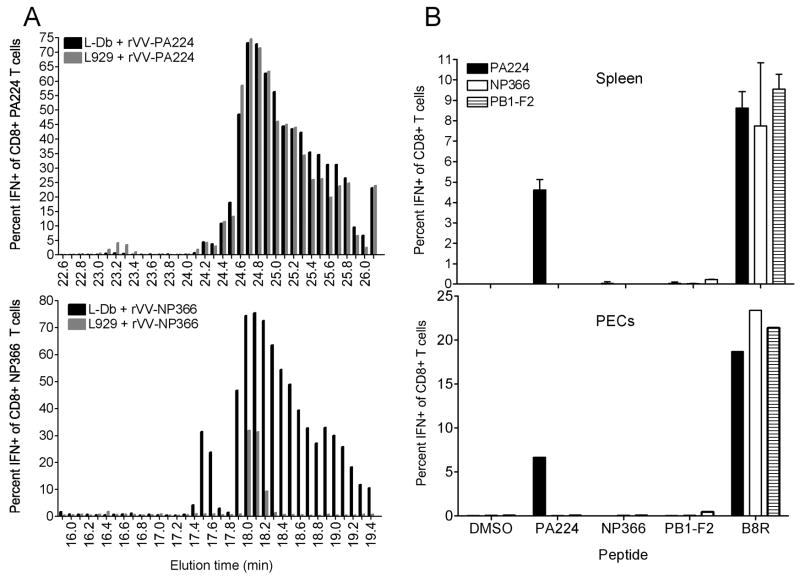
To characterize the cross-priming pool of PA224-233 in L929 cells, we measured antigenic peptides recovered from HPLC fractionated low molecular weight acid soluble material from homogenized cells infected with VV-encoded cytosolic PA224-233, or as a negative control, NP366-374. Activity was measured using CD8+ T cell populations expanded in vitro to each peptide. T cell populations demonstrated highly similar picomolar sensitivity to synthetic peptide (data not shown). Large amounts of NP366-374 were recovered from infected L-Db cells, with recovery of peak antigenic activity in fractions that co-elute with synthetic NP366-374 (Figure 7A). Antigenic activity in peak fractions could be detected at a dilution of 1/12500, while activity in infected L929 cells titrated at least 1000-fold lower amounts. This consistent with the classically reported rapid degradation of peptides lacking a high affinity class I receptor.
Figure 7.
Physical demonstration of the PA224-233 peptide pool in L929 cells.
(A, B) Acid soluble peptides recovered from L-Db or L929 cells infected for 5 h with VV-PA224-233 or VV-NP366-374 were separated by HPLC and fractions were tested for antigenic activity using CD8+ T cell lines raised to each peptide by ICS for IFN-γ.
(C) Cross-priming by rVV-minigene L-Db cells infected with ES-224(black), ES-366(white) and ES-PB1-F2 (striped) as described in Figure 6 legend.
PA224-233 was recovered from infected L-Db cells in even greater amounts than NP366-374 (Figure 7A). Peak activity co-eluted with synthetic PA224-233, but considerable amounts of activity spilled into slower eluting fractions. As with NP366-374, peak activity titrated to greater than 1/12,500. Most remarkably, essentially identical amounts of PA224-233 were recovered from the same fractions of infected L929 cells. This provides physical description of a large pool of PA224-233 that exists independently of Db. Indeed, Db expression has no discernible influence on the pool size, suggesting that only a small fraction of PA224-233 is present complexed with Db in infected L-Db cells.
All of our evidence points to the conclusion that the PA224-233 pool is present in the cytosol. We consider it extremely unlikely that the pool is present as a complex with a class Ia or Ib molecule expressed by L929 cells (particularly because the pool also exists in T2 cells, which in addition to lacking TAP, being of human origin, express a completely different set of class I molecules).
Based on the unprecedented nature of the pool, however, we thought it prudent to perform an additional experiment to characterize the possible contribution of class I protected peptides to cross-priming in the VV-system we employ.
To this end we immunized mice with L-Db cells infected with VV-NP366-374, VV-PB1-F262-70, or as a positive control VV-PA224-233. While cells expressing PA224-233 cross-primed as above, cells expressing the other minigene products did not (Figure 7B). Thus, despite the generation of very large number of Db complexed with NP366-374 or PB1-F262-70 peptides, protection by class I is not sufficient to enable cross-priming, implying that PA224-233 cross-priming occurs in a class I independent manner in the absence or even the presence of Db.
Discussion
Proteasomal protein cleavage generates oligopeptides of between 3 and 32 residues that are degraded by highly active cytosolic endopeptidases and aminopeptidases (Kisselev et al., 1998; Reits et al., 2004). The half-life of oligopeptides in this aggressively degradative environment is believed to be ~10 sec. Inasmuch as proteasome generated peptides are functionless, potentially dangerous agents that can interfere with cellular functions (including impeding the interaction of molecular chaperones with their substrates), the rapid removal of peptides and reutilization of amino acids is logical.
