Abstract
Dynamic Contrast Enhanced MRI (DCE-MRI) is today one of the most popular methods for tumor assessment. Several pharmacokinetic models have been proposed to analyze DCE-MRI. Most of them depend on an accurate arterial input function (AIF). We propose an automatic and versatile method to determine the AIF. The method has two stages, detection and segmentation, incorporating knowledge about artery structure, fluid kinetics, and the dynamic temporal property of DCE-MRI. We have applied our method in DCE-MRIs of four different body parts: breast, brain, liver and prostate. The results show that we achieve average 89.5% success rate for 40 cases. The pharmacokinetic parameters computed from the automatic AIF are highly agreeable with those from a manually derived AIF (R2=0.89, P(T<=t)=0.19) and a semiautomatic AIF (R2=0.98, P(T<=t)=0.01).
Keywords: DCE-MRI, AIF, tumor imaging
1 Introduction
In the U.S. for 2008 the American Cancer Society estimates 565,650 deaths due to cancer along with 437,180 new cases of cancer detected [1]. Excluding basal and squamous skin cancer, breast cancer has the highest incidence rate among women and is the second leading cause of cancer deaths in women after lung cancer [1]. In men prostate cancer has the highest incidence rate among men and is the leading cause of cancer death in men. Locating, identifying, and knowing the state of cancer tumors early is the best way to improve future prognosis.
Dynamic contrast enhanced MRI (DCE-MRI) is today one of the most popular methods for tumor assessment. Tumor growth beyond a certain size depends on the development of a vascular supply that meets the increased metabolic demand of neoplastic tissue. Studies show that tumor malignancy is highly correlated with increased vascularity. DCE-MRI allows the visualization of tumor vasculature.
The simplest pharmacokinetic model is the one-compartment model where it is assumed that all contrast agents are administered into a single blood vessel compartment [2]. Later a two-compartment model describing contrast agent exchange between the blood vessels and surrounding interstitium as a bi-directional process was proposed [3]. Most of these models such as the General Kinetic Model [4] are based on the indicator dilution theory and require the Arterial Input Function (AIF) to de-convolute with in order to find the concentration of blood (contrast agent) flowing into tissue. These models can [4] quantitatively characterize the permeability of tissue by calculating tracer dynamic parameters such as the extravasation rate (Ktrans), reflux rate (kep), and the extravascular extracellular distribution volume (ve). Examining these parameters can be used to monitor the effects of therapy on tumor and normal tissue. These models are currently being used in trials to examine breast, liver, brain, and prostate tumors at our institute [4, 5, 6].
Automating the process of finding the AIF can save valuable human-operator time. It also removes the inherent inter-operator variability when choosing an AIF and reduces the variability when comparing changes in pharmacokinetic parameters in follow-up studies during treatment therapy.
There are several methods already developed that automatically extract an AIF from DCE-MRIs. Most of them pick an AIF by looking at various characteristics of image voxel’s time-intensity curves, such as peak height, peak width, take-off time, and initial slope [7]. Parker et al. [8] proposed another method to automatically extract an AIF from various parts of the body in DCE-MRI; using a known characteristic of artery voxel time curves, high peak height, it chooses the top 5% of voxels that enhance the brightest and uses them to generate the AIF. However, this method may have some inaccuracy because voxels of an artery are not necessarily being used obtain an AIF. Also this method makes use of only 2d information and is not completely automatic because it requires an operator to pick one axial slice on which to find an AIF.
In this paper we propose a new method that automatically extracts an AIF in DCE-MRI by automatically segmenting the voxels of a major artery in 3d and using the averages of their time-intensity curves to define the AIF. The AIF can then be exported and used in pharmacokinetic models to calculate tumor permeability parameters. This method is versatile, able to extract an AIF from DCE-MRI of different body parts: brain, breast, liver, and prostate. We automatically detect the aorta near the breast and liver, the superior sagittal sinus in the brain and the iliac artery near the prostate and use them to computer AIF.
2 Materials and methods
2.1 Imaging Protocol
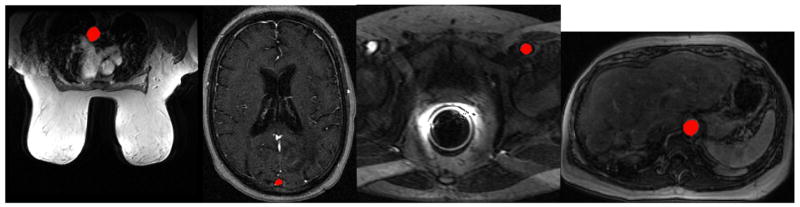
There are currently four different IRB approved DCE-MRI imaging protocols in our institution used to examine different human body parts: breast, brain, liver and prostate, as in Fig 3.
