Abstract
We have synthesized a stable and clinically relevant nanodevice (cRGD-BT-ND; ND for short) that exhibits superior binding to the biologic target αvβ3 integrins, when either compared to the same free cRGD peptide or to the biotinylated nanodevice without covalently attached peptides (BT-ND). Selective targeting of αvβ3 integrins was achieved by coupling cyclic cRGD peptides to the nanodevice (ND) surface, while biotin groups (BT) were used for amplified detection of bound cRGD-BT-ND by anti-biotin antibody or avidin linked to horseradish peroxidase after binding. The synthesis involved the following steps: the amino terminated ethylenediamine core generation 5 poly(amidoamine) (PAMAM_E5.NH2) dendrimer was first partially acetylated, then biotinylated, residual primary amine termini were converted to succinamic acid groups (SAH), some of which finally were conjugated with cRGD peptide residues through the amino group of the lysine side chain. The starting material and all derivatives were extensively characterized by polyacrylamide gel electrophoresis (PAGE), size exclusion chromatography (SEC), potentiometric acid-base titration, MALDI-TOF and NMR. Cytotoxicity of all dendrimer derivatives was examined in B16F10 melanoma cell cultures using the XTT colorimetric assay for cellular viability. Binding of nanodevices to the biological target was determined using plates coated with human αvβ3 integrin and αvβ3 receptor expressing human dermal microvascular endothelial cells (HDMECs). The PAMAM_E5.(NHAc)72(NHBT)8(NHSAH)35(NHSA-cRGD)4 nanodevice is non-toxic within physiologic concentration ranges and specifically binds to the αvβ3 integrins, apparently much stronger than the cyclic cRGD peptide itself.
INTRODUCTION
We are developing a novel Systemic Targeted Radiation Therapy (START) approach utilizing a dendrimer-based platform to detect and treat primary and metastatic cancers. As a first step using poly(amidoamine) (PAMAM) dendrimer we have synthesized the multifunctional nanodevice that selectively binds to αvβ3 integrins. PAMAM dendrimers are water soluble, well-defined polymers, built from symmetric dendrons, containing β-alanine subunits, emanating from a core (1,2). These polymers are nearly ideal delivery vehicles as they are not broken down by enzymes and are generally removed from the bloodstream by the filter organs (3–6). PAMAMs can be prepared to be non-immunogenic, non-mutagenic and non-toxic (see e.g. 7). Modifiable terminal functionalities of dendrimers offer a multipurpose mode to covalently attach drugs, diagnostic/imaging modules and targeting moieties (8). PAMAMs have been already used as delivery vehicles for oligonucleotides, antisense oligonucleotides, oligonucleotide arrays and chemotherapeutic cancer drugs (9–11). They have also been utilized as templates for composite nanoparticles, which allows them to host inorganic nanoclusters (12) thus providing a way to integrate desirable properties of inorganic materials and biofriendly polymers, and yielding several new imaging and therapy applications (13–15).
During the design of this ND, we considered the following. The αvβ3 integrins are highly overexpressed on the angiogenic endothelial cells present in tumors, and are not available for binding in normal blood vessels (16). This feature can be exploited to target nanodevices (NDs) to endothelial cells of tumor microvasculature by covalently attaching αvβ3 targeting peptides to the surface of each nanodevice. For this purpose four copies of a specific cyclic RGD peptide (Phe(f)-Lys-Arg-Gly-Asp, abbreviated to cRGD) were conjugated with dendrimer. We used a peptide that strongly binds to the αvβ3 integrins and is already in clinical trials (17), making the nanodevice potentially more clinically useful than other permutations of RGD peptides (10). Additionally, it’s a more stable cyclic RGD peptide compared to cyclization achieved via disulfide bond.
During the design, we have considered that toxicity of nanodevices, including dendrimers, often results from the presence of excessive positive charges (7, 11, 15). For this reason, we converted the unused primary amine surface groups that would have been positively charged (thus potentially toxic) at biologic pH values (pH=5.5–7.5), to carboxylate groups by reacting them with succinamic anhydride.
Exact measurement of biodistribution is a prerequisite for the construction of efficient medical nanodevices that carry therapeutic or imaging agents to cancer cells. In the present literature, dendrimer based nanodevices are often tracked by covalently attaching fluorescent dye molecules to the macromolecules. The apparent disadvantage of that method is that attaching large aromatic molecules to a dendrimer surface has to change the biodistribution of the dendrimer nanodevices. In our ND we used biotin as a label on the ND that can selectively be detected by labeled antibodies. We have conjugated eight biotin moieties per dendrimer to detect the presence of dendrimers sensitively and selectively after localization, as it is described in the experimental part below. Using nanodevices labeled with multiple biotin units results in considerable signal amplification, while it does not disturb the biodistribution of the nanodevice.
EXPERIMENTAL PROCEDURES
General
Ethylene-di-amino-core poly(amidoamine) PAMAM dendrimer of generation 5 (PAMAM_E5.NH2) was purchased from Dendritech Midland, MI, in methanol solution (14.17 wt%). Succinic anhydride, pyridine, acetic anhydride, biotin, bovine serum albumin, absolute methanol, dimethylsulfoxide (DMSO), 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide, 1-hydroxybenzotriazole were obtained from Aldrich and used as received. Cyclic RGD peptide (Phe(f)-Lys-Arg-Gly-Asp) was acquired from Peptides International (Louisville, KY) and characterized by HPLC, Electrospray Ionization-TOF MS, and amino acid analysis before use. αvβ3 integrin was purchased from Chemicon International (Temecula, CA). Immuno-Pure Avidin HRP and o-phenylene-diamine obtained form Pierce (Rockford, IL). Regenerated cellulose dialysis membranes (MWCO = 2000 and 10000) were acquired from Fisher. Water used in the experiments was purified by a Milli-Q Plus 185 water purification system (Millipore, Bedford, MA, USA) with resistivity higher than 18 MΩcm−1. Starting material and all derivatives were characterized by polyacrylamide gel electrophoresis (PAGE), size exclusion chromatography equipped with three detectors: multiangle laser light scattering, UV-vis diode-array and refractive index (SEC-MALLS-DAD-RI), potentiometric acid-base titration, matrix assisted laser desorption ionization - time of flight (MALDI-TOF) mass spectrometry and nuclear magnetic resonance (NMR).