Despite this, a considerable literature has accrued regarding the potential role of GRP94 and other abundant molecular chaperones in protecting peptides from degradation and promoting peptides immunogenicity upon transfer to pAPCs. There is compelling evidence that molecular chaperones have a bona fide role in activating innate immune receptors (Baker-LePain et al., 2002; Oizumi et al., 2007). The function of molecular chaperones, however, in specifically binding and delivering oligopeptides generated from biosynthesized proteins to pAPCs is controversial. Molecular chaperones exhibit low affinity for their substrates (on the order of 0.1 mM), consistent with their need to transiently interact with clients to promote folding (Flynn et al., 1989; Flynn et al., 1991; Fourie et al., 1994; Ying and Flatmark, 2006). With such a low affinity, peptides would be protected only fleetingly, achieve low steady state amounts in living cells, and dissociate rapidly upon release from cells and delivery to pAPCs. While there is limited evidence for recovery of defined peptides from GRP94 purified from cells, others have found only minute quantities of peptides bound to GRP94, and questioned the peptide chaperoning function of GRP94 in antigen presentation (Demine and Walden, 2005; Nicchitta et al., 2004; Wallin et al., 2002; Ying and Flatmark, 2006). Moreover, molecular chaperones are only weakly immunogenic relative to cell associated immunogens, even when loaded in vitro to the maximal extent attainable with synthetic peptides. Even non-cell associated heat aggregated viruses or proteins are more immunogenic than the most potent HSP preparations reported (Cho et al., 2003; Speidel et al., 1997).
On top of this, we and others have provided evidence strongly supporting a central role for intact proteins in cross-priming (Basta et al., 2005; Donohue et al., 2006; Freigang et al., 2007; Gasteiger et al., 2007; Norbury et al., 2004; Shen and Rock, 2004; Wolkers et al., 2004). Cellular oligopeptides generated via signal peptidase liberation, or following synthesis as cytosolic or ER-targeted minigene products display little to no cross-priming activity (Gasteiger et al., 2007; Norbury et al., 2004; Serna et al., 2003). The poor cross-priming capacity of minigene products, consistent with the lack of cross-presentation of minigene products observed in vitro by Serna et al. (2003), can be explained by their rapid degradation. Like naturally processed antigenic peptides, recovery of minigene encoded minimal peptides from cells is heavily dependent on the presence of a high affinity class I receptor (Anton et al., 1997). More recently, we precisely quantitated cell surface Kb-Ova257-264 complex expression in cells infected with VV-Ova257-264 using the TCR-like mAb 25-D1.16 (Princiotta et al., 2003). Upon abrogation of protein synthesis, delivery of Kb-Ova257-264 complexes to the cell surface ceased rapidly, consistent with the rapid degradation of Ova257-264.
In the present study we extend these findings to three additional defined peptides expressed as cytosolic or ER targeted minigenes. Using a novel panel of TCR-like Abs to measure cell surface Db-peptide complex expression, we found that for two of the peptides, NP366-374 and PB1-F262-70, the kinetics of shut down of complex delivery to the cell surface after inhibiting protein synthesis was virtually identical to those observed after BFA blockade of Db-peptide complex egress from the early secretory pathway, providing unequivocal evidence for the absence of a protected peptide pool capable of supplying the class I pathway. Crucially, these peptides also failed to exhibit substantial cross-priming activity when expressed in cells lacking Db.
In distinct contrast, while BFA rapidly abrogated cell surface delivery of PA224-233 complexes, delivery continued for hours following abrogation of protein synthesis, indicative of a sizable intracellular pool. This pool is independent of Db expression as demonstrated by the cross-priming activity of PA224-233 minigene products in L929 cells and also the recovery of large amounts of PA224-233 via acid extraction from the same cells. The TAP-independence of PA224-233 minigene product cross-priming points to a cytosolic location for the PA224-233 pool. Consistent with this interpretation, the PA224-233 pool and its cross-priming activity is abrogated by selectively interfering with cytosolic HSP90 function or expression, and is unaffected by ER GRP94 knock down. Further, we show that upon microinjection, a fluorescent derivative of PA224-233 diffuses more slowly in the cytosol than an equivalent form of PB1-F262-70, an effect that is rapidly abrogated by addition of a HSP90 inhibitor. This is consistent with the binding of PA224-233 to a HSP90 dependent cytosolic partner that retards its diffusion.
We were unable to recover PA224-233 using HSP90 specific antibodies to purify HSP90 from cytosol recovered from semi-intact cells or TX100 lysates (data not shown). Kunisawa and Shastri (2006) recovered numerous fragments of KOVAK from HSP90 antibody pull downs. It is uncertain, however, whether these peptides directly associate with HSP90, because HSP90 participates in large complexes containing other client binding-molecular chaperones. Two findings favor the idea that PA224-233 is only indirectly influenced by HSP90 activity. First, the PA224-233 pool persists for hours following protein synthesis inhibition, suggesting a high affinity interaction with its chaperone. Second, adding HSP90 inhibitors at the time of protein synthesis blockade did not deplete the pre-existing PA224-233 pool. This strongly suggests that pool formation is not directly based on association of PA224-233 with HSP90, because HSP90 inhibitors would be predicted to either cause the release of PA224-233 from HSP90 and rapid proteolysis, or the Ub-dependent degradation of HSP90 bound PA224-233 (Isaacs et al., 2003). In support of the rapid action of HSP90 inhibitors, we show that 17AAG rapidly increases the diffusion of HSP90α in living cells.
