Abstract
A comprehensive second-generation whole genome radiation hybrid (RH II), cytogenetic and comparative map of the horse genome (2n=64) has been developed using the 5000rad horse × hamster radiation hybrid panel and fluorescence in situ hybridization (FISH). The map contains 4,103 markers (3,816 RH, 1,144 FISH) assigned to all 31 pairs of autosomes and the X chromosome. The RH maps of individual chromosomes are anchored and oriented using 857 cytogenetic markers. The overall resolution of the map is one marker per 775 kilobase-pairs (kb), which represents a more than five-fold improvement over the first-generation map. The RH II incorporates 920 markers shared jointly with the two recently reported meiotic maps. Consequently the two maps were aligned with the RH II maps of individual autosomes and the X chromosome. Additionally, a comparative map of the horse genome was generated by connecting 1,904 loci on the horse map with genome sequences available for eight diverse vertebrates to highlight regions of evolutionarily conserved syntenies, linkages and chromosomal breakpoints. The integrated map thus obtained presents the most comprehensive information on the physical and comparative organization of the equine genome and will assist future assemblies of whole genome BAC fingerprint maps and the genome sequence. It will also serve as a tool to identify genes governing health, disease and performance traits in horses and assist us in understanding the evolution of the equine genome in relation to other species.
Keywords: radiation hybrid map, horse, comparative, whole genome
Introduction
High-resolution gene maps are essential for understanding the structure and organization of a genome, determining the location and relative order of genes and markers on chromosomes, obtaining detailed comparative information in relation to other genomes and isolating gene(s) governing traits of interest. In horses, traits of interest range from those governed by a single gene (e.g., coat color and a number of inherited disorders) to complex traits controlled by the interaction of several genes (e.g., allergies, disease resistance, athletic performance, reproduction, fertility). Horse (Equus caballus, ECA; 2n=64) whole-genome (WG) maps reported to date are low to medium density and contain ~700–800 markers distributed on various autosomes and the X chromosome. These maps include the first-generation WG radiation hybrid (RH) and comparative map (Chowdhary et al. 2003, denoted below as RH I), the latest iterations of the two linkage maps (IRFHP – Penedo et al. 2005; AHT – Swinburne et al. 2006) and cytogenetic maps (Milenkovic et al. 2002; Perrocheau et al. 2006). Though all these maps have been successfully used in the recent past to isolate genes governing some monogenic traits and to detect the mutation/variation responsible for the phenotype (see Chowdhary and Raudsepp 2008), their resolution is not sufficient to study the genetics of complex traits.
In recent years, medium to high density WG or single chromosome RH maps with a resolution of about 1 marker per megabase (Mb) have been generated for a range of livestock and pet species including cattle (Everts-van der Wind et al. 2004; Everts-van der Wind et al. 2005; Itoh et al. 2005; Jann et al. 2006; McKay et al. 2007), pig (Hamasima et al. 2003; Meyers et al. 2005), dog (Breen et al. 2004), etc. These maps are facilitating identification of genes for various traits in different species and are being used to compare genomes of distantly related mammals and study chromosome evolution (Murphy et al. 2005). The maps have also been instrumental in integrating synteny, cytogenetic and genetic linkage information into a single linearly ordered map, and have been useful in assembling the emerging WG sequence information (Rowe et al. 2003; Kwitek et al. 2004; Meyers et al. 2005; Jann et al. 2006; Snelling et al. 2007). Medium- to high-resolution gene maps have been reported for some of the 31 pairs of equine autosomes and the X chromosome (Lee et al. 2004; Raudsepp et al. 2004; Brinkmeyer-Langford et al. 2005; Gustafson-Seabury et al. 2005; Dierks et al. 2006; Wagner et al. 2006; Goh et al. 2007). Since then we have added markers to all chromosomes and produced a high-resolution second-generation map of the entire equine genome (denoted below as RH II), excluding the Y chromosome. This map should serve as a valuable tool for many types of equine genome analysis.
Material and Methods
Marker development and genotyping
Markers for RH mapping were developed using equine genome resources available from UCSC (http://genome.ucsc.edu/), NCBI (http://www.ncbi.nlm.nih.gov/), HorseMap (http://locus.jouy.inra.fr/cgibin/lgbc/mapping/horsemap/intro2.pl/), horse BES sequence databases (http://www.tiho-hannover.de/einricht/zucht/hgp/index.htm), and from published literature. Additionally, a number of gene specific markers were generated from conserved regions of orthologous mammalian genes using alignment (Chenna et al. 2003) of sequences from multiple species. The orthologous genes were chosen from the human genome sequence map at approximately every 1 Mb intervals as described earlier (Lee et al. 2004; Raudsepp et al. 2004; Goh et al. 2007). Primers were designed with Primer3 (http://www-genome.wi.mit.edu/cgibin/primer/primer3_www.cgi) or were obtained from publications. All PCR products amplified using heterologous primers were validated by sequencing. Details about all markers included in this study are available in Supplementary Table 2. All markers were genotyped in duplicate on the WG 5000rad horse × hamster RH panel (Chowdhary et al. 2002), resolved on 2% agarose gels and scored manually as described previously (Chowdhary et al. 2003; Brinkmeyer-Langford et al. 2005; Gustafson-Seabury et al. 2005; Wagner et al. 2006; Goh et al. 2007). Genotyping information available from previously published RH maps (Chowdhary et al. 2003; Raudsepp et al. 2004; Brinkmeyer-Langford et al. 2005; Gustafson-Seabury et al. 2005; Wagner et al. 2006; Goh et al. 2007) was included as part of the input for the map computation.
Map computation
Computations to analyze the genome-wide genotyping data and to construct RH maps for individual chromosomes were performed using the rh_tsp_map (Agarwala et al. 2000; Schäffer et al. 2007); ftp://ftp.ncbi.nih.gov/pub/agarwala/rhmapping/rh_tsp_map.tar), CONCORDE (Applegate et al. 2006); http://www.isye.gatech.edu/~wcook/rh/) and Qsopt (http://www.isye.gatech.edu/~wcook/qsopt) software packages using the same procedures as described in a recently published WG RH map for the cat (Murphy et al. 2007) and in the rh_tsp_map tutorial, but with increased automation (Schäffer et al. 2007). We used a consensus of three formulations of the maximum-likelihood (MLE) criterion (Agarwala et al. 2000). Markers were assigned to linkage groups by two-point analysis with a LOD score threshold of 7.6. Thresholds with one digit after the decimal between 7 and 8 were considered and LOD 7.6 gave the best balance between the competing objectives of: i) discarding fewer markers with inter-chromosomal scores above the threshold, and ii) unifying more linkage groups. Markers that did not have a score ≥ 7.6 with any other marker were removed from further analysis. The MLE-consensus maps passed a flips test at LOD threshold 0.5. Markers dropped from the MLE-consensus map were placed in an interval between consecutive markers if the best placement was at least 0.1 LOD units better than second best; multiple markers placed in the same interval passed a flips test. The MLE-consensus markers and placed markers were assigned cR positions by solving instances of a restricted traveling salesman problem. Remaining markers were binned if their best placements spanned at most three adjacent MLE-consensus intervals. The order and orientation of linkage groups on a chromosome were primarily determined by FISH and further verified using available genetic linkage maps (Penedo et al. 2005; Swinburne et al. 2006). Detailed information regarding the RH map for each chromosome (linkage group size, map distances, MLE-consensus, placements, binned markers) is available in Supplementary Table 3.
BAC library screening and cytogenetic mapping
The CHORI-241 BAC library was used to isolate clones containing markers pertinent for anchoring, ordering and orienting RH groups. Library screening by PCR and BAC DNA isolation followed procedures described earlier (Chowdhary et al. 2003). The BACs were individually labeled with biotin and/or digoxigenin and hybridized in pairs or triplets to horse metaphase or interphase chromosomes. DNA labeling, in situ hybridization, signal detection, microscopy, and image analysis were performed as previously described (Chowdhary et al. 2003).