Fig. 3.
Automatic artery segmentation colored in red on DCE-MRI of (a) breast, (b) brain, (c) prostate, and (d) liver.
In general the DCE-MRI image acquisition protocols were done using a 3D SPGRE sequence. The TR and TE are consistent with temporal and spatial resolution that varies between clinical studies (different body parts).
Axial Images were acquired 4–8 mm in slice thickness. The dynamic data sets were acquired during and after injection of 0.1mmol/Kg (typically between 12 and 20cc) of Gd-DTPA contrast at a rate of .2–3 cc/sec for a temporal resolution from 5–30 seconds; the values depend on imaging protocol used.
2.2 Flow chart
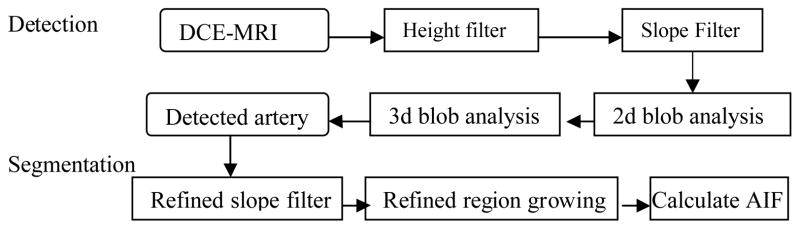
The flow chart of our AIF determination method is summarized in Fig. 1. It has a detection stage and a segmentation stage. A height filter is applied on the DCE-MRI to keep voxels with signal intensity greater than a threshold. Then a slope filter keeps only the voxels with a fast enough wash-in. A 2d blob labeling process is used to form distinguishable 2d-connected regions. The 3D overlapping region that is most cylindrical in shape is detected as the vessel. A refined slope filter and region growing algorithm is then conducted to segment the vessel. The AIF is calculated from the vessel’s intensity curves.
Fig. 1.
Flow Chart of AIF Determination Method
2.3 Artery Detection
In the detection phase, each voxel’s time intensity curve is examined. Major arteries have large amount of Gd-DTPA contrast agent flowing through them, causing the image voxels of the major arteries to brighten more than most other voxels. The peak height of an AIF is generally higher than the peak height of most others. Thus, a height filter is used to filter out voxels with small peak curves. The filter is written as,
| (1) |
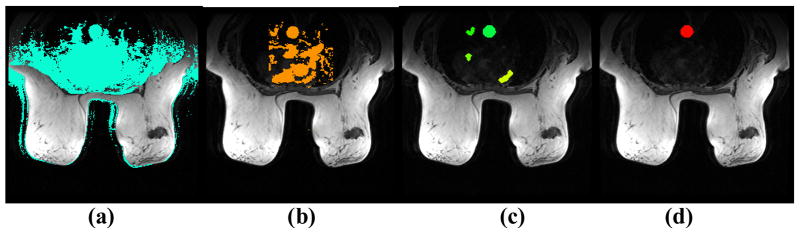
where tp is the time to peak, and μ is the mean peak value of all voxels in the image. The voxels passing the height filter are shown in Fig 2a.
Fig. 2. Illustration of AIF determination.
(a) Height Filter, (b) Initial Uptake Slope Filter, (c) 2d Blob regions, (d) Segmented Image.
The second filter is based on the fact that the signal intensity of major artery voxels brightens quickly from a fast wash-in of gadolinium contrast agent. This is represented by a steep uptake slope, thus a slope filter is used to keep the voxels with a slope greater than a threshold. The signal enhancement ratio of every time point is defined as
| (2) |
where R(t) is the ratio of signal enhancement from baseline to time t, S0 is the baseline signal which is computed as the average signal of the first three time points. The uptake slope is then computed as,
| (3) |
where tp is the signal peak time which is initially set to a time depending on the imaging protocol and body part. If the slope is greater than a preset threshold θ1, then it is kept as major artery candidate. The voxels passing the slope filter are shown in Fig 2b.