Notably, inactivation or depletion of HSP90 and GRP94 had little effect on the cross-priming capacity of B8R20-27 specific CD8+ T cells or polyclonal anti-VV CD8+ T cell responses. It is possible that other molecular chaperones are involved in cross-priming for these natural gene products. We believe it is unlikely, however, that this involves chaperoned peptides generated from these proteins.
Our findings demonstrate that oligopeptides are capable of cross-priming if they are spared from proteolysis. Thus, the inability of other minigene products or efficiently liberated peptides (such as signal sequences or oligopeptides released from Ub-fusion proteins by Ub-hydrolases) to cross-prime can be directly attributed to their evanescence, and not the general inability of oligopeptides to survive transfer to, and further processing in, pAPCs. This strongly implies that the poor immunogenicity of rapidly degraded model proteins, and presumably natural DRiPs as well, can be attributed to the rapid degradation of most proteasomal products, which simply do not achieve the steady state quantities needed for cross-priming. On the other hand, our finding that a subset of peptides is resistant to rapid proteolysis, would favor the possible contribution of to CD8+ T cell priming via oligopeptide transfer to pAPCs through gap junctions, which has been elegantly shown to function cultured cells by Neefjes and colleagues (Neijssen et al., 2005; Neijssen et al., 2007).
Materials and Methods
Mice
All experiments used 6- to 10-wk-old female C57BL/6J mice purchased from Taconic Farms. Mice were housed in the animal care facility at National Institute of Allergy and Infectious Diseases (NIAID) under specific pathogen-free conditions, maintained on standard mouse chow and water provided ad libitum and cared for with strict adherence to animal study.
Peptides, MHC complexes, reagents
Peptides were procured and characterized by the Biologic Resource Branch, NIAID (Rockville, MD). In each case, substances with the predicted mass constituted >95% of the material analyzed. Peptides were dissolved in DMSO at 10 mM and stored at −20°C. FITC labeled peptides (LSLRNPILV[K-ε-fluorescein]-OH, PA224-233 or SSLENFRAYV[K-ε-fluorescein]-OH, PB1-F262-70) were dissolved in DMSO at a concentration of 10 μg/μl and kept in −20°C until use. Immediately before microinjection, stock peptide solution was diluted to 100 ng/μl by in Dulbecco’s P with calcium and magnesium(DPBS). Biotinylated MHC monomers were provided by the NIAID tetramer facility (Emory University, Atlanta, GA). Cbz-Leu-Leu-Leucinal (zLLL), lactacystin were from Sigma (St. Louis, MO). Abs to hsp90 (rat IgM Ab 1R2D12p90), and grp94 (rat IgG2a Ab 9G10) were from StressGen (Victoria, British Columbia, Canada)
Cell culture and siRNA transfections
L929 and L-Db (L929 stably transfected with Db molecule) cells were cultured in DMEM containing 10% FBS, EL-4 thymoma cells (H-2b), TAP deficient T2 and T2-Db (T2 cells stably transfected with Db molecule) were cultured in RPMI 1640 containing 10% FBS, DC-like DC2.4 cells (H-2b) were cultured in IMDM containing 10% FBS. To generate stable GRP94 siRNA expressing HEK293 cell lines the sequence 5′-GGCUCAAGGACAGAUGAUGtt-3′ was cloned into the A pSilencer 2.0-U6 vector (Ambion). The pSilencer 2.0-U6-GRP94 siRNA and a control, non-targeting pSilencer 2.0-U6 siRNA vector (scrambled, obtained from Ambion) were transfected into HEK293 cells using Lipofectamine 2000 following the manufacturers protocol. Cells were selected thirty six hours post-transfection by addition of puromycin (1 microgram/ml) to the media. Puromycin resistant clones (both GRP94 siRNA and non-targeting siRNA) were expanded and clones screened for knockdown efficiency by immunoblotting, using GRP94 antibody DU120. Clones displaying greater than 90% knockdown were selected. Puromycin-resistant clones from the non-targeting siRNA were obtained in parallel and screened for normal GRP94 expression, by immunoblotting with DU120. HEK293 cells were cultured in DMEM containing 10% FBS. Media and FBS were all from Invitrogen Life Technologies. Transient siRNA transfections were carried out by transfecting 1–2×106 L-Db cells in V solution using Amaxa nucleofactor using T-24 program. The target sequences for HSP90 knock down were 5′-UUUCAGAGCUGUUGCGGUAUU-3′ and 5′-AGAGGUUGAUAGAGCGUUUUU- 3′ for HSP90α and 5′ –GGACAAGAUUCGAUAUGAGUU-3′ and 5′-GGUGUUAUGUAUUGUGGUUUU-3′ HSP90β (these sequences have been picked from Dharmacon on target sequences) used in a mixture of 10nM each. SiRNA specific for GRP94 and Neo were 5′-pGGCUCAAGGACAGAUGAUGtt and pAAUGAACUGCAGGACGAGGCAtt used at 10nM each. All siRNAs were purchased from Dharmacon, Lafayette, CO. For maximal knockdown transfection were repeated after four days and cells were harvested for experiments four days after the second transfection. Knockdown effectiveness was determined by immunoblotting using GRP94 and HSP90 specific antibodies.