Comparative analysis
Comparative information for equine orthologs of human, chimpanzee, dog, cattle, mouse, rat, opossum and chicken genes was retrieved from the UCSC Genome Browser (http://genome.ucsc.edu/). Homology between regions flanking equine microsatellites and the human genome sequence was obtained from published papers (Tozaki et al. 2007) and by BLAST. Homologies between human genome and equine BES were retrieved from (Leeb et al. 2006) and http://www.tiho-hannover.de/einricht/zucht/hgp/index.htm. Blocks of conserved synteny and conserved linkage were defined as described earlier (Nadeau and Sankoff 1998; Chowdhary et al. 2003, see Legends for Supplementary Figs. 1.1-1.X.). Comparative positions of centromeres and telomeres were retrieved from available sequence and cytogenetic maps as described below.
Construction of comparative maps
Comparative information was retrieved from the UCSC Genome Bioinformatics website (http://genome.ucsc.edu) using the following builds for each species: human- NCBI Build 36.1, browser March 2006; chimp - panTro2 Build 2 v1, browser March 2006; dog - canFam2 v2.0, browser May 2005; cattle - Baylor release Btau_4.0, browser September 2007; mouse - mm9 NCBI Build 37, browser July 2007; rat - rn4 version 3.4, browser November 2004; opossum - monDom4, browser January 2006 and chicken - galGal3 v2.1 draft assembly, browser May 2006. Sequence maps of individual species were used to identify Mb positions of the equine orthologs. Conserved syntenies and conserved linkages were manually demarcated following the convention laid out by (Nadeau and Sankoff 1998) and further explained by us (Chowdhary et al. 2003). A minimum of three markers sharing the same order in two species was considered as linked order shared between two species (i.e., conserved linkage). Within blocks of conserved linkages, flips up to 5 Mb were not considered as breakage in conservation because such variations could be attributed to e.g., statistical constraints, assembly errors and even marginal genotyping errors. Centromere positions of bi-armed chromosomes were retrieved from combined cytogenetic and sequence maps and were available only for human and chimp. Centromere positions for acrocentric chromosomes in cattle, dog, mouse and rat were determined as the lowest Mb position on their sequence map. Centromere positions were not available for opossum and chicken and for chimp chromosomes that have rearrangements compared to their human counterpart (Supplementary Table 5). Locations of telomeres were derived from comparative marker(s) located at or closest to 0 Mb and the highest Mb positions position on the sequence map for individual chromosomes in each species.
Results and Discussion
Radiation hybrid analysis and mapping
Markers and retention frequencies (RF)
A total of 4,493 markers were genotyped on the 5,000rad Equine WG RH-panel. During and following two-marker LOD score computations, 677 markers (15%) were discarded for one of the following reasons: i) marker retention frequency was below the designated threshold of 5%; ii) genotyping results were inconsistent between duplicate typing with the same marker; iii) the same marker was genotyped by different research groups under alias names; iv) markers had high LOD scores with other markers on at least two distinct chromosomes suggesting that primers do not recognize unique sequences; v) markers had no LOD score ≥ 7.6 with any other marker; and, vi) markers could not be reliably assigned to a multi-marker bin relative to the framework maps. The final RH map contains 3,816 markers (Table 1 and Supporting Information (SI) Tables 1 and 2.1-2.X for details) of which 1,917 are on maximum likelihood (MLE)-consensus (a.k.a. framework) maps, 1,311 are placed in relation to the MLE-consensus markers, and 588 are binned in intervals spanning at most four MLE-consensus markers (SI Tables 1 and 3).
Table 1.
Chromosome wise information on selected parameters of the integrated RH and comparative map
| Horse Chr. | Size (Mb) | Total size (cR) | RH groups | Marker density (Mb) | Loci on the RH map | FISH mapped loci | Meiotic map loci | Comparative loci | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | Type I | Type II | BES, STSs | Total | Anchor loci | |||||||
| ECA1 | 204 | 2506.3 | 13 | 0.78 | 260 | 136 | 120 | 4 | 100 | 70 | 71 | 147 |
| ECA2 | 135 | 1724.2 | 3 | 0.69 | 196 | 107 | 80 | 9 | 55 | 43 | 45 | 96 |
| ECA3 | 132 | 1795.8 | 4 | 0.92 | 143 | 72 | 68 | 3 | 46 | 30 | 26 | 81 |
| ECA4 | 120 | 2026.9 | 2 | 0.61 | 196 | 106 | 87 | 3 | 43 | 35 | 46 | 98 |
| ECA5 | 111 | 1588.1 | 4 | 0.63 | 177 | 102 | 69 | 6 | 48 | 32 | 32 | 102 |
| ECA6 | 108 | 1189.1 | 4 | 0.68 | 158 | 92 | 65 | 1 | 43 | 33 | 32 | 92 |
| ECA7 | 105 | 1430.3 | 5 | 0.88 | 120 | 45 | 72 | 3 | 70 | 41 | 23 | 62 |
| ECA8 | 105 | 1437.6 | 5 | 0.88 | 120 | 58 | 61 | 1 | 44 | 26 | 34 | 74 |
| ECA9 | 99 | 1354.9 | 2 | 0.9 | 110 | 44 | 65 | 1 | 22 | 14 | 37 | 44 |
| ECA10 | 96 | 1627.5 | 2 | 0.58 | 165 | 84 | 64 | 17 | 51 | 42 | 43 | 80 |
| ECA11 | 72 | 1827.9 | 4 | 0.56 | 129 | 56 | 65 | 8 | 46 | 33 | 31 | 59 |
| ECA12 | 63 | 679.1 | 2 | 1.15 | 55 | 25 | 30 | 0 | 12 | 9 | 14 | 27 |
| ECA13 | 60 | 805.1 | 3 | 0.63 | 95 | 47 | 45 | 3 | 23 | 19 | 23 | 43 |
| ECA14 | 120 | 1764.4 | 1 | 0.70 | 172 | 96 | 70 | 6 | 48 | 38 | 39 | 93 |
| ECA15 | 111 | 1393.3 | 2 | 0.82 | 136 | 64 | 68 | 4 | 40 | 32 | 38 | 65 |
| ECA16 | 108 | 1686.0 | 7 | 0.8 | 135 | 67 | 61 | 7 | 48 | 33 | 38 | 66 |
| ECA17 | 102 | 870.8 | 10 | 0.84 | 121 | 65 | 52 | 4 | 39 | 36 | 24 | 54 |
| ECA18 | 102 | 1245.6 | 3 | 0.96 | 106 | 55 | 46 | 5 | 21 | 17 | 27 | 52 |
| ECA19 | 96 | 1202.0 | 1 | 1.04 | 92 | 34 | 55 | 3 | 19 | 12 | 27 | 31 |
| ECA20 | 87 | 1321.0 | 2 | 0.65 | 134 | 54 | 66 | 14 | 38 | 24 | 32 | 53 |
| ECA21 | 81 | 875.6 | 3 | 0.70 | 116 | 70 | 44 | 2 | 37 | 26 | 27 | 66 |
| ECA22 | 72 | 911.4 | 1 | 0.54 | 133 | 82 | 50 | 1 | 52 | 45 | 26 | 71 |
| ECA23 | 69 | 711.8 | 3 | 0.91 | 76 | 26 | 43 | 7 | 21 | 15 | 28 | 28 |
| ECA24 | 63 | 830.7 | 2 | 1.07 | 59 | 19 | 38 | 2 | 19 | 14 | 24 | 21 |
| ECA25 | 60 | 662.0 | 1 | 1.33 | 45 | 19 | 26 | 0 | 16 | 12 | 13 | 21 |
| ECA26 | 63 | 741.9 | 1 | 0.81 | 78 | 34 | 32 | 12 | 16 | 13 | 19 | 33 |
| ECA27 | 57 | 564.0 | 3 | 0.81 | 70 | 28 | 36 | 6 | 12 | 11 | 23 | 26 |
| ECA28 | 54 | 456.2 | 1 | 0.95 | 57 | 30 | 27 | 0 | 16 | 12 | 16 | 29 |
| ECA29 | 51 | 626.3 | 1 | 0.96 | 53 | 28 | 23 | 2 | 9 | 8 | 15 | 25 |
| ECA30 | 48 | 594.1 | 1 | 1.09 | 44 | 26 | 18 | 0 | 9 | 9 | 14 | 26 |
| ECA31 | 45 | 381.7 | 1 | 1.05 | 43 | 18 | 19 | 6 | 11 | 9 | 10 | 17 |
| ECAX | 153 | 1529.8 | 5 | 0.69 | 222 | 148 | 72 | 2 | 70 | 64 | 23 | 122 |
| Overall | 2952 | 38,361.4 | 102 | 0.77 | 3816 | 1937 | 1737 | 142 | 1144 | 857 | 920 | 1904 |
Table 2.