After the two filters, a blob labeling process is run on the remaining voxels to form distinguishable 2d connected components (we call them blobs) from the candidate voxels in each axial slice. A size filter is used to keep only blobs that are big enough to possibly be part of the major artery (Fig. 2c). For each blob, the average peak time is calculated, and the earliest 20% time-to-peaks are averaged and set as the estimated major artery time-to-peak tp′. The slope filter is repeated using the estimated tp′. The next step is to calculate the compactness and circularity of each of the remaining blob as follows,
| (4) |
here P is the perimeter of the blob and A is the area.
The 2d blobs are then propagated in transverse direction to form 3d blobs. 2d blobs that overlap each other in axial slices above or below are considered to be part of the same object in 3d. The cylindricality of each 3d object is calculated by summing across the axial slices the circularity of each 2d blob belonging to the same 3d object.
| (5) |
CZ is the circularity of the object on the axial slice z. The most cylindrical object is selected as the artery to determine AIF. We recompute the new time-to-peak (tp″) using the detected artery and use it for the rest of the slope calculations. The center of the detected artery is used as the seed for the artery segmentation algorithm in the second stage.
2.4 Artery Segmentation and AIF determination
The detected artery is then segmented using a region growing technique. The slope is recalculated using the refined time-to-peak tp″. A refined slope filter using a slightly lower slope threshold (θ2) is conducted to ensure that the entire major artery is segmented.
Then on the middle axial slice we run the region growing algorithm using the seed obtained in the detection stage to segment the artery. The region growing is then propagated to every other axial slice using the center of the segmented artery on the previous slice as the seed. The segmented main artery is shown in Fig 2d.
For each axial slice of the object, the average time-intensity curve of voxels in the artery is computed as the AIF. To accommodate for possible flow artifacts, the AIF in the axial slice with the highest peak is omitted and the AIF that has the second highest peak is used for calculation in the pharmacokinetic models.
2.5 Setting the parameters and thresholds
There are several parameters in our method. The first one is time-to-peak tp. tp is the time over which to calculate the uptake slope. This parameter is initially set depending on the image protocol used. Initially tp=6, 4, 9 and 7 for breast, liver, brain and prostate respectively. tp, is later adaptively refined twice using the detected blobs and arteries.
The next parameter is the artery size threshold. A 2d blob region can not be too big or too small to be considered as potentially being part of the artery. The aorta near the breast and liver is larger than the superior sagittal sinus in the brain and iliac artery near the prostate so the breast and liver images are set to have a higher size threshold. The minimum size threshold is set to 50, 50, 30, and 20 voxels for the breast, liver, brain, and prostate respectively. The max size threshold is set to 1500 voxels.
The other 2 parameters are the slope thresholds used in the two stage slope filters. The first slope threshold θ1, is used to remove unwanted voxels in the artery detection phase. The second slope threshold θ2, is set lower than the first one to ensure the whole artery is segmented. The breast and liver DCE-MRI are set to a higher slope threshold than the brain and prostate. The reason for this is the breast and liver have a bigger artery (aorta) and more blood flow which allows faster contrast uptake in the breast and liver compared to the iliac artery near the prostate and the superior sagittal sinus in the brain. The default value of θ1 = 0.6, 0.9, 0.2 and 0.2 breast, liver, brain and prostate respectively, and that of θ2 = 0.6, 0.9, 0.2 and 0.1
3 Experiments and Results
3.1 Data sets
We have randomly selected 10 DCE-MRI studies for each body part in our experiments. Table 1 lists the study specifications, including number of axial slices, number of time points, time interval between images, and injection rate. Among the 10 breast studies, 5 have invasive breast cancer, 2 are invasive ductal carcinoma cases and 3 are some type of cancerous tumor.
Table 1.
DCE-MRI data.
| DCE-MRI scan | Studies | Slices | Time Points | Interval (sec). | Injection Rate |
|---|---|---|---|---|---|
| Breast | 10 | 30 | 16/18 | 30 | .3 cc/sec & 2cc/sec |
| Prostate | 10 | 10 & 12 | 56 | 5 | .3 cc/sec & 2cc/sec |
| Liver | 10 | 12 | 23 | 23 | .3 cc/sec & 2cc/sec |
| Brain | 10 | 12 | 30 | 30 | .3 cc/sec |
3.2 Segmentation success rate
We conducted experiments to evaluate the segmentation success rate. We checked the automatic artery segmentation on every axial slice and compared it with an operator manual segmentation. The automatic artery segmentation is determined to be a success if more than 80% of the artery segmentation agrees with each other. Table 2 shows the summary of success rate for the 4 different DCE-MRI studies. Examples of successful segmentation are shown in Figure 3.