rVVs infections
rVVs containing minigenes encoding PA224-233, NP366-374 and PB1-F262-70 peptides in the cytosol or targeted to the ER were generated as described for other minigene expressing rVVs (Lev et al., 2002) Cells were infected at a multiplicity of infection (MOI) of 10 for 20 min at 37°C with mixing every few minutes in balanced salt solution containing 0.1% bovine serum albumin (BSS/BSA). Cells were then incubated at 37°C in growth media for the remainder of the assay in the presence of 25 μg/ml cytosine arabinoside (Sigma, St. Louis, MO). Protein synthesis was inhibited by the addition of cycloheximide and emetine (Sigma, St. Louis, MO) to a final concentration of 25 μg/ml each. Brefeldin A (BFA) (Sigma, St. Louis, MO) was added to a final concentration of 10 μg/ml.
Phage Ab Generation
Selection of phage antibodies on biotinylated MHC complexes was preformed, as described (Lev et al., 2002). Briefly, a large human Fab library containing 4 × 1010 different Fab clones was used for the selection with decreasing amounts of biotinylated MHC-peptide complexes (25μg for the first round and 5μg for the following rounds). Streptavidin-coated magnetic beads (200 μl for the first round of selection, and 100 μl for the following rounds) were incubated with Phage mixture for 15 min at RT. The beads were washed extensively 12 times with PBS/0.1%Tween +2% milk, and additional two washes with PBS. Bound phages were then eluted with triethylamine (100 mM, 5 min at RT), followed by neutralization with Tris-HCl (1 M, pH 7.4), and used to infect E. coli TG1 cells (at OD600 = 0.5) for 30 min at 37°C. The diversity of the selected antibodies was determined by DNA fingerprinting using a restriction endonuclease (BstNI), which is a frequent cutter of antibody V gene sequences. The Fab DNA of different clones was PCR amplified using the primers pUC-reverse (5′-AGCGGATAACAATTTCACACAGG-3′) and fd-tet-seq (5′-TTTGTCGTCTTTCCAGACGTTAGT-3′), followed by digestion with BstNI (NEB, Beverly, MA) (2 h, 60°C) and analysis on agarose gel electrophoresis (data not shown).
Expression and purification of soluble recombinant Fab Abs
Fab antibodies were expressed and purified, as described. BL21 cells were grown to OD600 = 1.0 and induced to express the recombinant Fab antibody by the addition of 1 mM isopropyl β-D-thiogalactoside (IPTG) for 3 h at 30°C. Proteins were extracted using B-PER solution (Pierce, Rockford, IL), applied onto a pre-washed TALON column (BD, Palo Alto, CA), and bound Fabs were eluted using 0.5 ml of 100 mM imidazole in PBS. The eluted Fabs were dialyzed against PBS (overnight, 4°C) to remove residual imidazole.
ELISA with phage clones and purified Fab antibodies
The binding specificities of individual phage clones and soluble Fabs were determined by ELISA using biotinylated MHC-peptide complexes. ELISA plates (Falcon) were coated 1h at 37°C with BSA-biotin (1 ug/well). The plates were washed, and incubated (O.N at 4°C) with streptavidin (1 μg/well), washed extensively, and further incubated (1 h, RT) with 0.25 μg/well of MHC-peptide complexes. The plates were blocked for 30 min at RT with PBS/2% skim milk and subsequently were incubated for 1 h at RT with phage clones (~109 phages/well) or various concentrations of soluble purified Fab. After washing, the plates were incubated with HRP-conjugated/anti-human Fab antibody (for soluble Fabs) or HRP-conjugated anti-M13 phage (for phage-displayed Fabs). Detection was performed using tetramethylbenzidine reagent (Sigma-Aldrich, St. Louis, MO).
Flow Cytometry
Db-peptide amounts were determined by incubating cells for 30 min on ice with 20ug/ml purified Fab antibody. Cells were then washed and incubated for 20 min with Cy5 conjugated anti-human antibody. Cellular Cy5 amounts were determined on a FACScalibur cytofluorograph (Beckton Dickinson, San Jose, CA) using CellQuest (Beckton Dickinson) software or LSR II (Beckton Dickinson, San Jose, CA) using FACSDiva (BD) software, and data were analyzed using FlowJo (Tree Star, San Carlos, CA) software.