Discrepancies between RH II and the two recent linkage maps
| Marker | RH II, ECA | Penedo et al. 2005, ECA | Swinburne et al. 2006, ECA | FISH | Comparative, human HSA |
|---|---|---|---|---|---|
| COR006 | 1 | - | 20 | 1q25 | 14 |
| AHT113 | 1 | 25 | - | - | |
| TKY281 | 1 | 28 | 1 | - | |
| NVHEQ102 | 3 | 5 | - | - | |
| HMS56 | 4 | 10 | 10 | 4p15-p14 | |
| TKY916 | 9 | 1 | - | - | |
| AHT126 | 10 | 1 | 10 | - | |
| AHT016 | 11 | 15 | 15 | - | |
| TKY908 | 11 | 21 | - | - | |
| TKY286 | 12 | 28 | - | - | |
| SGCV03 | 13 | 13 | 22 | 13q12 | |
| AHT115 | 18 | 1 | - | - | |
| TKY520 | 21 | 22 | - | - | |
| TKY709 | 21 | 28 | - | ||
| TKY275 | 26 | - | 8 | - | |
| UM031 | 26 | 26 | 20 | - | 3 |
| UMNe107 | X | - | 13 | - |
Table 3.
Comparison of RH II and RH I (Chowdhary et al. 2003)
| Map size (cR) | No. of RH groups | Average marker density (kb) | No of markers | Type I | Type II | FISH anchors | Markers shared with linkage maps | Comparative loci | |
|---|---|---|---|---|---|---|---|---|---|
| RH II | 38,361.4 | 101 | 775 | 3816 | 1937 | 1879 | 857 | 920 | 1904 |
| RH I | 14,587.1 | 101 | 4,044 | 730 | 258 | 472 | 253 | - | 447 |
| Improvement | 2.6 | - | 5.2 | 5.2 | 7.5 | 3.9 | 3.4 | - | 4.3 |
The average retention frequency (RF) of markers in the panel is 19% (Supplementary Table 4). This ranges from 10.4% for ECA1 to 39.4% for ECA11, which contains the selectable TK1 marker preferentially retained in all cell hybrids. Markers with low RF are mainly found on the larger chromosomes, viz. ECA1, ECA17 and ECAX, while markers with RF above the genome average are present mainly on small chromosomes such as ECA29 and ECA30. This suggests that irradiation-induced breakages were fewer in the smaller chromosomes than the rest of the genome. Along the length of the chromosomes, the retention of markers is also slightly higher in the pericentromeric and telomeric region of many chromosomes, a trend that was also seen in RH I.
LOD score computations for all pairs of markers and subsequent single-linkage clustering partitioned the markers into 102 RH groups distributed over all horse autosomes and the X chromosome (Table 1, SI Figures 1.1-1.X). On average, there are three RH groups per chromosome. While ECA14, 19, 22, 25, 26, 28, 29, 30, and 31 contain only one RH group each, ECA1 and ECA17 have 13 and 10 RH groups, respectively which coincides with their lowest overall retention frequency (RF; Table 4). A break in the RH groups is typically observed at the centromeres of bi-armed chromosomes, except for ECA11 and ECAX (SI Figures 1.11 and 1.X). The large number of RH groups is influenced by regions of low RF as well as our decision to include ~25 small RH groups to avoid gaps in coverage. The overall size of the map calculated as the sum of the 102 RH group lengths is 38,361 cR. Considering the physical size of the horse genome to be somewhere between 2,462 Mb (UCSC EquCab1, http://genome.ucsc.edu/) and 2,952 Mb (Chowdhary et al. 2003; Supplementary Table 1), 1 cR in the 5000rad equine map correlates on average to ~64–76 kb.
Distribution and density of markers
RH II has an average density of 1 marker/775 kb – including all 3,816 markers and an average density of 1/915kb for the 3,228 markers assigned a cR position (using the genome length estimate of 2,952 Mb from Chowdhary et al. 2003 for ease of comparison). The marker density is highest on ECA22 with 1 marker/540 kb and lowest on ECA25 with 1 marker/1,330 kb (Table 1). The current map provides a greater than five-fold improvement compared to RH I, where the average density was 1 marker/4,044 kb, making it comparable to the recently reported 3000rad–7000rad WG RH maps in other species (Hamasima et al. 2003; Breen et al. 2004; Everts-van der Wind et al. 2005; Jann et al. 2006; McKay et al. 2007). In cattle, map resolution ranges from 1 marker/440 kb (Jann et al. 2006) to 1 marker/880 kb (Everts-van der Wind et al. 2005); in pig, the density is 1 marker/490kb (Hamasima et al. 2003); and in dog, the density is 1 marker/900kb (Breen et al. 2004).
The number and distribution of Type I (1,937) and Type II (1,737 microsatellite and 142 other STS) markers is fairly balanced on almost all chromosomes, with a slight bias towards genes on ECA5, 14 and X and towards microsatellites on ECA7, 19 and 24 (Table 1). The large number of polymorphic microsatellites makes RH II useful for genetic studies of horse traits. However, FISH-mapped markers and Type I markers were preferentially selected over others in computing the MLE-consensus map to enable better comparisons with high-resolution RH maps for domestic species that, are strongly biased towards genes (Breen et al. 2004; Everts-van der Wind et al. 2004).
Comparison of RH II with previously reported RH maps
RH II has improved almost all important map parameters compared to RH I (Table 3) both overall and on each chromosome. In recent years, medium- to high-resolution RH maps were generated for ten horse chromosomes or chromosomal regions (Lee et al. 2004; Raudsepp et al. 2004; Brinkmeyer-Langford et al. 2005; Gustafson-Seabury et al. 2005; Wagner et al. 2006; Goh et al. 2007), however, maps for all of these chromosomes/regions have been further improved by mapping and analyzing an additional set of markers. For example, the most recently published map for ECA14 (Goh et al. 2007) with 1 marker per 940 Mb has been further improved herein to a 1 marker per 700 kb.
FISH map
Cytogenetic anchoring of the RH map
The cytogenetic map contains 1,144 markers (Table 1, SI Figures 1.1-1.X). The majority of the 401 newly FISH-mapped markers were selected systematically from the ends of all RH groups and at regular intervals along the length of larger RH groups. Refined multicolor FISH in interphase nuclei using combinations of 3 markers was applied to resolve the position and orientation of all small RH groups. Altogether RH II contains 857 anchor loci (RH mapped or binned markers also present on the FISH map) that associate RH groups to chromosomes and confirm the computed marker order; 287 markers present only on the cytogenetic map contribute primarily to the comparative map.
The utility of FISH is particularly noted in the assignment of 19 small RH groups containing only 3–5 markers (on ECA1, 3, 6, 7, 13, 16, 17, 18, 23 and 27) and in correcting the location of some of the RH groups or individual markers compared to previously published data. For example, FISH mapping of LPL, SFTPC and CTSB showed these markers to be present on ECA2q and not on ECA9 as reported earlier (Milenkovic et al. 2002; Chowdhary et al. 2003). Similarly, new cytogenetic mapping of microsatellite AHT30 moved a small RH group (2 loci) from the previously reported location on ECA22q13 (Swinburne et al. 2000; Chowdhary et al. 2003) to ECA13q13; also, FISH localization of KNG, UMPS and ZNF148 showed that these loci are present on ECA19 and not on ECA16 (Godard et al. 2000; Milenkovic et al. 2002).
FISH also resolved discrepancies between the RH map and the most recent iteration of the two meiotic maps. For example, FISH corrects reverse orientation of the meiotic map for ECA26 by Penedo et al. (Penedo et al. 2005) (Supplementary Fig. 2.26) and both recent meiotic maps for ECA25 (Penedo et al. 2005; Swinburne et al. 2006) (Supplementary Fig. 2.25). With over one thousand FISH mapped markers and 857 anchor loci, the horse integrated RH/FISH map is one of the most comprehensive among domestic species, and is comparable only to the dog WG map that contains a total of 1,000 FISH markers and 851 anchor loci (Breen et al. 2004). The second-generation WG RH maps for other domestic species are not physically aligned to the chromosomes by FISH.
