Abstract
Rationale
Incorporation coefficients K* of arachidonic acid (AA) in brain are increased in a rat model of neuroinflammation, as are other markers of AA metabolism. Data also indicate that neuroinflammation contributes to Alzheimer disease (AD). On the basis of these observations, K* for AA was hypothesized to be elevated in AD patients.
Methods
Eight AD patients were studied with an average Mini-Mental State Examination (MMSE) score of 14.7 ± 8.4 (S.D.), aged 71.7 ± 11.2 years, as were 9 controls with a normal MMSE score, aged 68.7 ± 5.6 years. Each subject received a [15O]water positron emission tomography (PET) scan of regional cerebral blood flow (rCBF), followed after 15 min by a [1-11C]AA scan of regional K* for AA.
Results
In the AD compared to control subjects, global grey-matter K* for AA (corrected or uncorrected for the partial volume error, PVE) was significantly elevated, whereas only PVE-uncorrected global CBF was reduced significantly (p < 0.05). A False Discovery Rate (FDR) procedure indicated that PVE-corrected K* for AA was increased in 78 of 90 identified hemispheric grey-matter regions. PVE-corrected rCBF, although decreased in 12 regions at p < 0.01 by unpaired t-tests, did not survive the FDR procedure. The surviving K* increments were widespread in the neocortex, but were absent in caudate, pallidum and thalamic regions.
Conclusions
These preliminary results show that K* for AA is widely elevated in the AD brain, particularly in regions reported to have high densities of senile (neuritic) plaques with activated microglia. To the extent that the elevations represent upregulated AA metabolism associated with neuroinflammation, PET with [1-11C]AA could be used to examine neuroinflammation in patients with AD and other brain diseases.
Keywords: arachidonic, neuroinflammation, PET, Alzheimer, imaging
INTRODUCTION
The post-mortem Alzheimer disease (AD) brain is characterized by intracellular neurofibrillary tangles with paired helical filaments consisting of phosphorylated tau protein, and extracellular senile (neuritic) plaques containing β-amyloid fibrils. The senile plaques often are infiltrated by activated microglia that secrete inflammatory cytokines, release nitric oxide and express peripheral benzodiazepine receptors (1-4).
Inflammatory cytokines released from the microglia can bind to astrocytic cytokine receptors that are coupled to Ca2+-dependent enzymes, including cytosolic phospholipase A2 (cPLA2) and secretory sPLA2 (5). Each of these enzymes, when activated, hydrolyzes unesterified arachidonic acid (AA, 20:4n-6) from membrane phospholipids. The released nitric oxide also can promote AA hydrolysis from membrane by cPLA2, by stimulating glutamate release from nerve terminals and thereby increasing intracellular Ca2+ concentrations via post-synaptic ionotropic N-methyl-D-aspartate and other glutamatergic receptors (6-8). β-amyloid peptide in the AD brain also can provoke glutamate-induced excitotoxicity and well as PLA2 activation (9, 10). Consistent with this scenario, the AD brain demonstrates increased cytokine levels (3), increased expression of both cPLA2 and sPLA2, increased concentrations of inflammatory metabolites of AA (5, 11, 12), and increased glutamatergic markers and different forms of accumulated β-amyloid (9, 10, 13). Additionally, concentrations of isoprostane and isofuran metabolites of AA are elevated in cerebrospinal fluid (CSF) from AD patients (14).
In view of these observations, AA metabolism likely is elevated in the AD brain, and having a method to image this metabolism might help in examining neuroinflammation in AD in relation to disease course and therapy. We have developed such a method, which involves injecting radiolabeled AA intravenously, then determining regional brain AA incorporation coefficients K* (brain radioactivity normalized to integrated plasma radioactivity) using quantitative autoradiography in unanesthetized rodents or positron emission tomography (PET) in primates or humans. K* for AA represents metabolic loss of AA in brain, since once lost by metabolism, AA cannot be synthesized de novo nor converted from its precursor, linoleic acid (18:2n-6) (15). K* for AA is unaffected by changes in regional cerebral blood flow (rCBF), which makes it an ideal tracer for independently imaging brain metabolism (16-19).
Using this method, K* for AA was shown to be increased in wide areas of brain in unanesthetized rats in which bacterial lipopolysaccharide had been infused into the cerebral ventricles for 6 days, a model of chronic neuroinflammation. The elevations were accompanied by increased brain expression of cPLA2, increased turnover rates of AA in brain membrane phospholipids, increased formation of AA-derived eicosanoids such as prostaglandin E2 and, after a longer infusion time, formation of activated microglia (20-23). Furthermore, the increments in K* following lipopolysaccharide infusion could be reduced by chronic pre-treatment with lithium, which is reported to downregulate each of these markers of brain AA metabolism in control rats (24).
In this paper, the fatty acid method with PET and intravenous [1-11C]AA was used to image K* for AA, as a marker of neuroinflammation, in diagnosed AD patients and age-matched healthy controls. Methods for making measurements and calculations are reported elsewhere in detail (17-19, 25). PET and intravenous [15O]water also were employed to image rCBF as a marker of brain functional activity in the same subjects (26), and to co-register AA scans to anatomical magnetic resonance (MR) scans for regional data analysis and for correcting for the partial volume error (PVE).
MATERIALS AND METHODS
Subject Selection
The protocol was approved by the Institutional Review Board of the National Institute on Neurological Diseases and Stroke, and by the Radiation Safety Committee of the National Institutes of Health. Eight otherwise healthy male AD patients (mean age 71.1 ± 11.2 (S.D.) yr, range 51-87 yr) who met research criteria for AD and were mainly mildly-moderately (one severely) demented on the Mini-Mental State Examination (MMSE) (score 14.7 ± 8.4, range 4-27) (27) were studied, as were 9 healthy age-matched male controls (mean age 68.7 ± 5.6, range 60-76 yr, mean MMSE score 30). Exclusion criteria included a history of head trauma, hypertension or other cardiovascular disorder, diabetes, alcoholism, psychiatric or neurological disorder, or malignancy. AD and control subjects were normotensive and off centrally-acting medication for at least 2 weeks, aspirin for at least 2 days, and caffeine and alcohol for at least 12 hours before scanning.