Synthesis of PAMAM_E5.(NHAc)72(NHBT)8(NHSAH)35(NHSAcRGD)4
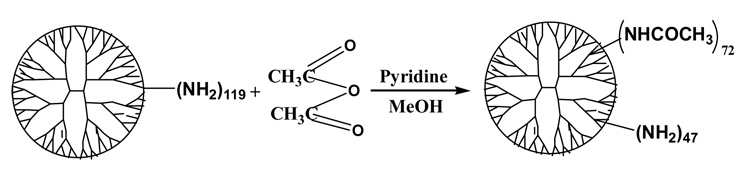
1) Synthesis of PAMAM_E5.(NAc)72(NH2)47 dendrimer (Scheme 1)
Scheme 1.
100 mL of methanol solution containing 3.08 g (n=1.13×10−4 mol) of PAMAM_E5.NH2 dendrimer was placed in a 250-mL round bottom flask. Then 100 mL of methanol solution with 1.53 g (n=1213×10−2 mol) pyridine and 0.98 g (n=9.60×10−3 mol) acetic anhydride was added. After 24 hours methanol was evaporated on a rotary evaporator. The residue was dissolved in water and dialyzed (using cellulose membrane with 2000 MWCO) against water for three days to remove byproducts and excess reactants. Obtained solution was filtered and lyophilized.
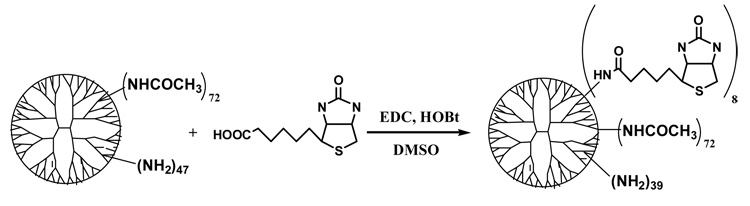
2) Synthesis of PAMAM_E5.(NHAc)72(NHBT)8(NH2)39 nanodevice (Scheme 2)
Scheme 2.
0.08 g biotin (n=3.28×10−4 mol) was dissolved in 50 mL of dimethylsulfoxide (DMSO) in a 250-mL round bottom flask then 0.13g (n=6.78×10−4 mol) of 1-(3-dimethylaminopropyl)-3-ethylcarbodiimide (EDC) and 0.09g (n=6.66×10−4 mol) 1-hydroxybenzotriazole (HOBt) was added and mixed for 0.5 h at room temperature to obtain active ester. Then 50 mL of DMSO solution containing 1.04 g (n=3.55×10−5 mol) of PAMAM_E5.(NAc)72(NH2)47 dendrimer (product obtained in the first step of the synthesis) was added to the mixture and stirred for an additional 24 h under nitrogen atmosphere. The reaction mixture was diluted with water and dialyzed for 3 days using cellulose membrane with 10000 MWCO, filtered and lyophilized.
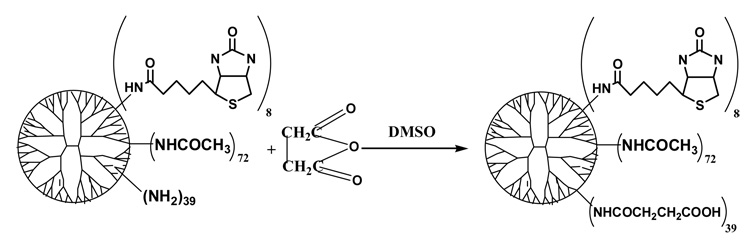
3) Synthesis of PAMAM_E5.(NHAc)72(NHBT)8(NHSAH)39 nanodevice (Scheme 3)
Scheme 3.
0.83341 g (n=2.73×10−5 mol) of PAMAM_E5.(NAc)72(NH-BT)8(NH2)39 (product obtained in the second step of the synthesis) was dissolved in 50 mL of DMSO solution and placed in a 250-mL round bottom flask. Then 0.68 g (n=6.73×10−3 mol 4 mol equivalents compared to the remaining terminal NH2 groups, number of amino termini was calculated based potentiometric titrations) of succinic anhydride was added with vigorous stirring. The reaction was stopped after 24 hours by diluting the reaction mixture with water. Byproducts and excess reactants were removed by dialysis using cellulose membrane with 10000 Daltons MWCO. The retentate was filtered, and the resulting solution was lyophilized.
4) Synthesis of PAMAM_E5.(NHAc)72(NHBT)8(NHSAH)35(NHSAcRGD)4 nanodevice
50 mL of DMSO stock solution containing 0.19 g (n=6.00×10−6 mol) of PAMAM_E5.(NHAc)72(NHBT)8(NHSAH)39 (product obtained in the previous step of synthesis) was placed in a 250-mL round bottom flask. Then 0.19 g of EDC and 0.08 g HOBt (4 mol equivalents compared to terminal COOH groups) was added stirring for 0.5 h to form active ester. Then 50 mL DMSO solution containing 0.03 g (n=4.97×10−5 mol) of c-RGD peptide was added. Reaction was stopped after 24 hours by diluting the reaction mixture with water. Byproducts and excess reactants were removed by dialysis using cellulose membrane with 10000 Daltons MWCO. The retentate was filtered, and the resulting solution was lyophilized.
Analytical Methods
Size Exclusion Chromatography (SEC)
Analysis was performed using an Alliance Waters 2690 separation module equipped with a Waters UV-Vis detector, a Wyatt Dawn laser photometer, Optilab interferometric refractometer and Waters ultrahydrogel columns. Phosphate buffer (0.05 M, pH 2.5) with 0.025% sodium azide was used as mobile phase. The flow rate was maintained at 0.6 mL/min. Sample concentration was kept at 2 mg/mL and 100 µL was injected. Data was elaborated using Astra and PeakFit software.
Potentiometry
Dendrimers were dissolved in NaCl (0.1 M) solution at concentration of 0.5 mg/mL, pH was set to around 3. In case of each sample the same number of moles (adjusted by the volume) was titrated. Titrations were performed at room temperature, under nitrogen atmosphere, using Molspin automated titration system, Mettler-Toledo combination InLab electrode and NaOH (0.1 M) as a titrant.
Polyacrylamide gel electrophoresis
PAGE of PAMAM dendrimers was performed using a vertical electrophoresis system (Model FB-VE10-1 FisherBiotech) with a commercial power supply (Model EC135-90; Thermo Electron Corporation), precast 4–20% gradient express gels for PAGE (BioExpress) and Tris-glycine (TG) native (pH = 8.3). Separations typically required 1.5 h min at 200 V. Into each sample well 2 µL of a sample solution composed of 2 or 3 µL 1mg/mL PAMAM dendrimer and 2 or 3 µL sucrose dye solutions (50% sucrose, 1% methylene blue) was injected. Developed gel was stained with 0.025% Comassie Blue R-250 in 40% methanol and 7% acetic acid aqueous solution overnight. The gels were destained with 7% (v/v) acetic acid and 5% (v/v) methanol in water.