Microscopy and FRAP analysis
Cells were examined and manipulated using a Leica TCS-SP5 DMI6000 with 8000 Hz resonant scanner and 40× 1.25 NA oil objective (Leica Microsystems, Germany). The microscope was equipped a Cube & the Box environmental chamber (Life Imaging Services, Switzerland). Cells were maintained at 37°C in in 95%/5% air/CO2 mixture. Peptides were microinjected into cells using an Eppendorf FemtoJet microinjector system (Eppendorf, NY). Cells were also transfected with eGFP or mCherry-HSP90α plasmids as controls using Amaxa nucleofactor with T-24 program. Prior to FRAP, a rectangular 9 μm × 35 μm area was positioned on a microinjected cell with a bleaching area rectangular 6 μm × 13 μm size placed in the center of the cells. 20 frames in 0.01 second were collected before bleaching was performed for 0.2 second at the maximum laser power of 458, 476 and 488 nm laser lines. Fluorescence recovery was monitored at a collection rate of 100 frames per 0.01 second. At least 50 cells for each condition were measured in more than three independent experiments. FRAP recovery half-times were calculated using Leica Application Suite Advanced Fluorescence software.
Extracting antigenic peptides from rVV-minigene infected cells, HPLC run and measure of CTL responses
5×108 L929, and 1.25 × 108 L-Db cells were infected with rVV- NP366-372 or rVV-PA224-233 at 20 pfu:cell. Cells were harvested three hours post-infection, snap frozen and resuspended in 8ml 0.1% TFA/H2O. After adding 3 mL of 1% TFA/H2O cells were disrupted by >20 strokes in a Dounce homogenizer, sonicated and rotated on wheel for 30 min at 4ºc. The lysate was pelleted by low speed centrifugation, and the supernatant passed through a 0.45μM syringe filter, and then a 3K cut-off filter (Macrosep™, filtron 3K, Cat OD003C36, Pall Filtron Corp. Northborough, MA). The filtrate was speed dried with vacuum to a volume of less then 400μl. The sample was filtered through a 0.45 μM micro-filter that was previously washed with 0.1% TFA solution before use and then directly loaded onto a HPLC C-18 column (Deltapack, Waters). Fractions were collected every tenth of a minute from 10 mins post-injection until 38.8 mins (3 × 96 well plates), and stored at 4ºC. ICS was performed as described for cross priming experiments using HPLC fractions to stimulate CD8+ T cell lines propagated in vitro by stimulation with NP366-374 or PA224-233.
Cross priming experiments with Intracellular cytokine staining (ICS)
L929 or HEK293 cells were infected at 10 MOI with rVVs expressing the IAV derived peptides as minigenes for 6h before UV irradiation. After washing 10 million cells were injected IP into each B6 mouse. Six days later splenic and peritoneal L929 cells from primed animals were suspended in RPMI 1640 medium plus 10% FBS and 10 mM HEPES buffer and seeded at 2 × 106 cells/well in U-bottom, 96-well plates. Peptides were added to wells to a final concentration of 0.5 μM. Cells were incubated initially for 1–2 h at 37°C and then for 3h with brefeldin A (BFA; Sigma-Aldrich, St. Louis, MO) at 10 μg/ml. Cells were then stained with CyChrome anti-CD8α mAb on ice for 30 min, washed, and fixed with 1% paraformaldehyde in PBS at room temperature for 20 min, then further stained with FITC-conjugated anti-IFN-γ in PBS containing 0.1% saponin (Sigma-Aldrich). Stained cells were analyzed on a FACSCalibur (BD Biosciences, Sunnyvale, CA) with live-gate on the CD8+ cells. A total of 150,000 cells were normally acquired and analyzed with FlowJo software (TreeStar, Ashland, OR).
Supplementary Material
Supplemental Figure 1- Specificity of recombinant TCR-like Fab antibodies selected on derived peptide-MHC complexes. (A) Specificity of Fabs shown by ELISA on immobilized H-2Db-peptide complexes. Binding of Fab clones was measured on Db-PA224, Db-NP366, Db-PB1-F262 and a mixture of Db molecules with control peptides. (B) Detection of H-2Db-peptide complexes on the surface of EL4 cells. EL4 cells were loaded with 10μM of specific (Black) or control (gray) IAV derived peptide for 2h at 37ºc. Peptide-loaded cells were then incubated with the specific soluble purified Fab antibodies followed by Cy5 conjugated anti-human antibody. (C) EL4 cells were pulsed for 2h at 37ºc with titrated amounts of peptides. Detection sensitivity of TCR-like fabs was then measured by flow cytometry.
Supplemental Figure 2- Schematic of rVVs. Mg-peptide: peptide is expressed as a cytosolic peptide with an additional initiating Met. ES-peptide. peptide is targeted to the ER by a NH2-termina signal sequence.