PET Procedure
[1-11C]AA was synthesized as reported (17, 28). The tracer was 97.6% pure on high performance liquid chromatography and its specific activity exceeded 100 mCi/μmol.
Indwelling radial vein and artery catheters were inserted prior to scanning, and the subject’s head was secured in a thermoplastic facemask fixed to the scanner bed. Scanning was performed with an Advance Tomograph (GE Healthcare, Waukesha, WI), which acquires 35 simultaneous slices with 4.25-mm separation and has in-plane and axial resolutions of 6-7 mm. Scans were obtained parallel to the orbitomeatal line and were conducted in a quiet dimly lit room, with the subject’s eyes open and ears unoccluded.
After performing a transmission scan for attenuation correction, 370 MBq (10 mCi) [15O]water was injected as an intravenous bolus to measure rCBF. A 60-s scan was acquired in the 3-dimensional (3D) mode after the bolus had reached the brain, and rCBF images were produced using the measured arterial input function (26). To quantify regional values of K* for AA and regional blood volumes Vb, 15 min after the [15O]water injection, 920 ± 115 MBq (24.9 ± 3.1 mCi) of [1-11C]AA was infused intravenously for 3 min at a constant rate, and serial dynamic 3D scans (30 s to 5 min) were acquired for one hour. Radial artery blood (1-3 mL) was sampled at fixed times, and radioactivity in whole blood and plasma was measured with a gamma counter. Details of the procedures are published (18, 19).
Motion Correction
Subject motion during the 60-min acquisition period was corrected with a mutual information registration of each time frame to a standard frame before attenuation correction. Based on calculated motion, the transmission images were resliced and projected for final attenuation correction, reconstruction, and realignment.
Plasma time-activity curves
Whole blood and plasma radioactivities were determined as a function of time after injecting [1-11C]AA. In 7 of the 9 controls, a rapid extraction procedure (18, 29) was used to determine radioactivity due to [1-11C]AA and [1-11C]CO2. In all the AD patients and in 2 of the controls, fractional metabolite measurements were not performed with this procedure. To analyze all PET scans together, therefore, the fractional data ([1-11C]AA and [1-11C]CO2) from the 7 controls having appropriate input function data were averaged to obtain mean fractional curves, which then were used to process individual radioactivity curves in each AD patient. The curves were multiplied by the group average fractional curve to produce time-activity curves of [1-11C]AA and [1-11C]CO2 in these patients and in the 2 remaining controls. To assess the effect of using average curves, data from the 7 controls also were analyzed with the averaged metabolite curves. T-tests showed that the results were not significantly affected by this procedure. To have a consistent statistical analysis, we therefore report data on all subjects processed with the averaged metabolite curves.
Modeling
On a pixel-by-pixel basis, reconstructed images were analyzed using the following equation to produce parametric images of K* for AA (incorporation coefficient of plasma AA into brain tissue, μL/min/mL brain) and of Vb (cerebral blood volume, (ml blood)/(ml brain)),
| [1] |
Ci(t), Cb(t), and Cp(t) are pixel, whole blood, and plasma [1-11C]AA time-activity curves, respectively; CCO2(t) is the predicted brain tissue concentration of [11C]CO2, and Δt is the delay between the brain and blood curves. The predicted tissue [11C]CO2 concentration was computed by measuring for each subject the plasma [11C]CO2 concentration and applying a one-tissue compartment model with published values of the gray matter influx rate constant K1 and the distribution volume for CO2 (30). Calculations were applied to the original radioactivity images and to images that had been corrected for the partial volume error (PVE) (18, 31).
Registration to MR images
For each subject, K* images derived from the original PET volumes following [1-11C]AA injection were registered to the CBF volume to correct for motion between the [1-11C]AA and [15O]water scans, using a 6-parameter transformation and a mutual information cost function (19, 32). CBF and MR volumes then were co-registered using the same algorithm. Images of K* for AA were transformed to MR space using the product of the two transformation matrices.
Partial Volume Error Correction
Brain radioactivity, corrected for the PVE, was calculated with a 3-compartment (gray matter, white matter, and CSF) MR-based approach (19, 33). Binary mask images for gray matter (mGM), white matter (mWM), and CSF (mCSF) first were produced using an adaptive fuzzy c-mean segmentation. The gray and white matter masks then were smoothed, based on a 6-mm full-width at half-maximum 3D Gaussian kernel that corresponded to the scanner’s resolution, to account for radioactivity spill-out and spill-in effects between gray and white matter and for spill-out of gray matter activity into CSF. White matter radioactivity was assumed uniform and CSF radioactivity was set at zero. For each voxel, the PVE-corrected value was calculated as:
| [2] |
where C3S is PVE-corrected gray matter radioactivity, C is measured PET radioactivity, CWM is estimated white matter radioactivity, and sGM and sWM are smoothed masks for gray and white matter, respectively. Cwm was calculated by an extrapolation method (19). For each frame, PET activity values of pixels with swm values > 0.99 were identified and then fitted as a linear function of swm. The fitted value at swm = 1 was used as Cwm.
Image Processing for Statistical Analysis
Individual rCBF images were aligned without smoothing to a stereotactic PET template using Statistical Parametric Mapping-2 software [http://www.fil.ion.ucl.ac.uk/spm/software] (19, 34). The transformation information obtained by alignment was used to resample the K* images from the [1-11C]AA scans, after they had been aligned to the CBF image in the same stereotactic space. The same transformation information was used to resample the gray matter volume into the stereotactic space. Region of interest (ROI) masks identifying 90 gray matter regions in the stereotactic space of the cerebral hemispheres (35), plus the resampled gray matter mask, then were applied to each subject’s spatially normalized rCBF and K* images to obtain 90 (45 bilateral) weighted mean rCBF and K* for AA values, respectively. Using the gray matter mask minimized mixing white matter values into the calculated ROI measurements.