Comparison of the FISH and RH maps
The observed discrepancies in marker order between the RH and FISH maps are minor and concern single or a few loci scattered over the genome. These are partly attributed to imprecise band designations reported in earlier FISH mapping studies. Further, most of the earlier FISH studies used single-color FISH which cannot precisely order closely locate loci. Some anomalies are also due to misidentification of the probes. In such cases, the BAC library was re-screened using published PCR primers, the amplicons were re-sequenced to verify gene identity, and the new BAC clones were again mapped by FISH. Examples of such corrections include the re-assignment of NFIA from ECA5q12-q13 (Milenkovic et al. 2002) to ECA7q12, BRCA2 from ECA17q22 (Milenkovic et al. 2002) to ECA17q14, and AR from ECAXq15-q16 (Milenkovic et al. 2002) to ECAXq12. Several of these discrepancies could also be attributed to isolation and FISH mapping of clones containing another member of the same gene family. For example, primers thought to be for NFIA actually correspond to NFIX.
Integration of the RH and linkage maps
The 766 markers in the IHRFP male linkage map (Penedo et al. 2005) and the 742 markers in the AHT sex-averaged meiotic map (Swinburne et al. 2006) were, to the extent possible, aligned with the 1,737 microsatellite markers present in RH II on all autosomes and the X chromosome. As a result, there are 920 markers shared between RH II and jointly the two meiotic maps. Alignments between the three maps demonstrate a general agreement in the order and orientation of markers and/or linkage groups (SI Figs. 2.1-2.X). The exceptions include i) minor flips on ECA5, 6, 10, 14, 16, 17, 20, and 21; ii) an evident difference in the relative order of HTG001 and ASB029 in the IHRFP map of ECA4 compared to their order in the other two maps (Supplementary Fig. 2.4); iii) reversals involving entire linkage groups (e.g., for ECA25 and ECA26, as described above); and, iv) 17 disparities involving assignment of markers to a different chromosome in one or both meiotic maps compared to RH II (Table 2). More differences were observed between the RH II and IHRFP maps than between the RH II and AHT maps.
Comparison of the spacing of framework markers between the linkage and RH maps reveals regions that have high or low recombination rates per cR. Typically recombination is reduced near centromeres and elevated at distal parts of the chromosomes (Rowe et al. 2003). For example, on ECA12 two pairs of markers - AHT027-TKY404 and COR058-UCDEQ497 are separated by similar distances on the RH map - >127.5 cR and 181.6 cR, respectively. However, their meiotic distances differ by more than 10 fold – 2.2 cM for the two pericentromeric markers AHT027-TKY404 and 23.4 cM for the two distal loci COR058-UCDEQ497 (Supplementary Fig. 2.12, Supplementary Table 3). The approximate genome-wide ratio of physical and genetic distances between the RH II and the IHRFP and the AHT linkage maps is 10.1 cR5000/cM and 13.7 cR5000/cM, respectively. This ratio varies between individual chromosomes but is clearly higher than those reported for cattle - from 4 cR5000/cM (Everts-van der Wind et al. 2004) to 7.5 cR7000/cM (Itoh et al. 2005) indicating reduced coverage of the genome between physical and genetic maps in the horse. The latter may be caused by the gaps between RH groups, as well as a lower number of shared markers between RH and linkage maps compared to cattle. On the whole, alignment of RH and linkage maps indirectly connects meiotic data with the cytogenetic and comparative information and facilitates integration of all available mapping information for the equine genome.
Comparative map
The 1,904 genes and BAC end sequences (BES) present in RH II enable a comparative overview of the organization of the horse genome in relation to eight sequenced vertebrate genomes representing eutherian mammals (human, chimp, dog, cattle, mouse, rat), marsupials (opossum) and birds (chicken). On average, comparative markers are distributed at 1.4 Mb intervals in RH II. Further, BLAST alignment of the flanking sequences of 766 equine microsatellite loci with the human genome sequence (Tozaki et al. 2007), provides additional comparative markers for these two genomes. The new map shows a four- to five-fold improvement in the number of comparative markers over RH I and extends the comparison of the horse genome from human and mouse to six additional species. Because the focus of this study is the high-resolution WG map for the horse, our remarks on map comparisons will be restricted to salient comparative features of the equine genome in relation to the eight sequenced genomes, without expanding on the putative common ancestor.
The comparative map presented in RH II map figures for 31 autosomes and the X chromosome (Supplementary Fig. 1.1-1.X) confirms and refines the boundaries of conserved syntenic segments known between the horse and human genomes (Raudsepp et al. 1996; Yang et al. 2004). For example, mapping 39 horse-human comparative loci on ECA27, 60 on ECA26 and 54 on ECA13 reaffirms synteny conservation and improves the previously known boundaries of correspondence with the human chromosomes HSA4/HSA8, HSA3/HSA21 and HSA7/HSA16, respectively. Mapping 14 HSA1 markers to the proximal region of ECA1q shows that the equine segment corresponds to the 225–229 Mb region on HSA1q and confirms recent Zoo-FISH findings (Yang et al. 2004). The map also reveals a previously undetected segment of homology between ECA2q and HSA8p. Furthermore, the new map refines the status of several previously reported horse-human conserved syntenies and corrects a number of single-locus-based homologies previously described between ECA1-HSA22, ECA2-HSA1, ECA3-HSA3, ECA5-HSA22 and ECA7-HSA19 (Milenkovic et al. 2002; Chowdhary et al. 2003).
An overview of conserved synteny or linkage between the horse and the eight compared genomes shows other interesting features. ECA11, ECA17, ECA22 and ECAX are the only chromosomes that share one-to-one homology with human and chimp chromosomes, but the conserved synteny does not translate into conserved gene order along the three autosomes. ECAX seems to be the only chromosome that shares conserved linkage with human, chimp and also dog counterparts, including the position of the centromere. Broadly, the conservation holds good also for pig (Raudsepp et al. 2004), but not for cattle, mouse and rat where there are several rearrangements (Supplementary Fig. 1.X).
The centromeres of many equine metacentric chromosomes represent sites of synteny breaks in the genomes of most of the compared species. Examples of such breaks are seen on ECA3, 6, 8 and 10 where the short and the long arms correspond to separate chromosomes in all species including opossum and chicken, suggesting that putative ancestral segments have fused at these points in the horse. Further, the position of equine centromeres and/or telomeres coincides at ~58% of the locations with human and chimp, at 37% of the locations with dog but at less than 20% of the locations of telomeres/centromeres in the remaining species (Supplementary Table 5). These observations, together with the overall size of syntenic segments in various species shared with the horse, indicate that the organization of the horse chromosomes resembles human/chimp more closely than other compared species.
Some other interesting aspects about comparative organization of the horse genome in relation to the eight sequenced genomes include:
- clustering of synteny breaks or rearrangements at a number of places in the eight genomes. Two such clusters can be seen on ECA1q14 and q15 where a distinct break in synteny is observed in almost all genomes. In cattle, where the break does not occur, a rearrangement is evident at the same spot. Another interesting example of such a reshuffle is seen at ECA9cen where an inversion is evident in all species sharing conserved synteny. Aggregation of these rearrangements or synteny breaks at specific spots in other genomes highlights some of the sites where fusion/reshuffle occurred during the formation of the horse chromosomes. Similar congregation of synteny breaks or rearrangements can be seen on ECA2cen, ECA3cen, ECA7, ECA21 and a number of other locations in relation to the horse genome as indicated by a red vertical line across the eight genomes (Supplementary Fig. 1.1-1.X).
- Overall, the human and chimp genomes exhibit an almost identical pattern of similarity with the horse genome. However, over the distal parts of ECA3q, 6q, 14q, and ECA21, the chimp genome shows inversions that are not seen in the human genome. These inversions were reported in previous human-chimp genome comparisons (Schmutz et al. 2004; Kehrer-Sawatzki et al. 2005a; Kehrer-Sawatzki et al. 2005b) and likely represent independent events that occurred during the evolution of the chimp chromosomes. However, chimp inversion breakpoints seen on ECA3 and ECA14 comparative maps might have broader evolutionary importance, as they coincide with synteny breaks in dog and rodents, respectively.