Acknowledgments
D. Tokarchick and K. Irvine provided outstanding technical assistance. We are grateful to I. Benhar and R. Azriel (Tel Aviv University) for their assistance during the phage display screening. We are grateful to P. Cresswell (Yale University) and K. Rock (University of Massachusetts Medical School) for their generous gift of T2 and DC2.4 cells, respectively, and J. Lukszo (NIAID) for synthetic peptide characterization.
References
- Andersen PS, Stryhn A, Hansen BE, Fugger L, Engberg L, Buus S. A recombinant antibody with the antigen-specific major histocompatibility complex-restricted specificity of T cells. Proc Natl Acad Sci USA. 1996;93:1820–1824. doi: 10.1073/pnas.93.5.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anton LC, Yewdell JW, Bennink JR. MHC class I-associated peptides produced from endogenous gene products with vastly different efficiencies. Journal of Immunology. 1997;158:2535–2542. [PubMed] [Google Scholar]
- Baker-LePain JC, Sarzotti M, Fields TA, Li CY, Nicchitta CV. GRP94 (gp96) and GRP94 N-terminal geldanamycin binding domain elicit tissue nonrestricted tumor suppression. J Exp Med. 2002;196:1447–1459. doi: 10.1084/jem.20020436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basta S, Stoessel R, Basler M, van den Broek M, Groettrup M. Cross-presentation of the long-lived lymphocytic choriomeningitis virus nucleoprotein does not require neosynthesis and is enhanced via heat shock proteins. J Immunol. 2005;175:796–805. doi: 10.4049/jimmunol.175.2.796. [DOI] [PubMed] [Google Scholar]
- Chen W, Pang K, Masterman KA, Kennedy G, Basta S, Dimopoulos N, Hornung F, Smyth M, Bennink JR, Yewdell JW. Reversal in the immunodominance hierarchy in secondary CD8+ T cell responses to influenza A virus: roles for cross-presentation and lysis-independent immunodomination. J Immunol. 2004;173:5021–5027. doi: 10.4049/jimmunol.173.8.5021. [DOI] [PubMed] [Google Scholar]
- Cho Y, Basta S, Chen W, Bennink JR, Yewdell JW. Heat-Aggregated Noninfectious Influenza Virus Induces a More Balanced CD8(+)-T-Lymphocyte Immunodominance Hierarchy Than Infectious Virus. J Virol. 2003;77:4679–4684. doi: 10.1128/JVI.77.8.4679-4684.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Haard HJ, van Neer N, Reurs A, Hufton SE, Roovers RC, Henderikx P, de Bruine AP, Arends JW, Hoogenboom HR. A Large Non-immunized Human Fab Fragment Phage Library That Permits Rapid Isolation and Kinetic Analysis of High Affinity Antibodies. J Biol Chem. 1999;274:18218–18230. doi: 10.1074/jbc.274.26.18218. [DOI] [PubMed] [Google Scholar]
- Demine R, Walden P. Testing the role of gp96 as peptide chaperone in antigen processing. Journal of Biological Chemistry. 2005 doi: 10.1074/jbc.M501233200. [DOI] [PubMed] [Google Scholar]
- Denkberg G, Reiter Y. Recombinant antibodies with T-cell receptor-like specificity: novel tools to study MHC class I presentation. Autoimmun Rev. 2006;5:252–257. doi: 10.1016/j.autrev.2005.07.004. [DOI] [PubMed] [Google Scholar]
- Donohue KB, Grant JM, Tewalt EF, Palmer DC, Theoret MR, Restifo NP, Norbury CC. Cross-priming utilizes antigen not available to the direct presentation pathway. Immunology. 2006;119:63–73. doi: 10.1111/j.1365-2567.2006.02406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenlohr LC, Huang L, Golovina TN. Rethinking peptide supply to MHC class I molecules. Nat Rev Immunol. 2007;7:403–410. doi: 10.1038/nri2077. [DOI] [PubMed] [Google Scholar]
- Falk K, R”tzschke O, Rammensee HG. Cellular peptide composition governed by major histocompatibility complex class I molecules. Nature. 1990;348:248–251. doi: 10.1038/348248a0. [DOI] [PubMed] [Google Scholar]
- Flynn GC, Chappell TG, Rothman JE. Peptide binding and release by proteins implicated as catalysts of protein assembly. Science. 1989;245:385–390. doi: 10.1126/science.2756425. [DOI] [PubMed] [Google Scholar]
- Flynn GC, Pohl J, Flocco MT, Rothman JE. Peptide-binding specificity of the molecular chaperone BiP. Nature. 1991;353:726–730. doi: 10.1038/353726a0. [DOI] [PubMed] [Google Scholar]
- Fourie AM, Sambrook JF, Gething MJ. Common and divergent peptide binding specificities of hsp70 molecular chaperones. J Biol Chem. 1994;269:30470–30478. [PubMed] [Google Scholar]