For each subject, global gray matter rCBF and K* for AA were calculated by averaging the 90 bilateral regional hemispheric values of rCBF and K*, respectively. Statistical comparisons were made between AD and control means. Data from an individual AD patient were not matched to data from an exact-age control, as PVE-corrected rCBF, K* for AA and regional glucose metabolism do not change with age in healthy nonhypertensive controls (19, 36), and the numbers of subjects were too limited to do this in any case. Group comparisons for global and PVE-corrected regional means were made using two-tailed t-tests. Additionally, a False Discovery Rate (FDR) procedure (37) was used to account for the increased risk of performing multiple comparisons. In this procedure, it was accepted that 5% of surviving differences between means were false positives. The FDR procedure applies to statistically independent as well as to positively correlated data (38).
RESULTS
Global values of CBF and K* for AA
Table 1 lists mean values of global grey matter CBF and K* for AA in the AD and control subjects, before and after a PVE correction. Without the correction, global CBF was reduced significantly by 28.6% in the AD (n = 6) compared to control (n = 7) subjects, whereas global K* for AA was elevated significantly by 18.3% in the AD (n = 8) compared to control (n = 9) subjects. There were fewer CBF and K* measurements because some of the CBF studies failed.
Table 1.
Global values of CBF and of K* for arachidonic acid in healthy control and Alzheimer disease subjects, before and after correction for partial volume error (PVE). Values were calculated from mean values from 90 hemispheric regions identified in a stereotactic atlas (35), and were compared by two-tailed t-tests
| Group | PVE-Uncorrected | PVE-Corrected | |||
|---|---|---|---|---|---|
| CBF | K* | CBF | K* | ||
| mL/100g/min | μL/min/mL | mL/100g/min | μL/min/mL | ||
| Control | Numbera | 7 | 9 | 7 | 9 |
| Mean ± SD CV % |
38.0 ± 5.3 (13.8) |
4.7 ± 0.6 (12.0) |
61.6 ± 10.1 (16.4) |
6.9 ± 0.8 (11.9) |
|
| AD | Numbera | 6 | 8 | 6 | 8 |
| Mean ± SD CV % |
27.1 ± 7.6* (28.1) |
5.5 ± 0.8* (15.2) |
48.3 ± 14.0 (28.9) |
8.7 ± 1.4** (15.7) |
|
| Diff % | -28.6 | 18.3 | -21.5 | 26.4 | |
Differs significantly from control mean by 2-tailed t-test, *p ≤ 0.05
p ≤ 0.01
Number of CBF scans is fewer than K* scans because of failure of some CBF scans
Since a PVE correction compensates for spill-out/spill-in effects by resolution and makes the measured tissue concentration approximate the true concentration (19), this correction increased global CBF and K* for AA. After the correction, differences between the AD patients and controls remained significant for global K* (p < 0.01) but not global CBF. PVE-corrected K* for AA was 26.4% higher in the AD patients than controls, 8.7 ± 1.4 μL/min/mL compared to 6.9 ± 0.8 μL/min/mL.
Comparison of regional PVE-corrected values of rCBF and K*
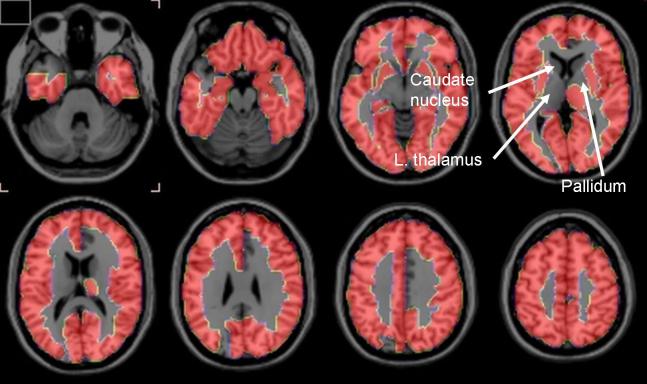
Two-tailed t-tests showed that PVE-corrected rCBF was less at p < 0.01 in the AD than control subjects in 12 of the 90 regions examined (data not shown). However, these regions, as well as the remaining 78, did not survive the FDR analysis. In contrast, 78 regions survived the FDR analysis with regard to PVE-corrected K* for AA, with all K*’s being higher in the AD than control subjects (Table 2). Regions that survived were largely in the neocortex, hippocampus, and amygdala. About half of the 12 regions that did not survive the FDR analysis were subcortical, and included the left thalamus, left and right pallidum, and left and right caudate nucleus. These differences are illustrated in the serial horizontal brain slices of Figure 1.
Table 2.
Brain regions where PVE-corrected K* for AA differed by the FDR procedure (bold type) between Alzheimer disease (AD) and control subjects. Regions defined in stereotactic atlas (35)
| Region name | Mean (SD) | Diff | Diff | p-value | Region name | Mean (SD) | Diff | Diff | p-value | ||
|---|---|---|---|---|---|---|---|---|---|---|---|
| AD | Control | % | (2-tail) | AD | Control | % | (2-tail) | ||||
| ParaHippocampal_R | 6.83(1.01) | 5.16(0.62) | 1.68 | 32.5 | 0.001 | Parietal_Inf_L | 9.16(1.49) | 7.32(0.97) | 1.84 | 25.2 | 0.008 |