Matrix-assisted laser desorption ionization-time of flight mass spectrometry
Spectra of dendrimer nanodevices were recorded on a Bruker Biflex IV spectrophotometer, using 2,5-dihydroxybenzoic acid (DHB) as a matrix. First 0. 5 µL of matrix was placed on target plated, and evaporated. Then 0.5 µL of the dendrimer (~1.5 mg/mL) was spotted over the matrix. Matrix and dendrimer were dissolved in 75% ACN and 0.1% TFA aqueous solution. For each spectrum 100 shots and 50% of laser power was applied. For each spectrum 100 shots and 50% of laser power was applied.
Nuclear Magnetic Resonance
Spectra were recorded using Bruker AMX 400 MHz, D2O as solvent and at sample concentrations 20 mg/mL.
Biological Studies
Toxicology Studies of the cRGD-BT-ND and the four intermediate materials leading up to its synthesis
Toxicity studies were conducted in vitro, using MatLyLu prostate cancer cells and B16F10 melanoma cells in tissue culture and 10, 100, 250, 500, 1000 and 2000 nM of ND. All samples were replicated in quintuplets. Tissue culture treated 96 well microtiter plates were coated with cells and incubated at 37 C, 5% CO2 for 24 hours. The following day, nanodevices in defined amounts were added to the plates, which were further incubated for 24 hours and 48 hours. To measure cell viability at different time points, plates were then treated with XTT (sodium 3′-[1-(phenylaminocarbonyl)-3,4-tetrazolium]-bis(4-methoxy-6-nitro)benzenesulfonic acid hydrate) according to manufacturer’s instructions (Roche Diagnostics Corporation, Indianapolis, IN). Briefly, at 0, 24 and 48 hour time points, cell culture medium was removed and an aliquot of 100 µL fresh medium was added to each well. Then an aliquot of 50 µL of XTT working solution prepared according to manufacturer instructions was added to each well using multichannel pipette. After 3 hour incubation in the dark at 37 °C, the absorbance at 490 nm was recorded using a plate reader.
Statistical analysis of the toxicity experiment
To assess differences among six dose levels at each time reading and differences among three time readings at each dose level, the Wilcoxon exact test was used. Tests for overall group differences were done at a 0.05 level of significance. If there was a significant difference for overall comparison, the Bonferroni adjusted test was used to perform pairwise comparisons. All statistical analyses were carried out using SAS (version 9.1) statistical software (Cary, NC).
Nanodevice binding to αvβ3 integrin in vitro
An aliquot of 100 µL of purified αvβ3 integrin receptor (Chemicon Int., Temecula, CA) 0.5 µg/mL in coating buffer (50 mM Tris, pH 7.4; 100 mM NaCl and 1 mM each of CaCl2,, MnCl2, and MgCl2,) was immobilized in 96-well microtiter plate overnight at 4 °C. The wells were washed once with binding buffer (50 mM Tris HCl, pH 7.4, 100 mM NaCl, 1 mM CaCl2, 1 mM MnCl2 and 1 mM MgCl2 and 35 mg/mL bovine serum albumin) and incubated with the same for 2 h at room temperature. The plate was then washed 3 times with binding buffer and an aliquot of 100 µL of BT-ND or cRGD-BT-ND (13.8 µM) added and incubated for 3 h at 4 °C. Competition assay was conducted adding 4, 40 and 400 fold excess of cRGD-peptide (Chemicon Int., Temecula, CA) simultaneously with the nanodevice. At the end of this incubation period, plate was washed five times with binding buffer. The bound biotinylated NDs were detected by Immuno-Pure Avidin conjugated with horseradish peroxidase (product # 21123; Pierce, Rockford, IL), which was subsequently developed using o-Phenylenediamine and the absorbance at 450 nm recorded according to the manufacturer’s directions (18).
Nanodevice binding to human dermal microvasculature cells (HDMECs)
HDMECs were seeded (50,000 cells per well) and grown on chamber slides (Lab Tek) in a conditioned endothelial culture medium EBM Bullet kit 2 (Cambrex, Walkersville, MD). After two days at 50% confluence, cells were treated with 1 µM nanodevices for 5 hours at 37 °C and washed 5 times with PBS buffer, fixed with cold acetone and dried. Cells were then freeze thawed (−80 °C) and blocked for 1 hour with 4 mg/mL BSA in PBS followed by 1 hour incubation with FITC labeled anti-biotin antibody (Vector Labs, Burlingame, CA) according to the manufacturer’s instruction at room temperature. Cells were washed 4 times with PBS buffer and DAPI counter stain was added.