Table 2.
Segmentation success rate.
| Success rate (%) | Std (%) | Worst (%) | Best (%) | |
|---|---|---|---|---|
| Breast | 95.5 | 11.5 | 70 | 100 |
| Prostate | 82.9 | 18.9 | 40 | 100 |
| Liver | 98.3 | 3.5 | 91. 7 | 100 |
| Brain | 81.3 | 18.9 | 50 | 100 |
|
| ||||
| Average | 89.5 | 13.2 | 62.9 | 100 |
3.3 Comparison of pharmacokinetic parameters
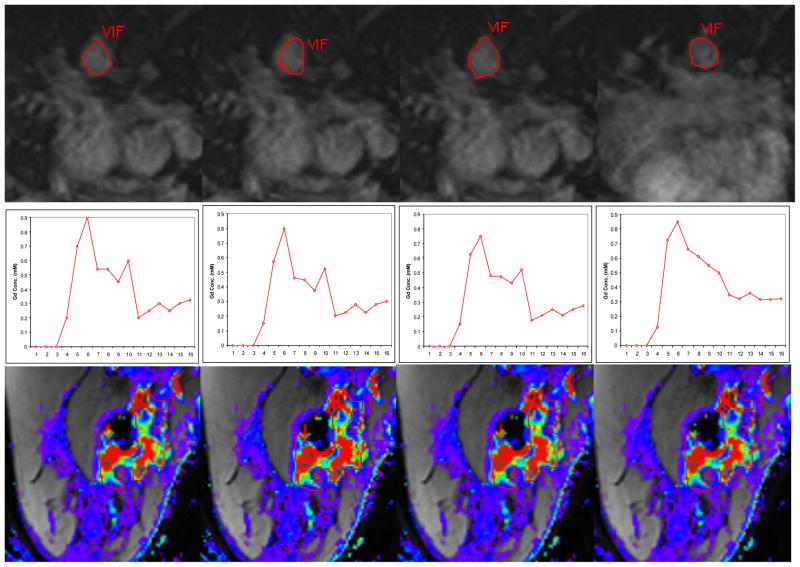
To further validate our method, we used the automatically determined AIF to compute pharmacokinetic parameters (Ktrans, kep, and ve) of the Generic Kinetic Model using a software called Cine Tool (GE Medical Systems) [4]. In addition to the automatically determined AIF, we also obtained AIFs from a manual segmentation of the artery on the same slice and two semi-automatic segmentations of the artery on the same and a different slice. For manual segmentation, the user manually traces the border of the artery. For semiautomatic segmentation, the user picks a seed point inside the artery and a region growing algorithm is run to segment the entire vessel (Cine Tool). Figure 4 shows examples of the four vessel segmentations, AIFs, and parameter maps. We used the 10 breast DCE-MRI cases for this experiment. For each case, we selected a biopsy-verified tumor region and evaluated the Ktrans, kep, and ve values of the tumor. Table 3 summarizes the differences between the tumor permeability kinetics values calculated using the four AIFs.
Fig. 4.
For each Row: (1st Row) Aorta segmentation, (2nd Row) AIF, (3rd Row) Color map of Ktrans values of a tumor ROI. Each Column: (a) Automatic segmentation on axial slice 11, (b) Manual Segmentation on axial slice 11, (c) Semi-Automatic Segmentation on axial slice 11 using Cine Tool, (d) Semi-Automatic Segmentation on axial slice 15 using Cine Tool.
Table 3.