- Freigang S, Eschli B, Harris N, Geuking M, Quirin K, Schrempf S, Zellweger R, Weber J, Hengartner H, Zinkernagel RM. A lymphocytic choriomeningitis virus glycoprotein variant that is retained in the endoplasmic reticulum efficiently cross-primes CD8(+) T cell responses. Proc Natl Acad Sci U S A. 2007;104:13426–13431. doi: 10.1073/pnas.0704423104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasteiger G, Kastenmuller W, Ljapoci R, Sutter G, Drexler I. Crosspriming of Cytotoxic T-cells Dictates Antigen Requisites for MVA Vector Vaccines. J Virol. 2007 doi: 10.1128/JVI.00903-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs JS, Xu W, Neckers L. Heat shock protein 90 as a molecular target for cancer therapeutics. Cancer Cell. 2003;3:213–217. doi: 10.1016/s1535-6108(03)00029-1. [DOI] [PubMed] [Google Scholar]
- Javid B, MacAry PA, Lehner PJ. Structure and Function: Heat Shock Proteins and Adaptive Immunity. 2007:2035–2040. doi: 10.4049/jimmunol.179.4.2035. [DOI] [PubMed] [Google Scholar]
- Kisselev AF, Akopian TN, Goldberg AL. Range of sizes of peptide products generated during degradation of different proteins by archaeal proteasomes. Journal of Biological Chemistry. 1998;273:1982–1989. doi: 10.1074/jbc.273.4.1982. [DOI] [PubMed] [Google Scholar]
- Koopmann JO, Albring J, Huter E, Bulbuc N, Spee P, Neefjes J, Hammerling GJ, Momburg F. Export of antigenic peptides from the endoplasmic reticulum intersects with retrograde protein translocation through the Sec61p channel. Immunity. 2000;13:117–127. doi: 10.1016/s1074-7613(00)00013-3. [DOI] [PubMed] [Google Scholar]
- Kunisawa J, Shastri N. Hsp90alpha chaperones large C-terminally extended proteolytic intermediates in the MHC class I antigen processing pathway. Immunity. 2006;24:523–534. doi: 10.1016/j.immuni.2006.03.015. [DOI] [PubMed] [Google Scholar]
- Lev A, Denkberg G, Cohen CJ, Tzukerman M, Skorecki KL, Chames P, Hoogenboom HR, Reiter Y. Isolation and characterization of human recombinant antibodies endowed with the antigen-specific, major histocompatibility complex-restricted specificity of T cells directed toward the widely expressed tumor T-cell epitopes of the telomerase catalytic subunit. Cancer Res. 2002;62:3184–3194. [PubMed] [Google Scholar]
- Neijssen J, Herberts C, Drijfhout JW, Reits E, Janssen L, Neefjes J. Cross-presentation by intercellular peptide transfer through gap junctions. Nature. 2005;434:83–88. doi: 10.1038/nature03290. [DOI] [PubMed] [Google Scholar]
- Neijssen J, Pang B, Neefjes J. Gap junction-mediated intercellular communication in the immune system. Progress in biophysics and molecular biology. 2007;94:207–218. doi: 10.1016/j.pbiomolbio.2007.03.008. [DOI] [PubMed] [Google Scholar]
- Nicchitta CV, Carrick DM, Baker-Lepain JC. The messenger and the message: gp96 (GRP94)-peptide interactions in cellular immunity. Cell Stress Chaperones. 2004;9:325–331. doi: 10.1379/CSC-62.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norbury CC, Basta S, Donohue KB, Tscharke DC, Princiotta MF, Berglund P, Gibbs J, Bennink JR, Yewdell JW. CD8+ T cell cross-priming via transfer of proteasome substrates. Science. 2004;304:1318–1321. doi: 10.1126/science.1096378. [DOI] [PubMed] [Google Scholar]
- Oizumi S, Strbo N, Pahwa S, Deyev V, Podack ER. Molecular and cellular requirements for enhanced antigen cross-presentation to CD8 cytotoxic T lymphocytes. J Immunol. 2007;179:2310–2317. doi: 10.4049/jimmunol.179.4.2310. [DOI] [PubMed] [Google Scholar]
- Picard D, Suslova E, Briand PA. 2-color photobleaching experiments reveal distinct intracellular dynamics of two components of the Hsp90 complex. Experimental cell research. 2006;312:3949–3958. doi: 10.1016/j.yexcr.2006.08.026. [DOI] [PubMed] [Google Scholar]
- Porgador A, Yewdell JW, Deng Y, Bennink JR, Germain RN. Localization, quantitation, and in situ detection of specific peptide-MHC class I complexes using a monoclonal antibody. Immunity. 1997;6:715–726. doi: 10.1016/s1074-7613(00)80447-1. [DOI] [PubMed] [Google Scholar]
- Princiotta MF, Finzi D, Qian SB, Gibbs J, Schuchmann S, Buttgereit F, Bennink JR, Yewdell JW. Quantitating protein synthesis, degradation, and endogenous antigen processing. Immunity. 2003;18:343–354. doi: 10.1016/s1074-7613(03)00051-7. [DOI] [PubMed] [Google Scholar]