- The mouse/rat genomes have undergone rapid karyotype evolution and therefore the total number of syntenic segments shared between them and other eutherian genomes is considerably higher than that seen for comparisons between non-rodent mammalian genomes. Although a similar trend is seen for horse-mouse/rat chromosome comparisons, some remarkably uninterrupted conserved syntenies and linkages spanning entire equine chromosomes are worth mentioning. For example, the conserved linkage shared between ECA11 and parts of MMU11 and RNO10 is not seen for corresponding chromosomal segments in the other six species. A similar trend is seen in conserved linkage shared between ECA24 and parts of MMU12/RNO6, and between ECA26 and parts of MMU16/RNO11. Additionally, ECA22, 27, 28, 29 and 30 share synteny conservation along their entire length with parts of (or complete) rat chromosomes, while ECA22, 27 and 30 share it with mouse chromosomes.
- While a number of segments from the chicken genome (particularly the macrochromosomes) individually correspond to parts of single equine chromosomes, the conserved linkage between ECA17 and part of GGA1, and the conserved synteny between ECA9, ECA18, ECA24, ECA28 and ECA31 with parts of GGA2, GGA7, GGA5, GGA1 and GGA3, respectively, are noteworthy because some of these segments most likely represent ancestral vertebrate regions.
- Among large equine autosomes, only ECA9 shares synteny and linkage conservation with the corresponding opossum chromosome (MDO3). Large blocks of synteny conservation are present also between other equine and marsupial chromosomes, but the gene order is usually rearranged resulting in shorter segments of conserved linkages. It is not yet clear whether lower degree of linkage conservation between eutherian and marsupial genomes is due to evolutionary divergence or an incomplete opossum genome assembly. Likewise, many synteny and linkage rearrangements observed between horse and cattle genomes can probably be attributed to the sequence assembly difficulties rather than to real differences.
- Finally, comparative maps of some horse chromosomes, e.g., ECA5q, 13qcen and 22q show discrepant synteny or linkage positions of some markers in all other species and most likely reflect inaccuracies in the horse RH map. Such discrepant markers tend to be located at the ends of RH groups and are shown in red font on RH maps (Supplementary Fig. 1.1-1.X).
RH map as a framework for WG BAC fingerprint and sequence assemblies
A WG physical map of BAC contigs based on fingerprinting 150,000 BAC clones from CHORI-241 library is currently under construction (O. Distl, unpublished). Since RH II contains over 500 markers that are derived from CHORI-241 BAC clones, they can be used to verify the BAC fingerprint assembly and anchor it to specific chromosomes. The majority of the 4,103 markers on the integrated map should also serve as an excellent framework with which the WG draft sequence assembly (currently at build 2) of the female horse Twilight (C. Wade, unpublished) could be validated. We used e-PCR (Schuler 1997) to locate the RH II markers on horse build 2 to quantify how much improvement is possible. Only 2,869 markers had a chromosomal location, and only 2,757 markers had a unique location. Thus, it may be possible to use the locations and primers of the other 1,000+ markers given herein to produce better horse genome assemblies. We anticipate that as for human (Lander et al. 2001; Olivier et al. 2001; Venter et al. 2001), mouse (Rowe et al. 2003), rat (Kwitek et al. 2004) and other species, the integrated WG RH II and comparative horse map presented in this study will serve as the main framework to support future efforts in both genome sequence and BAC contig assembly.
Supplementary Material
Supplementary Figures 1.1-1.X, 2.1-2.X, Legends for Supplementary Figures and Supplementary Tables 1, 2, 3, 4, 5.
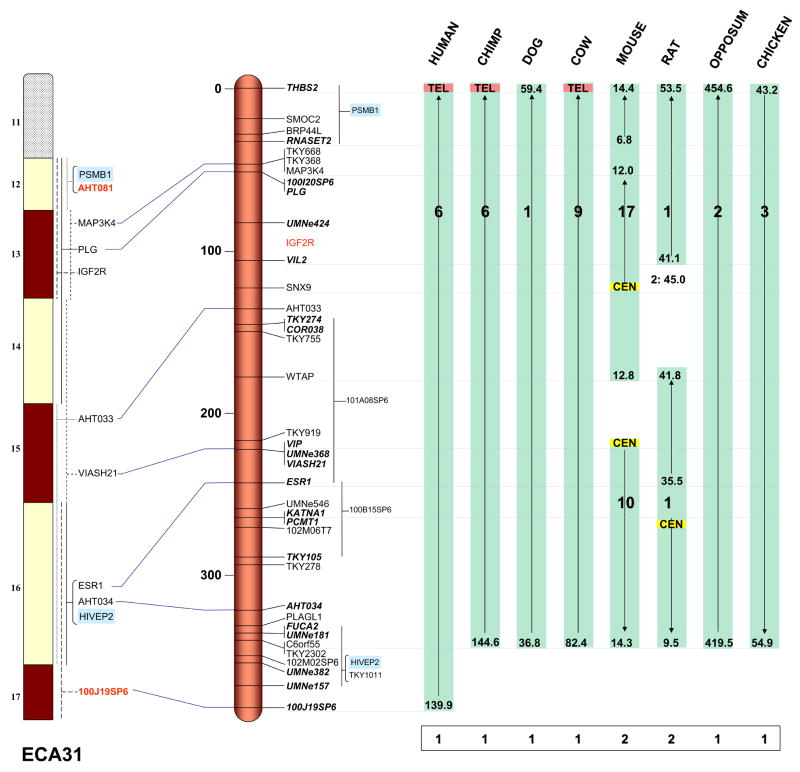
Figure 1.
Integrated map of ECA31: RH II (middle), cytogenetic map (left) and comparison with sequence maps of eight vertebrate species (right). Detailed legend and full size map for ECA31 are available in Legends for Supplementary Figures 1.1-1.X and Supplementary Fig. 1.31, respectively.
Acknowledgments
This work was supported by USDA/NRI grant 2003-03687 and 2006-04801, the Link Endowment, the Morris Animal Foundation, the Dorothy Russell Havemeyer Foundation, the Grayson Jockey Club and in part by the Intramural Research Program of the NIH, NLM.