Average percentage difference in GKM values compared to GKM values calculated using the semi-automatically determined AIF.
| % Difference | Manual | Automatic | Semi-Auto on different axial slices |
|---|---|---|---|
| Ktrans | 13.6±15.1 | 16.7±8.7 | 15.8±18.1 |
| kep | 11.9±9.6 | 9.08±10.4 | 18.9±10.9 |
| ve | 6.2±8.0 | 9.0±8.5 | 9.4±9.5 |
We further conducted paired t-tests comparing the average GKM Ktrans of the tumor region shown in Figure 4. Table 4 summarizes the comparison. The Ktrans values from the automatic AIF and the manually derived AIF are correlated though without statistical significance which is likely due to one outlier in the data set, giving validity that using the automated AIF can accurately calculate pharmacokinetic parameters. Comparison of Ktrans values derived from our automatic method and the semi-automatic AIF are statistically similar. The Ktrans values calculated from semi-automatic AIFs on different slices are also statistically similar (P=0.02); however, their difference is bigger than the difference between automatic AIF and the semi-auto on the same slice (P=0.01). This indicates that there is more variation when a different slice is used to generate the AIF than when picking a slightly different AIF on the same slice. Our automatic method will pick the same axial slice every time when choosing the AIF, making it more consistent than when two different operators possibly pick different axial slices to get the AIF on the same study.
Table 4.
Comparison of Ktrans values of one tumor, chi-squared and t-test.
| Correlation | Manual vs. Auto | Semi-Auto vs. Auto | Semi-Auto on different slices |
|---|---|---|---|
| R2 | 0.89 | 0.98 | 0.95 |
| P(T<=t) | 0.19 | 0.01 | 0.02 |
4 Conclusion and Discussion
We have proposed a versatile and robust method to automatically segment a major artery in various parts of the body to generate an AIF in DCE-MRI.
The manual and automatic artery segmentations are similar; however, small deviations in voxels used for the AIF can cause a change of GKM values up to 45%. Although there is variability, the GKM values calculated from the automatically derived AIF still reflect an estimate of the expected high tumor permeability. This shows validity that the automatically derived AIF can be used to calculate important values of pharmacokinetic models to assess tumors.
Our automatic method performed well in segmenting the aorta near the breast and liver. In some studies there was some over-segmentation when the aorta is close to the heart. The method had errors segmenting the major vessel in the brain in 3 out of the 10 studies due to over-segmentation of the branches off the main vessel. The prostate images were of worse quality and when the ureter runs adjacent to the iliac arteries, the method has a higher chance of failing by also segmenting the ureter. In the liver cases our method had the least problems in segmenting the artery. This is due to various factors; there were almost no motion artifacts; the aorta does not move even though surrounding organs may be moving, the aorta does not bend next to the liver; and there are no enhancing anatomical structures adjacent to the aorta.
We must point out that our method is not yet completely automatic for some cases. The two slope filter thresholds may needed to be adjusted manually to achieve optimal results. We are investigating adaptive techniques to fully automate these parameter settings.
References
- 1.Cancer Facts & Figures 2008 – American Cancer Society
- 2.Kety S. Cerebral circulation. In: Magoun H, editor. Neurophysiology. Handbook of physiology, vol III, sect 1. American Physiological Society; 1960. pp. 1751–1760. [Google Scholar]
- 3.Tofts P, et al. Estimating kinetic parameters from dynamic contrast-enhanced T1-weighted MRI of diffusible tracer: Standardized quantities and symbols. Journal of Magnetic Resonance Imaging. 1999;(10):223–232. doi: 10.1002/(sici)1522-2586(199909)10:3<223::aid-jmri2>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 4.Arpi Thukral, et al. Inflammatory Breast Cancer: Dynamic Contrast-enhanced MRI in Patients Receiving Bevacizumab-Initial Experience. Radiology. 2007;244:727–735. doi: 10.1148/radiol.2443060926. [DOI] [PubMed] [Google Scholar]
- 5.Harrer JU, Parker GJ, et al. Comparative study of methods for determining vascular permeability and blood volume in human gliomas. J Magn Reson Imaging. 2004 Nov;20(5):748–57. doi: 10.1002/jmri.20182. [DOI] [PubMed] [Google Scholar]
- 6.Choyke P, Dwyer A, Knopp M. Functional Tumor Imaging With Dynamic Contrast-Enhanced Magnetic Resonance Imaging. J Magn Reson Imaging. 2003 Jan;17:509–520. doi: 10.1002/jmri.10304. [DOI] [PubMed] [Google Scholar]
- 7.Morris ED, et al. Automated Determination of the Arterial Input Function for MR Perfusion Analysis; Presented at: 8th ISMRM; Denver, CO. April 3–7, 2000. [Google Scholar]
- 8.Parker GJ, Jackson A, Waterton JC, Buckley DL. Automated Arterial Input Function Extraction for T1-Weighted DCE-MRI. ISMRM; 2003. [Google Scholar]