| Hippocampus_L | 6.95(1.17) | 5.16(0.51) | 1.79 | 34.6 | 0.001 | Heschl_R | 9.33(1.52) | 7.52(0.93) | 1.81 | 24.1 | 0.009 |
| Temporal_Sup_R | 9.78(1.46) | 7.39(0.91) | 2.39 | 32.3 | 0.001 | Occipital_Mid_L | 10.54(2.17) | 7.87(1.49) | 2.67 | 33.9 | 0.009 |
| Thalamus_R | 8.6(1.17) | 6.68(0.77) | 1.93 | 28.9 | 0.001 | Olfactory_L | 6.24(1.31) | 4.81(0.58) | 1.43 | 29.8 | 0.009 |
| Frontal_Mid_Orb_R | 8.08(1.24) | 6.13(0.74) | 1.95 | 31.7 | 0.001 | Frontal_Sup_L | 8.31(1.27) | 6.65(1.04) | 1.66 | 25.0 | 0.010 |
| Hippocampus_R | 6.78(1.01) | 5.27(0.49) | 1.51 | 28.6 | 0.001 | Insula_R | 7.35(1.31) | 5.96(0.52) | 1.40 | 23.5 | 0.010 |
| Parietal_Sup_L | 9.89(1.48) | 7.45(1.08) | 2.44 | 32.7 | 0.001 | Frontal_Mid_R | 8.97(1.4) | 7.29(0.95) | 1.68 | 23.1 | 0.010 |
| Fusiform_R | 7.82(1.07) | 6.12(0.71) | 1.70 | 27.8 | 0.001 | Postcentral_R | 9.18(1.41) | 7.55(0.85) | 1.63 | 21.6 | 0.010 |
| Temporal_Mid_L | 9.99(1.94) | 7.22(0.89) | 2.78 | 38.5 | 0.001 | Paracentral_Lobule_L | 9.07(1.38) | 7.41(0.96) | 1.66 | 22.4 | 0.011 |
| Fusiform_L | 7.98(1.38) | 6.06(0.53) | 1.92 | 31.6 | 0.001 | SupraMarginal_L | 9.52(2) | 7.28(1.13) | 2.24 | 30.8 | 0.011 |
| Paracentral_Lobule_R | 9.52(1.17) | 7.5(1) | 2.02 | 26.9 | 0.002 | Angular_R | 9.46(1.9) | 7.33(1.12) | 2.13 | 29.0 | 0.012 |
| Olfactory_R | 6.29(0.91) | 4.84(0.67) | 1.45 | 29.8 | 0.002 | Parietal_Inf_R | 8.91(1.21) | 7.4(0.99) | 1.51 | 20.4 | 0.013 |
| Frontal_Mid_Orb_R | 10.8(1.53) | 8.25(1.28) | 2.54 | 30.8 | 0.002 | Frontal_Sup_Orb_R | 10.22(1.49) | 8.25(1.39) | 1.97 | 23.8 | 0.013 |
| Temporal_Sup_L | 9.62(1.89) | 7.02(0.87) | 2.60 | 37.0 | 0.002 | Frontal_Inf_Tri_R | 9.43(1.9) | 7.51(0.76) | 1.92 | 25.5 | 0.014 |
| Rolandic_Oper_R | 8.81(1.07) | 7.09(0.85) | 1.72 | 24.3 | 0.002 | Frontal_Inf_Orb_R | 10.06(1.93) | 7.97(1.09) | 2.09 | 26.2 | 0.014 |
| Temporal_Inf_R | 8.77(1.29) | 6.88(0.79) | 1.88 | 27.4 | 0.002 | Putamen_L | 8.54(2.19) | 6.48(0.49) | 2.06 | 31.9 | 0.015 |
| Precuneus_L | 9.35(1.31) | 7.4(0.85) | 1.94 | 26.2 | 0.002 | Frontal_Mid_L | 8.85(1.51) | 7.1(1.11) | 1.76 | 24.7 | 0.015 |
| Lingual_R | 8.84(1.06) | 7(1.01) | 1.84 | 26.3 | 0.002 | Angular_L | 9.76(2.05) | 7.49(1.35) | 2.27 | 30.3 | 0.016 |
| Precuneus_R | 9.25(1.35) | 7.28(0.82) | 1.96 | 27.0 | 0.002 | Occipital_Mid_R | 10.38(2.24) | 7.92(1.47) | 2.47 | 31.2 | 0.016 |
| Temporal_Mid_R | 9.67(1.52) | 7.42(0.99) | 2.25 | 30.4 | 0.002 | Rectus_R | 8.41(1.73) | 6.61(0.94) | 1.80 | 27.2 | 0.016 |
| Frontal_Sup_Medial_L | 8.15(1.16) | 6.39(0.83) | 1.75 | 27.4 | 0.003 | Occipital_Inf_R | 9.88(2.24) | 7.35(1.59) | 2.53 | 34.4 | 0.016 |
| Frontal_Mid_Orb_L | 8.12(1.3) | 6.19(0.9) | 1.92 | 31.1 | 0.003 | Frontal_Inf_Oper_L | 8.9(1.81) | 7.12(0.79) | 1.78 | 25.0 | 0.017 |
| Supp_Motor_Area_L | 8.57(1.28) | 6.77(0.79) | 1.81 | 26.7 | 0.003 | Calcarine_L | 10.07(1.32) | 8.25(1.48) | 1.83 | 22.2 | 0.017 |
| Temporal_Inf_L | 8.8(1.78) | 6.52(0.72) | 2.29 | 35.1 | 0.003 | Rectus_L | 8.11(1.19) | 6.63(1.1) | 1.48 | 22.3 | 0.018 |
| Insula_L | 7.51(1.29) | 5.75(0.74) | 1.76 | 30.7 | 0.003 | Frontal_Inf_Orb_L | 10.31(2.42) | 7.92(1.11) | 2.39 | 30.1 | 0.018 |
| Amygdala_L | 5.77(1.42) | 4.01(0.47) | 1.76 | 43.8 | 0.003 | SupraMarginal_R | 9.07(1.51) | 7.43(1.01) | 1.63 | 22.0 | 0.018 |
| Occipital_Inf_L | 10.27(2.17) | 7.24(1.38) | 3.04 | 42.0 | 0.003 | Cuneus_L | 10.04(1.56) | 8.27(1.17) | 1.76 | 21.3 | 0.018 |
| Calcarine_R | 10(1.39) | 7.96(1.07) | 2.05 | 25.7 | 0.004 | Temporal_Pole_Mid_R | 9.11(1.34) | 7.53(1.12) | 1.58 | 21.0 | 0.018 |
| Frontal_Mid_Orb_L | 10.84(1.75) | 8.28(1.35) | 2.56 | 30.8 | 0.004 | Occipital_Sup_R 5 | 10.33(1.88) | 8.18(1.49) | 2.15 | 26.3 | 0.019 |
| Lingual_L | 8.93(1.21) | 7.2(0.89) | 1.73 | 24.1 | 0.004 | Frontal_Inf_Tri_L | 9.39(2.13) | 7.38(1.02) | 2.01 | 27.3 | 0.023 |
| Postcentral_L | 9.54(1.44) | 7.49(1.05) | 2.04 | 27.3 | 0.004 | Precentral_R | 8.82(1.19) | 7.6(0.88) | 1.22 | 16.1 | 0.028 |
| Amygdala_R | 5.32(0.96) | 4.09(0.53) | 1.23 | 30.2 | 0.004 | Putamen_R | 8.6(2.09) | 6.86(0.7) | 1.75 | 25.5 | 0.031 |
| Precentral_L | 9.14(1.4) | 7.24(0.92) | 1.90 | 26.2 | 0.004 | Cingulum_Ant_L | 6.46(1.32) | 5.28(0.69) | 1.18 | 22.4 | 0.032 |
| Temporal_Pole_Sup_R | 8.04(0.88) | 6.62(0.88) | 1.42 | 21.4 | 0.005 | Regions which did not survive FDR analysis | |||||
| Cuneus_R | 10.38(1.55) | 8.24(1.11) | 2.14 | 26.0 | 0.005 | Occipital_Sup_L | 10.52(2.4) | 8.39(1.65) | 2.13 | 25.4 | 0.047 |