References
- Agarwala R, Applegate DL, Maglott D, Schuler GD, Schäffer AA. A fast and scalable radiation hybrid map construction and integration strategy. Genome Res. 2000;10:350–364. doi: 10.1101/gr.10.3.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applegate DL, Bixby R, Chvátal VWC. The Traveling Salesman Problem: A Computational Study. Princeton University Press; New Jersey: 2006. [Google Scholar]
- Breen M, Hitte C, Lorentzen TD, Thomas R, Cadieu E, Sabacan L, Scott A, Evanno G, Parker HG, Kirkness EF, Hudson R, Guyon R, Mahairas GG, Gelfenbeyn B, Fraser CM, Andre C, Galibert F, Ostrander EA. An integrated 4249 marker FISH/RH map of the canine genome. BMC Genomics. 2004;5:65. doi: 10.1186/1471-2164-5-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkmeyer-Langford C, Raudsepp T, Lee E-J, Goh G, Schäffer AA, Agarwala R, Wagner ML, Tozaki T, Skow LC, Womack JE, Mickelson JR, Chowdhary BP. A high-resolution physical map of equine homologs of HSA19 shows divergent evolution compared with other mammals. Mamm Genome. 2005;16:631–649. doi: 10.1007/s00335-005-0023-1. [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD. Multiple sequence alignment with the Clustal series of programs. Nucl Acids Res. 2003;31:3497–3500. doi: 10.1093/nar/gkg500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhary BP, Raudsepp T, Honeycutt D, Owens EK, Piumi F, Guérin G, Matise TC, Kata SR, Womack JE, Skow LC. Construction of a 5000rad whole-genome radiation hybrid panel in the horse and generation of a comprehensive and comparative map for ECA11. Mamm Genome. 2002;13:89–94. doi: 10.1007/s00335-001-2089-8. [DOI] [PubMed] [Google Scholar]
- Chowdhary BP, Raudsepp T, Kata SR, Goh G, Millon LV, Allan V, Piumi F, Guérin G, Swinburne J, Binns M, Lear TL, Mickelson J, Murray J, Antczak DF, Womack JE, Skow LC. The first-generation whole-genome radiation hybrid map in the horse identifies conserved segments in human and mouse genomes. Genome Res. 2003;13:742–751. doi: 10.1101/gr.917503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhary BP, Raudsepp T. The Horse Genome Derby: racing from map to whole genome sequence. Chromosome Res. 2008;16:109–127. doi: 10.1007/s10577-008-1204-z. [DOI] [PubMed] [Google Scholar]
- Dierks C, Mömke S, Drögemuller C, Leeb T, Chowdhary BP, Distl O. A high-resolution comparative radiation hybrid map of equine chromosome 4q12-q22. Anim Genet. 2006;37:513–517. doi: 10.1111/j.1365-2052.2006.01510.x. [DOI] [PubMed] [Google Scholar]
- Everts-van der Wind A, Kata SR, Band MR, Rebeiz M, Larkin DM, Everts RE, Green CA, Liu L, Natarajan S, Goldammer T, Lee JH, McKay S, Womack JE, Lewin HA. A 1463 gene cattle-human comparative map with anchor points defined by human genome sequence coordinates. Genome Res. 2004;14:1424–1437. doi: 10.1101/gr.2554404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everts-van der Wind A, Larkin DM, Green CA, Elliott JS, Olmstead CA, Chiu R, Schein JE, Marra MA, Womack JE, Lewin HA. A high-resolution whole-genome cattle-human comparative map reveals details of mammalian chromosome evolution. Proc Natl Acad Sci U S A. 2005;102:18526–18531. doi: 10.1073/pnas.0509285102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godard S, Vaiman A, Schibler L, Mariat D, Vaiman D, Cribiu E-P, Guérin G. Cytogenetic localization of 44 new coding sequences in the horse. Mamm Genome. 2000;11:1093–1097. doi: 10.1007/s003350010206. [DOI] [PubMed] [Google Scholar]
- Goh G, Raudsepp T, Durkin K, Wagner ML, Schäffer AA, Agarwala R, Tozaki T, Mickelson JR, Chowdhary BP. High-resolution gene maps of horse chromosomes 14 and 21: additional insights into evolution and rearrangements of HSA5 homologs in mammals. Genomics. 2007;89:89–112. doi: 10.1016/j.ygeno.2006.06.012. [DOI] [PubMed] [Google Scholar]
- Gustafson-Seabury A, Raudsepp T, Goh G, Kata SR, Wagner ML, Tozaki T, Mickelson JR, Womack JE, Skow LC, Chowdhary BP. High-resolution RH map of horse chromosome 22 reveals a putative ancestral vertebrate chromosome. Genomics. 2005;85:188–200. doi: 10.1016/j.ygeno.2004.10.012. [DOI] [PubMed] [Google Scholar]
- Hamasima N, Suzuki H, Mikawa A, Morozumi T, Plastow G, Mitsuhashi T. Construction of a new porcine whole-genome framework map using a radiation hybrid panel. Anim Genet. 2003;34:216–220. doi: 10.1046/j.1365-2052.2003.00984.x. [DOI] [PubMed] [Google Scholar]
- Itoh T, Watanabe T, Ihara N, Mariani P, Beattie CW, Sugimoto Y, Takasuga A. A comprehensive radiation hybrid map of the bovine genome comprising 5593 loci. Genomics. 2005;85:413–424. doi: 10.1016/j.ygeno.2004.12.007. [DOI] [PubMed] [Google Scholar]
- Jann OC, Aerts J, Jones M, Hastings N, Law A, McKay S, Marques E, Prasad A, Yu J, Moore SS, Floriot S, Mahe MF, Eggen A, Silveri L, Negrini R, Milanesi E, Ajmone-Marsan P, Valentini A, Marchitelli C, Savarese MC, Janitz M, Herwig R, Hennig S, Gorni C, Connor EE, Sonstegard TS, Smith T, Drogemuller C, Williams JL. A second generation radiation hybrid map to aid the assembly of the bovine genome sequence. BMC Genomics. 2006;7:283. doi: 10.1186/1471-2164-7-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H, Sandig C, Chuzhanova N, Goidts V, Szamalek JM, Tanzer S, Muller S, Platzer M, Cooper DN, Hameister H. Breakpoint analysis of the pericentric inversion distinguishing human chromosome 4 from the homologous chromosome in the chimpanzee (Pan troglodytes) Hum Mutat. 2005a;25:45–55. doi: 10.1002/humu.20116. [DOI] [PubMed] [Google Scholar]
- Kehrer-Sawatzki H, Sandig CA, Goidts V, Hameister H. Breakpoint analysis of the pericentric inversion between chimpanzee chromosome 10 and the homologous chromosome 12 in humans. Cytogenet Genome Res. 2005b;108:91–97. doi: 10.1159/000080806. [DOI] [PubMed] [Google Scholar]
- Kwitek AE, Gullings-Handley J, Yu J, Carlos DC, Orlebeke K, Nie J, Eckert J, Lemke A, Andrae JW, Bromberg S, Pasko D, Chen D, Scheetz TE, Casavant TL, Soares MB, Sheffield VC, Tonellato PJ, Jacob HJ. High-density rat radiation hybrid maps containing over 24,000 SSLPs, genes, and ESTs provide a direct link to the rat genome sequence. Genome Res. 2004;14:750–757. doi: 10.1101/gr.1968704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, Gage D, Harris K, Heaford A, Howland J, Kann L, Lehoczky J, LeVine R, McEwan P, McKernan K, Meldrim J, Mesirov JP, Miranda C, Morris W, Naylor J, Raymond C, Rosetti M, Santos R, Sheridan A, Sougnez C, Stange-Thomann N, Stojanovic N, Subramanian A, Wyman D, Rogers J, Sulston J, Ainscough R, Beck S, Bentley D, Burton J, Clee C, Carter N, Coulson A, Deadman R, Deloukas P, Dunham A, Dunham I, Durbin R, French L, Grafham D, Gregory S, Hubbard T, Humphray S, Hunt A, Jones M, Lloyd C, McMurray A, Matthews L, Mercer S, Milne S, Mullikin JC, Mungall A, Plumb R, Ross M, Shownkeen R, Sims S, Waterston RH, Wilson RK, Hillier