RESULTS AND DISCUSSION
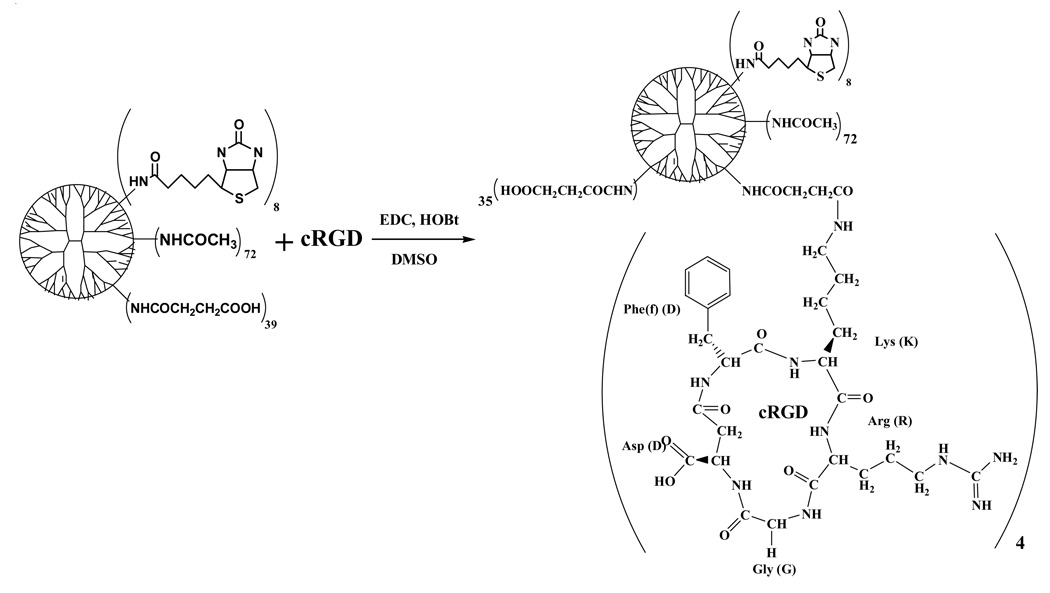
We have synthesized the targeted dendrimer nanodevice (cRGD-BT-ND) as follows. The starting material, PAMAM_E5.NH2 amino terminated dendrimer (abbreviated as E5.NH2) was partially acetylated (1 - NHAc), then biotinylated (2 - NHBT), followed by converting the rest of primary amine termini to succinamic acid groups (3 - NHSAH), and finally conjugated with four cRGD peptides (4 - NHSAcRGD) as described by the following reaction sequence (in which subscripts correspond to number of measured functional groups):
E5.(NH2)119 => E5.(NHAc)72(NH2)47
E5.(NHAc)72(NH2)47 => E5.(NHAc)72(NHBT)8(NH2)39
E5.(NHAc)72(NHBT)8(NH2)39 => E5.(NHAc)72(NHBT)8(NHSAH)39
E5.(NHAc)72(NHBT)8(NHSAH)39 => E5.(NHAc)72(NHBT)8(NHSAH)35(NHSAcRGD)4
The acetylation was performed first to obtained more uniform distribution of the functional groups within nanodevice (ND) molecules and decrease their toxicity since positively charged (amino terminated) PAMAM dendrimers are know to be toxic (7). Biotin (BT) was selected as reporter moiety for ND detection using labeled antibodies. In addition it is much smaller compared to commonly used fluorescent dyes that can potentially affect biological activity of NDs. Lastly carboxyl groups of succinamic acid were used for cRGD peptides conjugation via amid bound with lysine side chain amino group, which is not involved in binding of the peptide to αvβ3 integrins. Derivatives in each synthetic step were purified and characterized by polyacrylamide gel electrophoresis (PAGE), size exclusion chromatography equipped with three detectors: multiangle laser light scattering, UV-vis diode-array and refractive index (SEC-MALLS-DAD-RI), potentiometric acid-base titration, matrix assisted laser desorption ionization - time of flight (MALDI-TOF) mass spectrometry and nuclear magnetic resonance (NMR), providing complementary results for thorough analysis.
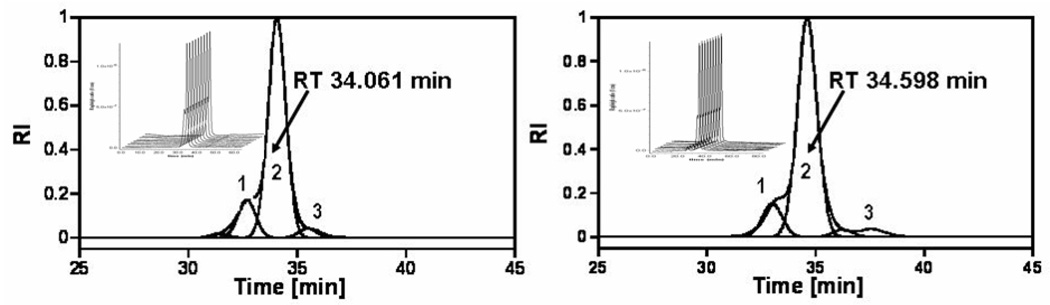
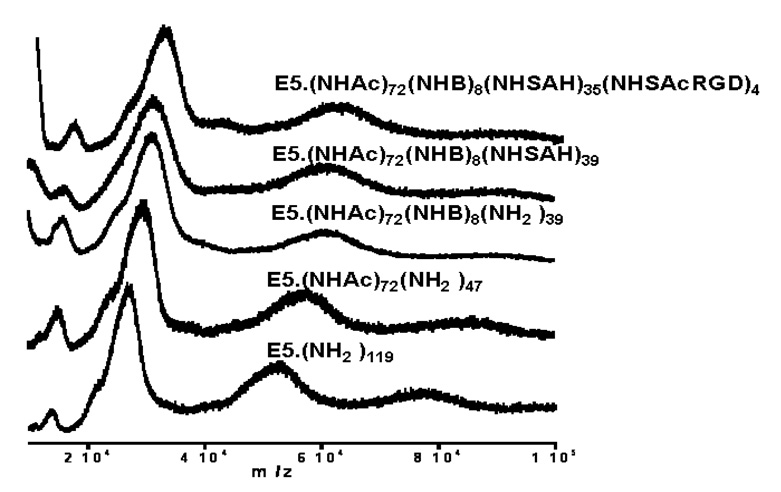
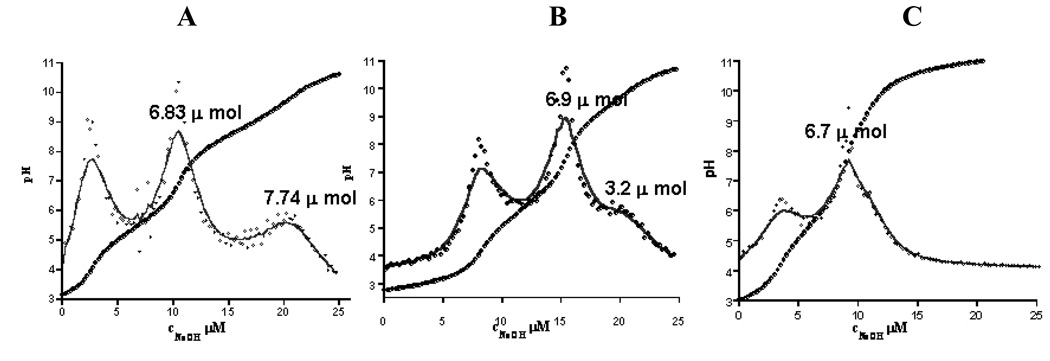
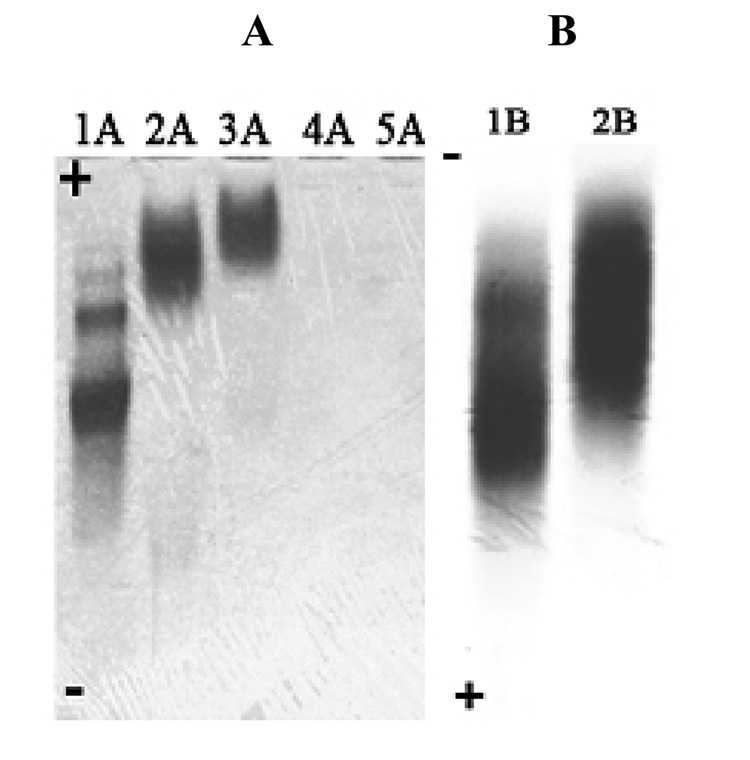
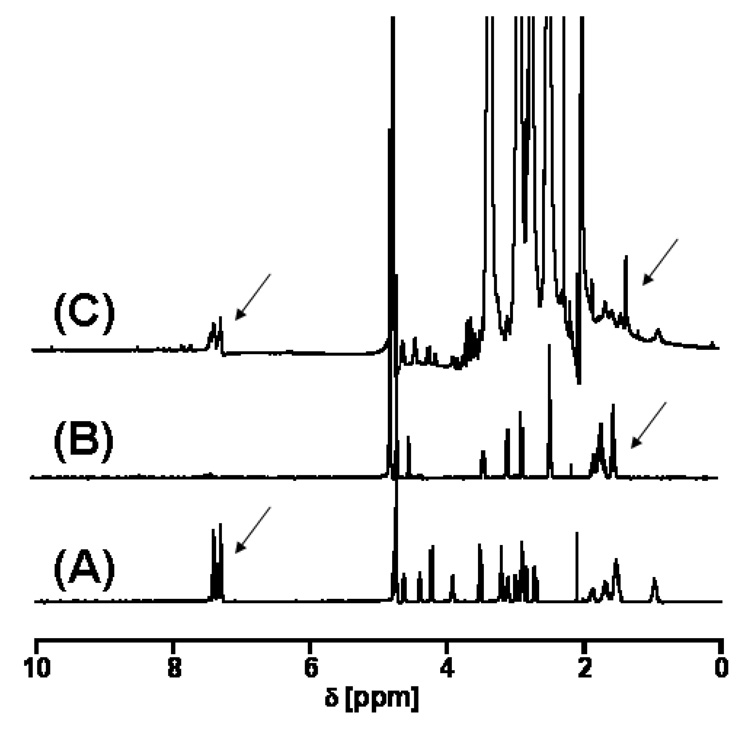
SEC was used to determine the composition, average molecular mass and polydispersity of the nanodevices. Figure 1 compares the SEC data of the (A) starting material and of the (B) final product (cRGD-BT-ND). The value of polydispersity index (Mw/Mn) changed only by 0.007, indicating that all components of the starting material were equally modified. Light scattering data confirmed the corresponding increase of the molecular mass throughout the synthesis, even though the hydrodynamic volume slightly decreased as indicated by longer retention times (as pointed out in the graph). Unmodified and fully protonated PAMAM_E5NH2 dendrimer is in an extended conformation at acidic pH (pH=2.5 for SEC-MALLS). This extended structure reduces in size after its terminal amino groups are substituted, accompanied by the decrease of charge of the nanodevice. Figure 2 presents successive mass spectra of all analyzed compounds. The shift in the spectra towards higher molecular masses after each synthesis step indicates successful modifications of intermediates with the selected terminal functional groups during the construction of the required ND structure. Figure 3 shows examples of titration curves and their derivatives for the (A) PAMAM_E5.