| Parietal_Sup_R | 9.98(2.1) | 7.4(1.06) | 2.58 | 34.8 | 0.005 | Cingulum_Mid_R | 7.7(1.48) | 6.55(0.66) | 1.15 | 17.6 | 0.051 |
| Heschl_L | 9.18(1.79) | 7.02(0.88) | 2.17 | 30.9 | 0.006 | Thalamus_L | 8.21(1.77) | 6.95(0.56) | 1.26 | 18.1 | 0.060 |
| Frontal_Sup_Medial_R | 8.28(1.07) | 6.68(0.98) | 1.60 | 23.9 | 0.006 | Temporal_Pole_Sup_L | 7.99(2.24) | 6.38(1) | 1.61 | 25.3 | 0.069 |
| Supp_Motor_Area_R | 8.58(1.19) | 7.02(0.82) | 1.56 | 22.2 | 0.006 | Temporal_Pole_Mid_L | 9.17(2.58) | 7.41(1.21) | 1.76 | 23.7 | 0.087 |
| Frontal_Sup_R | 8.39(1.09) | 6.84(0.93) | 1.55 | 22.6 | 0.006 | Cingulum_Post_L | 7.75(1.71) | 6.64(0.61) | 1.11 | 16.7 | 0.087 |
| Frontal_Inf_Oper_R | 9.01(1.4) | 7.34(0.73) | 1.67 | 22.7 | 0.007 | Cingulum_Ant_R | 6.29(1.27) | 5.45(0.73) | 0.83 | 15.3 | 0.113 |
| Frontal_Sup_Orb_L | 10.06(1.24) | 8.14(1.28) | 1.92 | 23.6 | 0.007 | Pallidum_L 7021 | 6.69(1.49) | 5.83(0.66) | 0.86 | 14.8 | 0.136 |
| Rolandic_Oper_L | 8.67(1.59) | 6.81(0.78) | 1.86 | 27.3 | 0.007 | Cingulum_Post_R | 7.45(2.39) | 6.17(0.58) | 1.28 | 20.7 | 0.140 |
| ParaHippocampal_L | 6.92(1.47) | 5.21(0.73) | 1.71 | 32.9 | 0.007 | Pallidum_R | 6.5(1.51) | 5.71(1.04) | 0.79 | 13.8 | 0.226 |
| Cingulum_Mid_L | 7.77(1.3) | 6.33(0.5) | 1.44 | 22.7 | 0.008 | Caudate_R | 6.02(1.92) | 5.33(0.74) | 0.68 | 12.8 | 0.338 |
| Caudate_L | 5.8(2.15) | 5.33(0.76) | 0.47 | 8.9 | 0.544 | ||||||
Figure 1.
Horizontal brain slices showing brain regions in which partial volume error-corrected K* for arachidonic acid survived a False Discovery Rate procedure that separated Alzheimer disease and control subjects at the 5% level of significance. Regions in the 1-mm thick T1 MRI slices are defined in Table 2 from a stereotactic atlas (35).
Normalized regional values of rCBF and K* for AA
After PVE-corrected rCBF was normalized to global PVE-corrected CBF, seven group mean differences survived the FDR procedure. Reductions were noted in the right temporal inferior and middle gyri, the left precuneous, and the right supramarginal gyrus, whereas increases occurred in the left frontal superior medial gyrus, left putamen and right thalamus. No group difference of normalized PVE-corrected K* for AA survived the FDR procedure.
Regional blood volume
Significant differences in regional blood volume Vb (Eq. 1) between AD and control subjects occurred in only a few regions, and were not in a consistent direction (data not shown).
DISCUSSION
PET with intravenously injected [1-11C]AA was used to show that mean values of PVE-uncorrected and -corrected grey matter global hemispheric K* for AA were increased by 18% (p < 0.05) and 26% (p < 0.01), respectively, in otherwise healthy, mildly-moderately (one severely) demented unmedicated AD patients, compared with age-matched healthy controls. Of the 90 hemispheric regions studied (35), the FDR procedure showed that mean PVE-corrected regional K* for AA was elevated in 78 regions, largely in the cerebral cortex. The 12 regions in which PVE-corrected regional K* did not survive the FDR procedure included right and left caudate, right and left pallidum, and left thalamus (Table 2 and Figure 1).
PVE-uncorrected but not PVE-corrected mean global CBF was significantly less by 26% (p < 0.05) in the AD than control subjects (Table 1). Of the 90 regions examined, PVE-corrected rCBF was reduced at p < 0.01 on a two-tailed t-test in 12 of them, but none of these reductions survived the FDR procedure. The procedure did indicate that PVE-corrected rCBF, when normalized to the global CBF, was reduced in 4 regions and increased in 3, of which two were the left putamen and right thalamus. Significant differences in regional blood volume Vb between the two groups occurred in a few regions but were not in the same direction and were uninformative.
The mean PVE-corrected global gray matter CBF in the controls, 61.0 ± 10.1 (CV = 16.4%) ml/100 g/min (Table 1) is similar to a reported gray matter mean in older healthy subjects, 66 (CV = 13%) ml/100 g/min (19). The mean PVE-corrected global K* for AA, 6.9 ± 0.8 (CV = 11.9%) μL/min/mL (Table 1), corresponds to a published global gray matter mean, 7.03 (CV = 8%) μL/min/mL in older healthy subjects (19). Regional PVE-corrected means of rCBF and K* for AA also correspond to published values.