- Reits E, Griekspoor A, Neijssen J, Groothuis T, Jalink K, van Veelen P, Janssen H, Calafat J, Drijfhout JW, Neefjes J. Peptide Diffusion, Protection, and Degradation in Nuclear and Cytoplasmic Compartments before Antigen Presentation by MHC Class I. Immunity. 2003;18:97–108. doi: 10.1016/s1074-7613(02)00511-3. [DOI] [PubMed] [Google Scholar]
- Reits E, Neijssen J, Herberts C, Benckhuijsen W, Janssen L, Drijfhout JW, Neefjes J. A major role for TPPII in trimming proteasomal degradation products for MHC class I antigen presentation. Immunity. 2004;20:495–506. doi: 10.1016/s1074-7613(04)00074-3. [DOI] [PubMed] [Google Scholar]
- Roelse J, Gromm‚ M, Momburg F, H„mmerling G, Neefjes J. Trimming of TAP-translocated peptides in the endoplasmic reticulum and in the cytosol during recycling. Journal of Experimental Medicine. 1994;180:1591–1597. doi: 10.1084/jem.180.5.1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serna A, Ramirez MC, Soukhanova A, Sigal LJ. Cutting edge: efficient MHC class I cross-presentation during early vaccinia infection requires the transfer of proteasomal intermediates between antigen donor and presenting cells. The Journal of Immunology. 2003;171:5668–5672. doi: 10.4049/jimmunol.171.11.5668. [DOI] [PubMed] [Google Scholar]
- Shen L, Rock KL. Cellular protein is the source of cross-priming antigen in vivo. Proc Natl Acad Sci USA. 2004;101:3035–3040. doi: 10.1073/pnas.0308345101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L, Rock KL. Priming of T cells by exogenous antigen cross-presented on MHC class I molecules. Curr Opin Immunol. 2006;18:85–91. doi: 10.1016/j.coi.2005.11.003. [DOI] [PubMed] [Google Scholar]
- Speidel K, Osen W, Faath S, Hilgert I, Obst R, Braspenning J, Momburg F, Hammerling GJ, Rammensee HG. Priming of cytotoxic T lymphocytes by five heat-aggregated antigens in vivo: conditions, efficiency, and relation to antibody responses. European Journal of Immunology. 1997;27:2391–2399. doi: 10.1002/eji.1830270938. [DOI] [PubMed] [Google Scholar]
- Srivastava P. Interaction of heat shock proteins with peptides and antigen presenting cells: chaperoning of the innate and adaptive immune responses. Annu Rev Immunol. 2002;20:395–425. doi: 10.1146/annurev.immunol.20.100301.064801. [DOI] [PubMed] [Google Scholar]
- Wallin RP, Lundqvist A, More SH, von Bonin A, Kiessling R, Ljunggren HG. Heat-shock proteins as activators of the innate immune system. Trends Immunol. 2002;23:130–135. doi: 10.1016/s1471-4906(01)02168-8. [DOI] [PubMed] [Google Scholar]
- Wolkers MC, Brouwenstijn N, Bakker AH, Toebes M, Schumacher TN. Antigen bias in T cell cross-priming. Science. 2004;304:1314–1317. doi: 10.1126/science.1096268. [DOI] [PubMed] [Google Scholar]
- Yewdell JW. Plumbing the sources of endogenous MHC class I peptide ligands. Curr Opin Immunol. 2007;19:79–86. doi: 10.1016/j.coi.2006.11.010. [DOI] [PubMed] [Google Scholar]
- Yewdell JW, Bennink JR. Brefeldin A specifically inhibits presentation of protein antigens to cytotoxic T lymphocytes. Science. 1989;244:1072–1075. doi: 10.1126/science.2471266. [DOI] [PubMed] [Google Scholar]
- Ying M, Flatmark T. Binding of the viral immunogenic octapeptide VSV8 to native glucose-regulated protein Grp94 (gp96) and its inhibition by the physiological ligands ATP and Ca2+ Febs J. 2006;273:513–522. doi: 10.1111/j.1742-4658.2005.05084.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1- Specificity of recombinant TCR-like Fab antibodies selected on derived peptide-MHC complexes. (A) Specificity of Fabs shown by ELISA on immobilized H-2Db-peptide complexes. Binding of Fab clones was measured on Db-PA224, Db-NP366, Db-PB1-F262 and a mixture of Db molecules with control peptides. (B) Detection of H-2Db-peptide complexes on the surface of EL4 cells. EL4 cells were loaded with 10μM of specific (Black) or control (gray) IAV derived peptide for 2h at 37ºc. Peptide-loaded cells were then incubated with the specific soluble purified Fab antibodies followed by Cy5 conjugated anti-human antibody. (C) EL4 cells were pulsed for 2h at 37ºc with titrated amounts of peptides. Detection sensitivity of TCR-like fabs was then measured by flow cytometry.
Supplemental Figure 2- Schematic of rVVs. Mg-peptide: peptide is expressed as a cytosolic peptide with an additional initiating Met. ES-peptide. peptide is targeted to the ER by a NH2-termina signal sequence.