LW, McPherson JD, Marra MA, Mardis ER, Fulton LA, Chinwalla AT, Pepin KH, Gish WR, Chissoe SL, Wendl MC, Delehaunty KD, Miner TL, Delehaunty A, Kramer JB, Cook LL, Fulton RS, Johnson DL, Minx PJ, Clifton SW, Hawkins T, Branscomb E, Predki P, Richardson P, Wenning S, Slezak T, Doggett N, Cheng JF, Olsen A, Lucas S, Elkin C, Uberbacher E, Frazier M, Gibbs RA, Muzny DM, Scherer SE, Bouck JB, Sodergren EJ, Worley KC, Rives CM, Gorrell JH, Metzker ML, Naylor SL, Kucherlapati RS, Nelson DL, Weinstock GM, Sakaki Y, Fujiyama A, Hattori M, Yada T, Toyoda A, Itoh T, Kawagoe C, Watanabe H, Totoki Y, Taylor T, Weissenbach J, Heilig R, Saurin W, Artiguenave F, Brottier P, Bruls T, Pelletier E, Robert C, Wincker P, Smith DR, Doucette-Stamm L, Rubenfield M, Weinstock K, Lee HM, Dubois J, Rosenthal A, Platzer M, Nyakatura G, Taudien S, Rump A, Yang H, Yu J, Wang J, Huang G, Gu J, Hood L, Rowen L, Madan A, Qin S, Davis RW, Federspiel NA, Abola AP, Proctor MJ, Myers RM, Schmutz J, Dickson M, Grimwood J, Cox DR, Olson MV, Kaul R, Shimizu N, Kawasaki K, Minoshima S, Evans GA, Athanasiou M, Schultz R, Roe BA, Chen F, Pan H, Ramser J, Lehrach H, Reinhardt R, McCombie WR, de la Bastide M, Dedhia N, Blocker H, Hornischer K, Nordsiek G, Agarwala R, Aravind L, Bailey JA, Bateman A, Batzoglou S, Birney E, Bork P, Brown DG, Burge CB, Cerutti L, Chen HC, Church D, Clamp M, Copley RR, Doerks T, Eddy SR, Eichler EE, Furey TS, Galagan J, Gilbert JG, Harmon C, Hayashizaki Y, Haussler D, Hermjakob H, Hokamp K, Jang W, Johnson LS, Jones TA, Kasif S, Kaspryzk A, Kennedy S, Kent WJ, Kitts P, Koonin EV, Korf I, Kulp D, Lancet D, Lowe TM, McLysaght A, Mikkelsen T, Moran JV, Mulder N, Pollara VJ, Ponting CP, Schuler G, Schultz J, Slater G, Smit AF, Stupka E, Szustakowski J, Thierry-Mieg D, Thierry-Mieg J, Wagner L, Wallis J, Wheeler R, Williams A, Wolf YI, Wolfe KH, Yang SP, Yeh RF, Collins F, Guyer MS, Peterson J, Felsenfeld A, Wetterstrand KA, Patrinos A, Morgan MJ, de Jong P, Catanese JJ, Osoegawa K, Shizuya H, Choi S, Chen YJ. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Lee E-J, Raudsepp T, Kata SR, Adelson D, Womack JE, Skow LC, Chowdhary BP. A 1.4-Mb interval RH map of horse chromosome 17 provides detailed comparison with human and mouse homologues. Genomics. 2004;83:203–215. doi: 10.1016/j.ygeno.2003.07.002. [DOI] [PubMed] [Google Scholar]
- Leeb T, Vogl C, Zhu B, de Jong PJ, Binns MM, Chowdhary BP, Scharfe M, Jarek M, Nordsiek G, Schrader F, Blocker H. A human-horse comparative map based on equine BAC end sequences. Genomics. 2006;87:772–776. doi: 10.1016/j.ygeno.2006.03.002. [DOI] [PubMed] [Google Scholar]
- McKay SD, Schnabel RD, Murdoch BM, Aerts J, Gill CA, Gao C, Li C, Matukumalli LK, Stothard P, Wang Z, Van Tassell CP, Williams JL, Taylor JF, Moore SS. Construction of bovine whole-genome radiation hybrid and linkage maps using high-throughput genotyping. Anim Genet. 2007;38:120–125. doi: 10.1111/j.1365-2052.2006.01564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers SN, Rogatcheva MB, Larkin DM, Yerle M, Milan D, Hawken RJ, Schook LB, Beever JE. Piggy-BACing the human genome II. A high-resolution, physically anchored, comparative map of the porcine autosomes. Genomics. 2005;86:739–752. doi: 10.1016/j.ygeno.2005.04.010. [DOI] [PubMed] [Google Scholar]
- Milenkovic D, Oustry-Vaiman A, Lear TL, Billault A, Mariat D, Piumi F, Schibler L, Cribiu E, Guérin G. Cytogenetic localization of 136 genes in the horse: comparative mapping with the human genome. Mamm Genome. 2002;13:524–534. doi: 10.1007/s00335-001-2137-4. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Larkin DM, Everts-van der Wind A, Bourque G, Tesler G, Auvil L, Beever JE, Chowdhary BP, Galibert F, Gatzke L, Hitte C, Meyers SN, Milan D, Ostrander EA, Pape G, Parker HG, Raudsepp T, Rogatcheva MB, Schook LB, Skow LC, Welge M, Womack JE, O’Brien SJ, Pevzner PA, Lewin HA. Dynamics of mammalian chromosome evolution inferred from multispecies comparative maps. Science. 2005;309:613–617. doi: 10.1126/science.1111387. [DOI] [PubMed] [Google Scholar]
- Murphy WJ, Davis B, David VA, Agarwala R, Schäffer AA, Pearks Wilkerson AJ, Neelam B, O’Brien SJ, Menotti-Raymond M. A 1.5-Mb-resolution radiation hybrid map of the cat genome and comparative analysis with the canine and human genomes. Genomics. 2007;89:189–196. doi: 10.1016/j.ygeno.2006.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadeau JH, Sankoff D. Counting on comparative maps. Trends Genet. 1998;14:495–501. doi: 10.1016/s0168-9525(98)01607-2. [DOI] [PubMed] [Google Scholar]
- Olivier M, Aggarwal A, Allen J, Almendras AA, Bajorek ES, Beasley EM, Brady SD, Bushard JM, Bustos VI, Chu A, Chung TR, De Witte A, Denys ME, Dominguez R, Fang NY, Foster BD, Freudenberg RW, Hadley D, Hamilton LR, Jeffrey TJ, Kelly L, Lazzeroni L, Levy MR, Lewis SC, Liu X, Lopez FJ, Louie B, Marquis JP, Martinez RA, Matsuura MK, Misherghi NS, Norton JA, Olshen A, Perkins SM, Perou AJ, Piercy C, Piercy M, Qin F, Reif T, Sheppard K, Shokoohi V, Smick GA, Sun WL, Stewart EA, Fernando J, Tejeda Tran NM, Trejo T, Vo NT, Yan SC, Zierten DL, Zhao S, Sachidanandam R, Trask BJ, Myers RM, Cox DR. A high-resolution radiation hybrid map of the human genome draft sequence. Science. 2001;291:1298–1302. doi: 10.1126/science.1057437. [DOI] [PubMed] [Google Scholar]
- Penedo MCT, Millon LV, Bernoco D, Bailey E, Binns M, Cholewinski G, Ellis N, Flynn J, Gralak B, Guthrie A, Hasegawa T, Lindgren G, Lyons LA, Roed KH, Swinburne JE, Tozaki T. International equine gene mapping workshop report: a comprehensive linkage map constructed with data from new markers and by merging four mapping resources. Cytogenet Genome Res. 2005;111:5–15. doi: 10.1159/000085664. [DOI] [PubMed] [Google Scholar]
- Perrocheau M, Boutreux V, Chadi S, Mata X, Decaunes P, Raudsepp T, Durkin K, Incarnato D, Iannuzzi L, Lear TL, Hirota K, Hasegawa T, Zhu B, de Jong P, Cribiu EP, Chowdhary BP, Guérin G. Construction of a medium-density horse gene map. Anim Genet. 2006;37:145–155. doi: 10.1111/j.1365-2052.2005.01401.x. [DOI] [PubMed] [Google Scholar]
- Raudsepp T, Frönicke L, Scherthan H, Gustavsson I, Chowdhary BP. Zoo-FISH delineates conserved chromosomal segments in horse and man. Chromosome Res. 1996;4:218–225. doi: 10.1007/BF02254963. [DOI] [PubMed] [Google Scholar]