The significant reduction in the mean global CBF uncorrected for the PVE (Table 1) also agrees with the literature (39), whereas the absence of a significant difference or of a difference that survived the FDR procedure in PVE-corrected CBF or rCBF, respectively, likely reflected the few (six) subjects in which rCBF was measured, the high coefficient of variation of the flow measurements (Table 1), and noise introduced by the PVE correction (40).
The results support the hypothesis (see Introduction) that regional AA incorporation coefficients K* would be increased in AD patients, as a marker of upregulated AA metabolism accompanying neuroinflammation. This hypothesis was based on showing elevated brain AA incorporation coefficients K* and other markers of AA metabolism in a rodent lipopolysaccharide-infusion model of neuroinflammation, and on evidence of neuroinflammation associated with senile (neuritic) plaques surrounded by activated microglia and of accumulated β-amyloid in the AD brain (see Introduction). The more common distribution of senile plaques in cortical than subcortical brain regions (41, 42) may explain why PVE-corrected K* for AA in the left thalamus and in bilateral caudate nucleus and pallidum did not survive the FDR analysis (Table 2, Figure 1).
Comparing K* for AA with rCBF in this study was useful for several reasons. The [15O]water PET scans helped to co-register a subject’s [1-11C]AA PET scan with his anatomic MR scan, helping to identify regions in the K* images and to perform the PVE correction (32). K*’s independence of rCBF was confirmed by finding that PVE-corrected values for global and regional K* were elevated, whereas uncorrected global CBF was decreased and PVE-corrected rCBF tended to decrease. Flow independence of K* has been demonstrated in preclinical studies (17), and exists because the unesterified AA that is highly bound to circulating albumin acts as an “infinite” and rapidly available source of for AA delivery to brain (16).
The pattern of AD-related differences in K* for AA in this study might be compared to differences found with other relevant PET compounds. For example, brain uptake of [11C](R)-PK11195, a ligand for peripheral benzodiazepine receptors on activated brain microglia, was increased in cortical but not subcortical regions in AD compared with control subjects (43, 44), as was uptake of [11C]PIB, a ligand for β-amyloid (45). Comparisons with [11C]PIB imaging may be particularly important in the course of AD, as β-amyloid peptide alone can stimulate cytokine formation and activate PLA2 before senile plaques accumulate (9, 10). Glucose metabolism, to which rCBF is coupled, is reduced in AD more in neocortical than subcortical regions (31, 46).
The results of this paper should be considered preliminary. Future studies should include more subjects, and perhaps consider additional methods (e.g. SPM, correlation approaches) to evaluate the data. Doing so should help to identify significant changes in rCBF and to relate rCBF and K* for AA in AD patients to each other and to dementia severity. Individual metabolite-corrected input functions to calculate K* also should be employed. We could not do this in the AD patients and in two of the controls in this paper, but comparable significant changes in K* using the average input function after correcting for plasma metabolites indicated that this did not affect our conclusions.
Regional rCBF and K* for AA were analyzed after correcting for the PVE, to address the effect of atrophy and estimate actual parenchymal rates (19, 31). With this correction, the estimated tracer concentration in high uptake areas is increased, which translates into a non-uniform increase in K* for AA or rCBF. Furthermore, the correction increases noise in the data (40). However, the results and the analyses of global values suggest that this correction did not affect group separation with regard to K*.
In summary, regional PVE-corrected AA incorporation coefficients K*, a measure of regional brain AA consumption, were elevated in widespread cortical areas of mildly-moderately (one severely) demented but otherwise healthy AD patients, compared with age-matched controls. These results support the hypothesis, which was based on evidence of upregulated markers of brain AA metabolism in a rat model of neuroinflammation, and of the presence of neuroinflammation in AD, that K* for AA would be elevated in AD. If such elevations are confirmed in follow-up studies on larger groups of patients and controls, we might then use intravenous [1-11C]AA with PET to image AA’s involvement in neuroinflammation in AD and other brain diseases.
ACKNOWLEDGEMENTS
We thank Dr. Madhav Thambisetty for his helpful comments about this manuscript. This work was supported by the intramural programs of National Institute on Aging, the National Institute on Neurological Disease and Stroke, and the PET Department of the Clinical Center at the National Institutes of Health, Bethesda, Maryland. No author has a conflict of interest with regard to this manuscript.