(NH2)119 dendrimer, (B) its partially acetylated derivative (PAMAM_E5.(NHAc)72(NH2)47) and (C), the E5.(NHAc)72(NHBT)8(NHSAH)39 intermediate. Degree of acetylation can be precisely measured by acid-base titration, because the acetamide termini cannot be protonated within the investigated pH range. The inflection point related to deprotonation of primary amino groups shifts from 7.74 (Fig. 3A) to 3.2 mol equivalent (Fig. 3B) of the titrant, indicating that 60% of initial amino groups were acetylated. However, position of end points of titration of tertiary amines does not change (Fig. 3A 6.83 µmol and Fig. 3B 6.9 µmol, observed 0.07 µmol difference is within the experimental error). Similarly, number of conjugated BT molecules and succinamic acid termini can be determined since amide groups formed upon modification of terminal NH2 groups with these functionalities can not be protonated, eventually leading to the disappearance of the deprotonation peak as shown in the titration curve of the intermediate obtained after succinamination (Figure 4C). In polyacrylamide gel electrophoresis (PAGE) (Fig. 4) all synthetic products exhibited different electrophoretic mobilities as expected, indicating the decrease of positive charge due to increasing substitution of free terminal amine groups of NDs. After conversion of the primary amines to succinamic acid termini, the resulting intermediates become negatively charged, and migrate towards the positive electrode (image B in Figure 4). Change of polarity indicates that substitution of remaining amino groups of E5.(NHAc)72(NHB)8(NH2)39 biotinylated dendrimer, with succinamic acid was successful. 1H NMR confirmed the presence of all functional groups in the cRGD-BT-ND (Figure 5). Average numbers of functional groups in the consecutive products were determined by analyzing all characterization data collected in Table 1. It must be pointed out that increase of the molecular mass of the nanodevice after succinamination, as measured by MALDI and MALLS, indicates conversion of only 10 –;11 primary amino groups, which is most likely due to structural changes occurring upon modification (decrease of hydrodynamic radius changing dn/dc values). However, potentiometric titration indicated the conversion of all terminal NH2 groups into succinamic acid (Figure 3C), which conclusion is further confirmed by PAGE analysis (Figure 4B). In this scenario, the number of succinamic acid moieties must equal to 39, the number of available amine groups.
Figure 1.