- Raudsepp T, Lee E-J, Kata SR, Brinkmeyer C, Mickelson JR, Skow LC, Womack JE, Chowdhary BP. Exceptional conservation of horse-human gene order on X chromosome revealed by high-resolution radiation hybrid mapping. Proc Natl Acad Sci U S A. 2004;101:2386–2391. doi: 10.1073/pnas.0308513100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe LB, Barter ME, Kelmenson JA, Eppig JT. The comprehensive mouse radiation hybrid map densely cross-referenced to the recombination map: a tool to support the sequence assemblies. Genome Res. 2003;13:122–133. doi: 10.1101/gr.858103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäffer AA, Rice ES, Cook W, Agarwala R. rh_tsp_map 3.0: End-to-end radiation hybrid mapping with improved speed and quality control. Bioinformatics. 2007;23:1156–1158. doi: 10.1093/bioinformatics/btm077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Martin J, Terry A, Couronne O, Grimwood J, Lowry S, Gordon LA, Scott D, Xie G, Huang W, Hellsten U, Tran-Gyamfi M, She X, Prabhakar S, Aerts A, Altherr M, Bajorek E, Black S, Branscomb E, Caoile C, Challacombe JF, Chan YM, Denys M, Detter JC, Escobar J, Flowers D, Fotopulos D, Glavina T, Gomez M, Gonzales E, Goodstein D, Grigoriev I, Groza M, Hammon N, Hawkins T, Haydu L, Israni S, Jett J, Kadner K, Kimball H, Kobayashi A, Lopez F, Lou Y, Martinez D, Medina C, Morgan J, Nandkeshwar R, Noonan JP, Pitluck S, Pollard M, Predki P, Priest J, Ramirez L, Retterer J, Rodriguez A, Rogers S, Salamov A, Salazar A, Thayer N, Tice H, Tsai M, Ustaszewska A, Vo N, Wheeler J, Wu K, Yang J, Dickson M, Cheng JF, Eichler EE, Olsen A, Pennacchio LA, Rokhsar DS, Richardson P, Lucas SM, Myers RM, Rubin EM. The DNA sequence and comparative analysis of human chromosome 5. Nature. 2004;431:268–274. doi: 10.1038/nature02919. [DOI] [PubMed] [Google Scholar]
- Schuler GD. Sequence mapping by electronic PCR. Genome Res. 1997;7:541–550. doi: 10.1101/gr.7.5.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snelling WM, Chiu R, Schein JE, Hobbs M, Abbey CA, Adelson DL, Aerts J, Bennett GL, Bosdet IE, Boussaha M, Brauning R, Caetano AR, Costa MM, Crawford AM, Dalrymple BP, Eggen A, Everts-van der Wind A, Floriot S, Gautier M, Gill CA, Green RD, Holt R, Jann O, Jones SJ, Kappes SM, Keele JW, de Jong PJ, Larkin DM, Lewin HA, McEwan JC, McKay S, Marra MA, Mathewson CA, Matukumalli LK, Moore SS, Murdoch B, Nicholas FW, Osoegawa K, Roy A, Salih H, Schibler L, Schnabel RD, Silveri L, Skow LC, Smith TP, Sonstegard TS, Taylor JF, Tellam R, Van Tassell CP, Williams JL, Womack JE, Wye NH, Yang G, Zhao S. A physical map of the bovine genome. Genome Biol. 2007;8:R165. doi: 10.1186/gb-2007-8-8-r165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinburne J, Gerstenberg C, Breen M, Aldridge V, Lockhart L, Marti E, Antczak D, Eggleston-Stott M, Bailey E, Mickelson J, Roed K, Lindgren G, von Haeringen W, Guerin G, Bjarnason J, Allen T, Binns M. First comprehensive low-density horse linkage map based on two 3-generation, full-sibling, cross-bred horse reference families. Genomics. 2000;66:123–134. doi: 10.1006/geno.2000.6207. [DOI] [PubMed] [Google Scholar]
- Swinburne JE, Boursnell M, Hill G, Pettitt L, Allen T, Chowdhary B, Hasegawa T, Kurosawa M, Leeb T, Mashima S, Mickelson JR, Raudsepp T, Tozaki T, Binns M. Single linkage group per chromosome genetic linkage map for the horse, based on two three-generation, full-sibling, crossbred horse reference families. Genomics. 2006;87:1–29. doi: 10.1016/j.ygeno.2005.09.001. [DOI] [PubMed] [Google Scholar]
- Tozaki T, Swinburne J, Hirota K, Hasegawa T, Ishida N, Tobe T. Improved resolution of the comparative horse-human map: investigating markers with in silico and linkage mapping approaches. Gene. 2007;392:181–186. doi: 10.1016/j.gene.2006.12.018. [DOI] [PubMed] [Google Scholar]
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, Amanatides P, Ballew RM, Huson DH, Wortman JR, Zhang Q, Kodira CD, Zheng XH, Chen L, Skupski M, Subramanian G, Thomas PD, Zhang J, Gabor Miklos GL, Nelson C, Broder S, Clark AG, Nadeau J, McKusick VA, Zinder N, Levine AJ, Roberts RJ, Simon M, Slayman C, Hunkapiller M, Bolanos R, Delcher A, Dew I, Fasulo D, Flanigan M, Florea L, Halpern A, Hannenhalli S, Kravitz S, Levy S, Mobarry C, Reinert K, Remington K, Abu-Threideh J, Beasley E, Biddick K, Bonazzi V, Brandon R, Cargill M, Chandramouliswaran I, Charlab R, Chaturvedi K, Deng Z, Di Francesco V, Dunn P, Eilbeck K, Evangelista C, Gabrielian AE, Gan W, Ge W, Gong F, Gu Z, Guan P, Heiman TJ, Higgins ME, Ji RR, Ke Z, Ketchum KA, Lai Z, Lei Y, Li Z, Li J, Liang Y, Lin X, Lu F, Merkulov GV, Milshina N, Moore HM, Naik AK, Narayan VA, Neelam B, Nusskern D, Rusch DB, Salzberg S, Shao W, Shue B, Sun J, Wang Z, Wang A, Wang X, Wang J, Wei M, Wides R, Xiao C, Yan C, Yao A, Ye J, Zhan M, Zhang W, Zhang H, Zhao Q, Zheng L, Zhong F, Zhong W, Zhu S, Zhao S, Gilbert D, Baumhueter S, Spier G, Carter C, Cravchik A, Woodage T, Ali F, An H, Awe A, Baldwin D, Baden H, Barnstead M, Barrow I, Beeson K, Busam D, Carver A, Center A, Cheng ML, Curry L, Danaher S, Davenport L, Desilets R, Dietz S, Dodson K, Doup L, Ferriera S, Garg N, Gluecksmann A, Hart B, Haynes J, Haynes C, Heiner C, Hladun S, Hostin D, Houck J, Howland T, Ibegwam C, Johnson J, Kalush F, Kline L, Koduru S, Love A, Mann F, May D, McCawley S, McIntosh T, McMullen I, Moy M, Moy L, Murphy B, Nelson K, Pfannkoch C, Pratts E, Puri V, Qureshi H, Reardon M, Rodriguez R, Rogers YH, Romblad D, Ruhfel B, Scott R, Sitter C, Smallwood M, Stewart E, Strong R, Suh E, Thomas R, Tint NN, Tse S, Vech C, Wang G, Wetter J, Williams S, Williams M, Windsor S, Winn-Deen E, Wolfe K, Zaveri J, Zaveri K, Abril JF, Guigo R, Campbell MJ, Sjolander KV, Karlak B, Kejariwal A, Mi H, Lazareva B, Hatton T, Narechania A, Diemer K, Muruganujan A, Guo N, Sato S, Bafna V, Istrail S, Lippert R, Schwartz R, Walenz B, Yooseph S, Allen D, Basu A, Baxendale J, Blick L, Caminha M, Carnes-Stine J, Caulk P, Chiang YH, Coyne M, Dahlke C, Mays A, Dombroski M, Donnelly M, Ely D, Esparham S, Fosler C, Gire H, Glanowski S, Glasser K, Glodek A, Gorokhov M, Graham K, Gropman B, Harris M, Heil J, Henderson S, Hoover J, Jennings D, Jordan C, Jordan J, Kasha J, Kagan L, Kraft C, Levitsky A, Lewis M, Liu X, Lopez J, Ma D, Majoros W, McDaniel J, Murphy S, Newman M, Nguyen T, Nguyen N, Nodell M, Pan S, Peck J, Peterson M, Rowe W, Sanders R, Scott J, Simpson M, Smith T, Sprague A, Stockwell T, Turner R, Venter E, Wang M, Wen M, Wu D, Wu M, Xia A, Zandieh A, Zhu X. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Wagner ML, Raudsepp T, Goh G, Agarwala R, Schäffer AA, Dranchak PK, Brinkmeyer-Langford C, Skow LC, Chowdhary BP, Mickelson JR. A 1.3-Mb interval map of equine homologs of HSA2. Cytogenet Genome Res. 2006;112:227–234. doi: 10.1159/000089875. [DOI] [PubMed] [Google Scholar]
- Yang F, Fu B, O’Brien PCM, Nie W, Ryder OA, Ferguson-Smith MA. Refined genome-wide comparative map of the domestic horse, donkey and human based on cross-species chromosome painting: insight into the occasional fertility of mules. Chromosome Res. 2004;12:65–76. doi: 10.1023/b:chro.0000009298.02689.8a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figures 1.1-1.X, 2.1-2.X, Legends for Supplementary Figures and Supplementary Tables 1, 2, 3, 4, 5.