Abbreviations
- AA
arachidonic acid
- AD
Alzheimer disease
- CBF
global cerebral blood flow
- rCBF
regional cerebral blood flow
- CSF
cerebrospinal fluid
- CV
coefficient of variation
- FDR
False Discovery rate
- MMSE
Mini-Mental State Examination
- MR
magnetic resonance
- PET
positron emission tomography
- cPLA2
cytosolic phospholipase A2
- sPLA2
secretory PLA2
- PVE
partial volume error
- ROI
region of interest
- SPM
statistical parametric mapping
REFERENCES
- 1.Banati RB, Myers R, Kreutzberg GW. PK (‘peripheral benzodiazepine’)--binding sites in the CNS indicate early and discrete brain lesions: microautoradiographic detection of [3H]PK11195 binding to activated microglia. J Neurocytol. 1997 Feb;26(2):77–82. doi: 10.1023/a:1018567510105. [DOI] [PubMed] [Google Scholar]
- 2.Kreutzberg GW. Microglia: a sensor for pathological events in the CNS. Trends Neurosci. 1996 Aug;19(8):312–318. doi: 10.1016/0166-2236(96)10049-7. [DOI] [PubMed] [Google Scholar]
- 3.McGeer PL, McGeer EG. Glial cell reactions in neurodegenerative diseases: pathophysiology and therapeutic interventions. Alzheimer Dis Assoc Disord. 1998;12(Suppl 2):S1–6. [PubMed] [Google Scholar]
- 4.Pahan K, Sheikh FG, Namboodiri AM, Singh I. Lovastatin and phenylacetate inhibit the induction of nitric oxide synthase and cytokines in rat primary astrocytes, microglia, and macrophages. J Clin Invest. 1997;100(11):2671–2679. doi: 10.1172/JCI119812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sun GY, Horrocks LA, Farooqui AA. The roles of NADPH oxidase and phospholipases A2 in oxidative and inflammatory responses in neurodegenerative diseases. J Neurochem. 2007 Oct;103(1):1–16. doi: 10.1111/j.1471-4159.2007.04670.x. [DOI] [PubMed] [Google Scholar]
- 6.Dennis EA. Diversity of group types, regulation, and function of phospholipase A2. J Biol Chem. 1994;269:13057–13060. [PubMed] [Google Scholar]
- 7.Weichel O, Hilgert M, Chatterjee SS, Lehr M, Klein J. Bilobalide, a constituent of Ginkgo biloba, inhibits NMDA-induced phospholipase A2 activation and phospholipid breakdown in rat hippocampus. Naunyn Schmiedebergs Arch Pharmacol. 1999 Dec;360(6):609–615. doi: 10.1007/s002109900131. [DOI] [PubMed] [Google Scholar]
- 8.Basselin M, Chang L, Bell JM, Rapoport SI. Chronic lithium chloride administration attenuates brain NMDA receptor-initiated signaling via arachidonic acid in unanesthetized rats. Neuropsychopharmacology. 2006;31(8):1659–1674. doi: 10.1038/sj.npp.1300920. [DOI] [PubMed] [Google Scholar]
- 9.Mattson MP, Chan SL. Neuronal and glial calcium signaling in Alzheimer’s disease. Cell Calcium. 2003 Oct-Nov;34(45):385–397. doi: 10.1016/s0143-4160(03)00128-3. [DOI] [PubMed] [Google Scholar]
- 10.Lehtonen JY, Holopainen JM, Kinnunen PK. Activation of phospholipase A2 by amyloid beta-peptides in vitro. Biochemistry. 1996 Jul 23;35(29):9407–9414. doi: 10.1021/bi960148o. [DOI] [PubMed] [Google Scholar]
- 11.Stephenson DT, Lemere CA, Selkoe DJ, Clemens JA. Cytosolic phospholipase A2 (cPLA2) immunoreactivity is elevated in Alzheimer’s disease brain. Neurobiol Dis. 1996 Feb;3(1):51–63. doi: 10.1006/nbdi.1996.0005. [DOI] [PubMed] [Google Scholar]
- 12.Bazan NG, Colangelo V, Lukiw WJ. Prostaglandins and other lipid mediators in Alzheimer’s disease. Prostaglandins Other Lipid Mediat. 2002 Aug;68-69:197–210. doi: 10.1016/s0090-6980(02)00031-x. [DOI] [PubMed] [Google Scholar]
- 13.Greenamyre JT, Maragos WF, Albin RL, Penney JB, Young AB. Glutamate transmission and toxicity in Alzheimer’s disease. Prog Neuropsychopharmacol Biol Psychiatry. 1988;12:421–430. doi: 10.1016/0278-5846(88)90102-9. [DOI] [PubMed] [Google Scholar]
- 14.Montine KS, Quinn JF, Zhang J, et al. Isoprostanes and related products of lipid peroxidation in neurodegenerative diseases. Chem Phys Lipids. 2004 Mar;128(12):117–124. doi: 10.1016/j.chemphyslip.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 15.DeMar JC, Jr., Lee HJ, Ma K, et al. Brain elongation of linoleic acid is a negligible source of the arachidonate in brain phospholipids of adult rats. Biochim Biophys Acta. 2006 Sep;1761(9):1050–1059. doi: 10.1016/j.bbalip.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 16.Robinson PJ, Noronha J, DeGeorge JJ, Freed LM, Nariai T, Rapoport SI. A quantitative method for measuring regional in vivo fatty-acid incorporation into and turnover within brain phospholipids: Review and critical analysis. Brain Res Rev. 1992;17:187–214. doi: 10.1016/0165-0173(92)90016-f. [DOI] [PubMed] [Google Scholar]
- 17.Chang MCJ, Arai T, Freed LM, et al. Brain incorporation of [1-11C]-arachidonate in normocapnic and hypercapnic monkeys, measured with positron emission tomography. Brain Res. 1997;755:74–83. doi: 10.1016/s0006-8993(97)00088-7. [DOI] [PubMed] [Google Scholar]
- 18.Giovacchini G, Chang MC, Channing MA, et al. Brain incorporation of [11C]arachidonic acid in young healthy humans measured with positron emission tomography. J Cereb Blood Flow Metab. 2002 Dec;22(12):1453–1462. doi: 10.1097/01.WCB.0000033209.60867.7A. [DOI] [PubMed] [Google Scholar]
- 19.Giovacchini G, Lerner A, Toczek MT, et al. Brain incorporation of 11C-arachidonic acid, blood volume, and blood flow in healthy aging: a study with partial-volume correction. J Nucl Med. 2004 Sep;45(9):1471–1479. [PubMed] [Google Scholar]
- 20.Rosenberger TA, Villacreses NE, Hovda JT, et al. Rat brain arachidonic acid metabolism is increased by a 6-day intracerebral ventricular infusion of bacterial lipopolysaccharide. J Neurochem. 2004;88:1168–1178. doi: 10.1046/j.1471-4159.2003.02246.x. [DOI] [PubMed] [Google Scholar]
- 21.Richardson RL, Kim EM, Gardiner T, O’Hare E. Chronic intracerebroventricular infusion of lipopolysaccharide: effects of ibuprofen treatment and behavioural and histopathological correlates. Behav Pharmacol. 2005 Nov;16(7):531–541. doi: 10.1097/01.fbp.0000179278.03868.96. [DOI] [PubMed] [Google Scholar]