(A) Molecular mass distributions of the starting material E5.NH2 dendrimer and (B), the E5.(NHAc)72(NHB)8(NHSAH)35(NHSAcRGD)4 nanodevice shown by their RI signals (insert - MALLS data). Fit of RI signals was performed using Gaussian deconvolution of SEC chromatograms using PeakFit software. The starting material contains (A1) (E5.NH2)2 dimers (16.42%); (A2) E5.NH2 generation five dendrimers (80.42%), and (A3) a small amount of trailing generation E4.NH2 (3.15%). Average molecular mass of Mn=27280, Mw=30280 with a polydispersity of Mw/Mn = 1.110 were obtained for this material. The final product (B) contains modified dimers (B1=16.32%); modified E5 derivatives (B2=80.22%), and a small amount of modified trailing generation (B3=3.46%). (Respective molecular masses calculated from PeakFit are Mn=34080, Mw=38067, and Mw/Mn = 1.117).
Figure 2.
MALDI-TOF spectra of starting dendrimer, of the intermediates and the final nanodevice using 2,5-dihydroxy-benzoic acid (DHB) as a matrix.
Figure 3.
Potentiometric titration curves of (A) the PAMAM_E5.NH2 amino terminated dendrimer (starting material), (B) its acetylated derivative obtained after the first step of synthetic path, clearly showing shift of the end point of titration of primary amino groups upon acetylation and (C) E5.(NHAc)72(NHBT)8(NHSAH)39 showing disappearance of NH2 groups.
Figure 4.
PAGE of starting dendrimer, nanodevice intermediates and the final product:
(1A) E5.(NH2)119
(2A) E5.(NHAc)72(NH2)47
(3A) E5.(NHAc)72(NHBT)8(NH2)39;
(4A) E5.(NHAc)72(NHBT)8(NHSAH)39
(5A) E5.(NHAc)72(NHBT)8(NHSAH)35-(NHSAcRGD)4
(1B) E5.(NHAc)72(NHBT)8(NHSAH)39
(2B) E5.(NHAc)72(NHBT)8(NHSAH)35(NHSAcRGD)4.
Figure 5.
1HNMR spectra of (A) c-RGD, (B) Biotin, (C) cRGD-BT-ND revealing the presence of c-RGD peptide and biotin in the sample of cRGB-BT-ND well seen in the aromatic range of the spectrum pointed by arrows.
Table 1.
Characterization of dendrimer nanodevices
| # | Functional Groups | # of groups measured by potentiometry | # of groups measured by SEC | # of groups measured by MALDI | Mass measured by SEC | Mass measured by MALDI* |
|---|---|---|---|---|---|---|
| 1 | NH2 | 119 | -------- | -------- | 27180 | 26572 |
| 2 | NHAc | 72 | 51 | 55 | 29300 | 28887 |
| 3 | NHBT | 8 | 5 | 6 | 30530 | 30369 |
| 4 | NHSAH | 39 | 11 | 10 | 31650 | 31416 |
| 5 | NHSAcRGD | ---------- | 4 | 2 | 34110 | 32691 |
1 - PAMAM_E5.(NH2)119 (starting material); 2 - PAMAM_E5.(NHAc)72(NH2)47 (partially acetylated derivative); 3 - PAMAM_E5.(NHAc)72(NHB)8(NH2)39 (biotinylated and partially acetylated derivative); 4 - PAMAM_E5.(NHAc)72(NHB)8(NHSAH)39 (partially acetylated, biotinylated and succinaminated compound); 5 - PAMAM_E5.(NHAc)72(NHB)8(NHSAH)35(NHSAcRGD)4 (cRGD peptides conjugated to compound, for short RGD targeted and biotinylated nanodevice.
Molecular mass of the highest frequency fragment in the major peak on Figure 2.
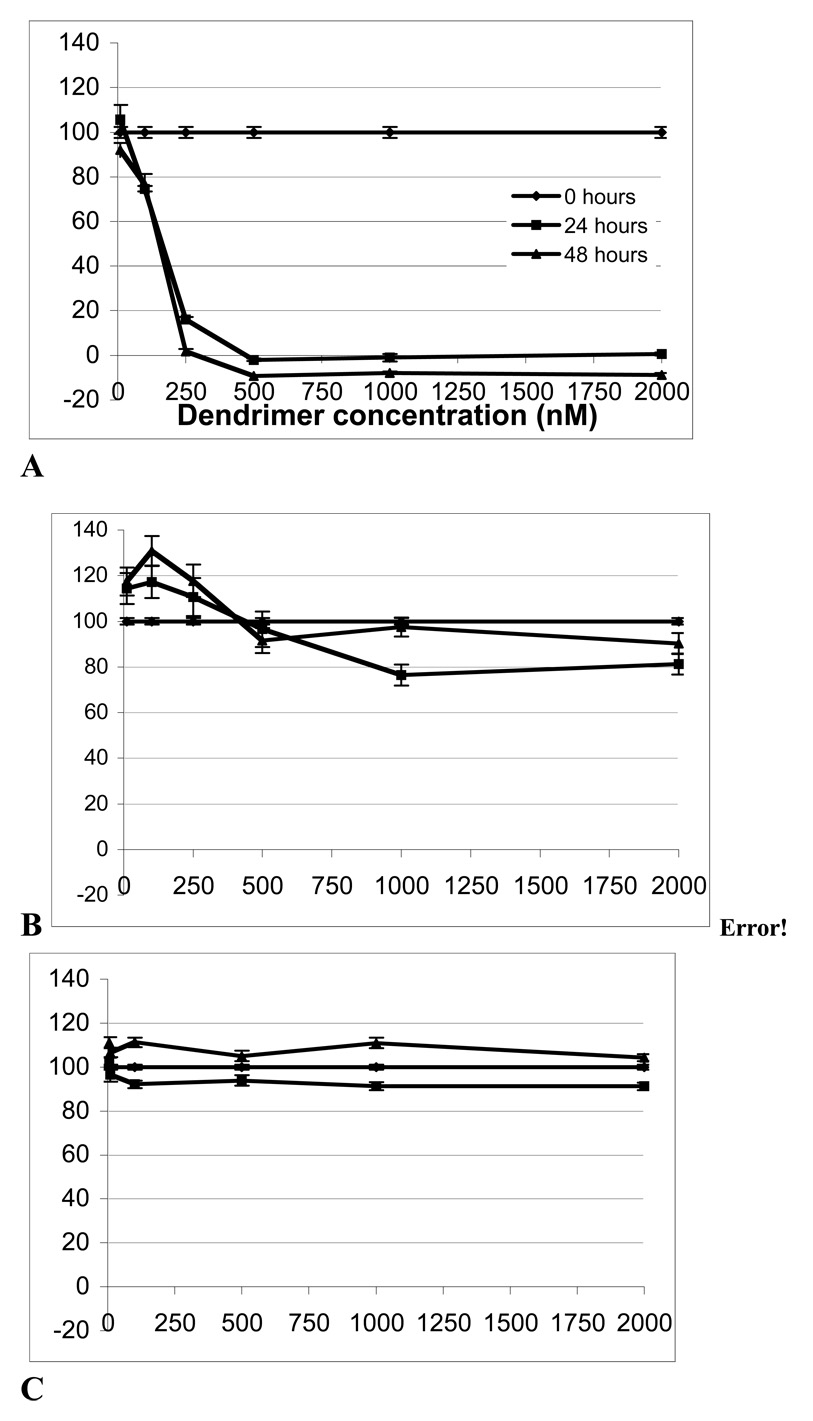
We next carried out biologic characterization of the NDs, namely in vitro toxicity and binding to integrins. Cytotoxicity of all dendrimer derivatives, concentrations varying from 10 – 2000 nM was examined in B16F10 melanoma cell cultures using the XTT colorimetric assay for cellular viability. Only the positively charged surfaced starting material PAMAM_E5.NH2, was toxic to B16F10 cells at or above 250 nM (Fig. 6A) killing the cells after 24 hour incubation. cRGD-BT-ND did not show any significant reduction in cell viability up to a device concentration of 500 nM. At very high concentrations, at or above 1000 nM, we observed ~20% decrease in the cell viability after 24 hour post treatment compare to untreated cells (statistical analysis by Wilcoxon test indicated p=0.0003 compare to untreated). The cell viability was recovered to the levels of the untreated cell after two days post treatment at 2000 nM concentration (p=0.2991). The cRGD-BT-ND was also non-toxic to MatLyLu prostate cancer cells at all concentrations tested, with proliferation rates close to that of control cells (data not shown). We also tested the effect of cRGD-BT-ND on human dermal microvasculature cells (HDMEC). The cRGD-BT-ND did not show any significant toxicity towards HDMECs (Fig. 6C). At 24 h the statistical test gave a significant (p=0.0283) but biologically irrelevant (<10%) difference, and by 48 h post treatments there was no significant different in the overall value for all six dose levels tested (p=0.0977),
Figure 6.
B16 cell viability in the presence of (A) PAMAM_E5.NH2 and (B) cRGD-BT-ND and (C) HDMEC viability in the presence of cRGD-BT-ND at 0, 10, 100, 250, 500, 1000, 2000 nM concentrations and 0, 1 and 2 days.
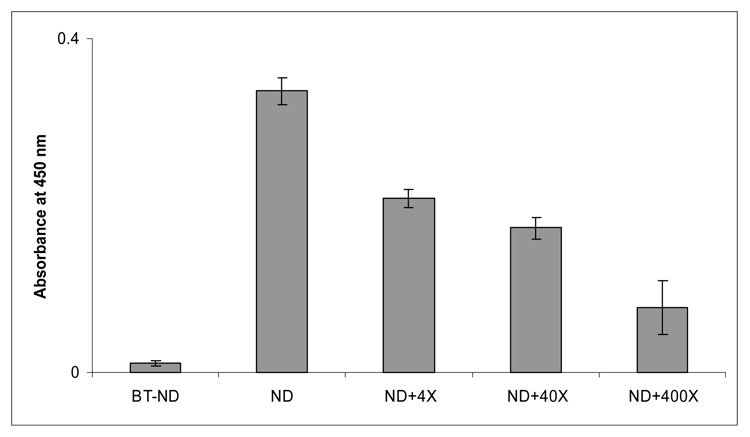
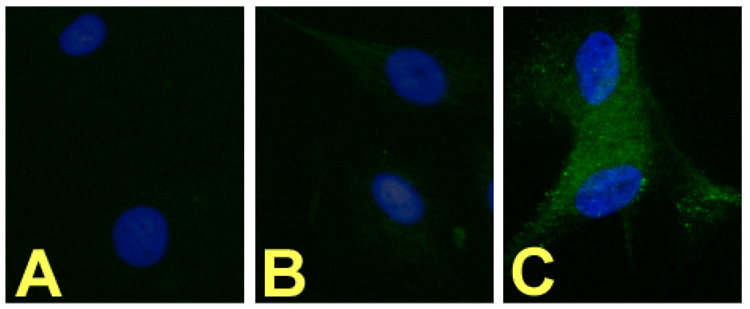
The cRGD-BT-NDs were then shown to specifically bind in vitro to plates coated with human αvβ3 integrin. The cRGD targeted ND displayed superior specific binding compared to the control (biotinylated ND without cRGD, i.e., E5.(NHAc)72(NHBT)8(NHSAH)39). Even a 400 fold excess of free cyclic cRGD peptide was unable to displace over 25% bound RGD-BT-ND (Figure 7) in a competitive binding experiment. The binding of NDs to cultured cells was performed using αvβ3 receptor expressing human dermal microvascular endothelial cells (HDMECs). Our results show that only the cRGD coupled biotinylated NDs notably bind to HDMECs (Fig. 8C). This is the first in vitro data showing specific ligand (cRGD) - αvβ3 receptor binding by these NDs.
Figure 7.
Nanodevice Binding to Integrin Receptors in an in vitro plate binding assay: Plates coated with αvβ3 integrins were treated with the respective nanodevices (Eq. 4, i.e., with and without covalently bound cRGD targeting moieties) and the cRGD competitor peptide. The bound NDs were detected by their biotin functions using avidin linked to horseradish peroxidase and the color was developed with o-phenylene-diamine. The labels 4X, 40X, and 400X denote 4, 40, and 400 times of molar excess of cRGD mixed with the cRGD-BT-ND nanodevice, respectively competing for the binding sites.
Figure 8.
Fluorescence microscopy of αvβ3 receptor overexpressing HDMEC cells exposed to cRGDBT-ND: FITC labeled anti-biotin antibody (green) labels the RGD-BT-ND, and DAPI (blue) stains the nuclei. A: No treatment, B: BT-ND treated, C: cRGD-BT-ND treated.
In summary, we have synthesized biotinylated and cRGD peptide conjugated carboxylate terminated dendrimer NDs from commercially available amino terminated PAMAM_E5NH2 dendrimer. Importantly, we attached a form of cRGD that is already in clinical trial as an anti-angiogenic agent and therefore may have more applicability in humans than other forms. Analytical data indicate successful modifications of dendrimers according to designed terminal functionalities. We have determined the composition of the ND (cRGD-BT-ND) by comparing results of different analytical methods to provide insight into the complex structure of these dendrimer based NDs. Detailed characterization is a crucial factor in understanding their biological response. The final ND is non-toxic within physiologic concentration ranges and shows superior binding to the biologic target αvβ3. Strong binding is a prerequisite for constructing targeted NDs that can carry therapeutic or imaging agents to cancer cells (19).