- 22.Lee H, Villacreses NE, Rapoport SI, Rosenberger TA. In vivo imaging detects a transient increase in brain arachidonic acid metabolism: a potential marker of neuroinflammation. J Neurochem. 2004 Nov;91(4):936–945. doi: 10.1111/j.1471-4159.2004.02786.x. [DOI] [PubMed] [Google Scholar]
- 23.Lee HJ, Rao JS, Chang L, Rapoport SI, Bazinet RP. Chronic N-methyl-D-aspartate administration increases the turnover of arachidonic acid within brain phospholipids of the unanesthetized rat. J Lipid Res. 2007 Oct 23; doi: 10.1194/jlr.M700406-JLR200. [DOI] [PubMed] [Google Scholar]
- 24.Basselin M, Villacreses NE, Lee HJ, Bell JM, Rapoport SI. Chronic lithium administration attenuates up-regulated brain arachidonic acid metabolism in a rat model of neuroinflammation. J Neurochem. 2007 Aug;102(3):761–772. doi: 10.1111/j.1471-4159.2007.04593.x. [DOI] [PubMed] [Google Scholar]
- 25.Esposito G, Giovacchini G, Der M, et al. Imaging signal transduction via arachidonic acid in the human brain during visual stimulation, by means of positron emission tomography. Neuroimage. 2007 Feb 15;34(4):1342–1351. doi: 10.1016/j.neuroimage.2006.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Herscovitch P, Markham J, Raichle ME. Brain blood flow measured with intravenous H2(15)O. I. Theory and error analysis. J Nucl Med. 1983 Sep;24(9):782–789. [PubMed] [Google Scholar]
- 27.Folstein MF, Folstein SE, McHugh PR. “Mini-Mental State.” A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 28.Channing MA, Simpson N. Radiosynthesis of 1-[11C]polyhomoallylic fatty acids. J Labeled Compounds Radiopharmacol. 1993;33:541–546. [Google Scholar]
- 29.Belfrage P, Vaughan M. Simple liquid-liquid partition system for isolation of labeled oleic acid from mixtures with glycerides. J Lipid Res. 1969;10:341–344. [PubMed] [Google Scholar]
- 30.Brooks DJ, Lammertsma AA, Beaney RP, et al. Measurement of regional cerebral pH in human subjects using continuous inhalation of 11CO2 and positron emission tomography. J Cereb Blood Flow Metab. 1984;4:458–465. doi: 10.1038/jcbfm.1984.65. [DOI] [PubMed] [Google Scholar]
- 31.Ibanez V, Pietrini P, Alexander GE, et al. Regional glucose metabolic abnormalities are not the result of atrophy in Alzheimer’s disease. Neurology. 1998;50:1585–1593. doi: 10.1212/wnl.50.6.1585. [DOI] [PubMed] [Google Scholar]
- 32.Jenkinson M, Smith S. A global optimisation method for robust affine registration of brain images. Med Image Anal. 2001 Jun;5(2):143–156. doi: 10.1016/s1361-8415(01)00036-6. [DOI] [PubMed] [Google Scholar]
- 33.Muller-Gartner HW, Links JM, Prince JL, et al. Measurement of radiotracer concentration in brain gray matter using positron emission tomography: MRI-based correction for partial volume effects. J Cereb Blood Flow Metab. 1992;12:571–583. doi: 10.1038/jcbfm.1992.81. [DOI] [PubMed] [Google Scholar]
- 34.Frackowiak RSJ, Friston KJ, Frith CD, et al. Human Brain Function. 2nd ed. Academic Press; San Diego: 2004. [Google Scholar]
- 35.Tzourio-Mazoyer N, Landeau B, Papathanassiou D, et al. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage. 2002 Jan;15(1):273–289. doi: 10.1006/nimg.2001.0978. [DOI] [PubMed] [Google Scholar]
- 36.Ibanez V, Pietrini P, Furey ML, et al. Resting state brain glucose metabolism is not reduced in normotensive healthy men during aging, after correction for brain atrophy. Brain Res Bull. 2004;63:147–154. doi: 10.1016/j.brainresbull.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 37.Benjamini Y, Hochberg Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J Royal Statistical Soc Ser B. 1995;57(1):239–300. [Google Scholar]
- 38.Benjamini Y, Drai D, Elmer G, Kafkafi N, Golani I. Controlling the false discovery rate in behavior genetics research. Behav Brain Res. 2001 Nov 1;125(12):279–284. doi: 10.1016/s0166-4328(01)00297-2. [DOI] [PubMed] [Google Scholar]
- 39.Jagust W. Cerebral blood flow and metabolism in dementia: regional patterns and the biology of Alzheimer’s disease. Dev Brain Dysfunct. 1994;7:306–314. [Google Scholar]
- 40.Rousset OG, Ma Y, Evans AC. Correction for partial volume effects in PET: principle and validation. J Nucl Med. 1998 May;39(5):904–911. [PubMed] [Google Scholar]
- 41.DeCarli CS, Atack JR, Ball MJ, et al. Post-mortem regional neurofibrillary tangle densities but not senile plaque densities are related to regional cerebral metabolic rates for glucose during life in Alzheimer’s disease patients. Neurodegeneration. 1992;1:113–121. [Google Scholar]
- 42.Giannakopoulos P, Hof PR, Mottier S, Michel JP, Bouras C. Neuropathological changes in the cerebral cortex of 1258 cases from a geriatric hospital: retrospective clinicopathological evaluation of a 10-year autopsy population. Acta Neuropathol (Berl) 1994;87(5):456–468. doi: 10.1007/BF00294172. [DOI] [PubMed] [Google Scholar]
- 43.Cagnin A, Brooks DJ, Kennedy AM, et al. In-vivo measurement of activated microglia in dementia. Lancet. 2001 Aug 11;358(9280):461–467. doi: 10.1016/S0140-6736(01)05625-2. [DOI] [PubMed] [Google Scholar]
- 44.Versijpt JJ, Dumont F, Van Laere KJ, et al. Assessment of neuroinflammation and microglial activation in Alzheimer’s disease with radiolabelled PK11195 and single photon emission computed tomography. A pilot study. Eur Neurol. 2003;50(1):39–47. doi: 10.1159/000070857. [DOI] [PubMed] [Google Scholar]
- 45.Kemppainen NM, Aalto S, Wilson IA, et al. Voxel-based analysis of PET amyloid ligand [11C]PIB uptake in Alzheimer disease. Neurology. 2006 Nov 14;67(9):1575–1580. doi: 10.1212/01.wnl.0000240117.55680.0a. [DOI] [PubMed] [Google Scholar]
- 46.Kumar A, Schapiro MB, Grady C, et al. High-resolution PET studies in Alzheimer’s disease. Neuropsychopharmacology. 1991;4:35–46. [PubMed] [Google Scholar]