Scheme 4.
Acknowledgment
This research was funded with Federal Funds from the National Institutes of Health, National Cancer Institute (NCI) (# R01-CA-104479-01).
Abbreviations
- PAMAM_E5.NH2
amine-terminated, ethylenediamine core generation 5 poly(amidoamine) dendrimer
- ND
nanodevice
- BT
biotin
References
- 1.Tomalia D, Dvornic P. Starburst polyamidoamine dendrimers. Polymeric Materials Encyclopedia. 1996;3:1814–1830. CRC Press. [Google Scholar]
- 2.Mansfield M, Klushin L. Monte Carlo studies of dendrimer macromolecules. Macromolecules. 1993;26:4262–4268. [Google Scholar]
- 3.Malik N, Evagorou E, Duncan R. Dendrimer-platinate: a novel approach to cancer chemotherapy. Anticancer Drugs. 1999;10:767–776. [PubMed] [Google Scholar]
- 4.Malik N, Wiwattanapatapee R, Klopsch R, Lorenz K, Frey H, Weener JW, Meijer EW, Paulus W, Duncan R. Dendrimers: Relationship between structure and biocompatibility in vitro and preliminary studies on the biodistribution of 125I-labelled PAMAM dendrimers in vivo. J. Control Release. 2000;65:133–148. doi: 10.1016/s0168-3659(99)00246-1. [DOI] [PubMed] [Google Scholar]
- 5.El-Sayed M, Kiani M, Naimark M, Hikal AH, Ghandehari H. Extravasation of poly(amidoamine) (PAMAM) dendrimers across microvascular network endothelium. Pharm. Res. 2001;18:23–28. doi: 10.1023/a:1011066408283. [DOI] [PubMed] [Google Scholar]
- 6.Nigavekar SS, Sung LY, Llanes M, El-Jawahri A, Lawrence TS, Becker CW, Balogh L, Khan MK. 3H dendrimer nanoparticle organ/tumor distribution Pharm. Res. Pharm. Res. 2004;21:476–483. doi: 10.1023/B:PHAM.0000019302.26097.cc. [DOI] [PubMed] [Google Scholar]
- 7.Roberts JC, Bhalgat MK, Zera RT. Preliminary biological evaluation of polyamidoamine (PAMAM) Starburst™ dendrimers. J. Biomed. Mater Res. 1996;30:53. doi: 10.1002/(SICI)1097-4636(199601)30:1<53::AID-JBM8>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 8.Bielinska A, Kukowska-Latallo JF, Johnson J, Tomalia DA, Baker JR., Jr Regulation of in vitro gene expression using antisense oligonucleotides or antisense expression plasmids transfected using starburst PAMAM dendrimers. Nucleic Acids Res. 1996;24:2176–2182. doi: 10.1093/nar/24.11.2176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Majoros IJ, Thommey PT, Chandan BM, Baker JR., Jr Dendrimer-Based Multifunctional Engineered Nanodevice for Cancer Therapy. J. Med. Chem. 2005;48:5892–5899. doi: 10.1021/jm0401863. [DOI] [PubMed] [Google Scholar]
- 10.Shukla R, Thomas TP, Peters J, Kotlyar A, Myc A, Baker JR., Jr Tumor angiogenic vasculature targeting with PAMAM dendrimer-RGD conjugates. Chem. Commun. 2005:5739–5749. doi: 10.1039/b507350b. [DOI] [PubMed] [Google Scholar]
- 11.Eichman J, Bielinska A, Kukowska-Latallo J, Baker JR., Jr The Use of PAMAM Dendrimers in the Efficient Transfer of Genetic. Pharmaceutical Science and Technology Today. 2000;3:232–245. doi: 10.1016/s1461-5347(00)00273-x. [DOI] [PubMed] [Google Scholar]
- 12.Balogh L, Laverdure KS, Gido SP, Mott AG, Miller MJ, Ketchel BP, Tomalia DA. Organic/Inorganic Hybrid Materials. Mat. Res. Soc. Symp. Proc. 1999;576:69–75. [Google Scholar]
- 13.Balogh L, Bielinska A, Eichman JD, Valluzzi R, Lee I, Baker JR, Lawrence TE, Khan MK. Dendrimer nanocomposites in medicine. Chimica Oggi (Chemistry Today) 2002;20:35–40. [Google Scholar]
- 14.Balogh L, Nigavekar S, Cook A, Minc L, Khan MK. Dendrimer-Gold Radioactive Nanocomposites to Treat Cancer Microvasculature. Pharma. Chem. 2003;2:94–99. [Google Scholar]
- 15.Khan MK, Nigavekar SS, Minc LD, Kariapper MST, Nair BM, Lesniak WG, Balogh LP. Host-Guest Nanodevices: In Vivo Biodistribution of Dendrimers and Dendrimer Nanocomposites - Implications for Cancer Imaging and Therapy. TCRT 4. 2005:603–613. doi: 10.1177/153303460500400604. [DOI] [PubMed] [Google Scholar]
- 16.Stromblad S, Cheresh DA. Integrins, angiogenesis and vascular cell survival. Chem. Biol. 1996;3:881–885. doi: 10.1016/s1074-5521(96)90176-3. [DOI] [PubMed] [Google Scholar]
- 17.Ragusea J, Gatha H, Biera J, Riess H, Oettle H. Predictive factors and distribution of lymph node metastasis in lip cancer patients and their implications on the treatment of the neck. Oral Oncology. 2004;40:228–230. doi: 10.1016/j.oraloncology.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 18.Smith J, Vestal D, Irwin S, Burke T, Cheresh D. Purification and functional characterization of integrin αvβ5: an adhesion receptor for vitronectin. J. Biol. Chem. 1990;265:11008–11013. [PubMed] [Google Scholar]
- 19.Kukowska-Latallo JF, Kimberly A, Zhengyi C, Nigavekar SS, Majoros IJ, Thomas TP, Balogh LP, Khan MK, Baker JR., Jr Nanoparticle targeting of anticancer drug improves therapeutic response in animal model of human epithelial cancer. Cancer Res. 2005;65:5317–5324. doi: 10.1158/0008-5472.CAN-04-3921. [DOI] [PubMed] [Google Scholar]