Summary
Saccharomyces cerevisiae RecQ helicase, Sgs1, and XPF-family endonuclease, Mus81-Mms4, are implicated in processing joint molecule (JM) recombination intermediates. We show that cells lacking either enzyme frequently experience chromosome segregation problems during meiosis and when both enzymes are absent attempted segregation fails catastrophically. In all cases, segregation appears to be impeded by unresolved JMs. Analysis of the DNA events of recombination indicates that Sgs1 limits aberrant JM structures that result from secondary strand-invasion events and often require Mus81-Mms4 for their normal resolution. Aberrant JMs contain high levels of single Holliday junctions and include intersister JMs, multi-chromatid JMs comprising three and four chromatids, and newly identified recombinant JMs containing two chromatids, one of which has undergone crossing-over. Despite persistent JMs in sgs1 mms4 double mutants, crossover and noncrossover products still form at high levels. We conclude that Sgs1 and Mus81-Mms4 collaborate to eliminate aberrant JMs whereas as-yet-unidentified enzymes process normal JMs.
Introduction
Template-directed repair of broken chromosomes occurs via homologous recombination (HR). HR is also an intrinsic part of the meiotic program, where it facilitates the pairing and disjunction of homologous chromosomes (homologs) at the first meiotic division (MI)(Hunter, 2006). Meiotic HR is initiated by DNA double-strand-breaks (DSBs), catalyzed by the transesterase, Spo11 (Neale et al., 2005). DSB-ends are resected to form long 3’-single-stranded tails which then assemble into nucleoprotein filaments together with homologous-pairing and strand-exchange proteins, Rad51 and Dmc1 (Shinohara and Shinohara, 2004). Strand-exchange with a homologous chromosome forms joint molecule (JM) intermediates from which repair synthesis can ensue. Meiotic DSB repair can occur with or without an associated crossover (a reciprocal exchange of chromosome arms). The crossover or noncrossover fate is highly regulated and is thought to be designated shortly after initial JMs form (Allers and Lichten, 2001a; Bishop and Zickler, 2004; Borner et al., 2004; Hunter and Kleckner, 2001)
In budding yeast, Saccharomyces cerevisiae, two prominent JM types have been identified in vivo. Strand-invasion by one DSB-end gives rise to a Single End Invasion (SEI) (Hunter and Kleckner, 2001). Subsequent interaction with the second DSB-end, together with recombination-associated DNA synthesis, leads to formation of a double-Holliday Junction (dHJ) (Allers and Lichten, 2001b; Lao et al., 2008; Schwacha and Kleckner, 1995). Available evidence indicates that both SEIs and dHJs are primarily crossover precursors (Allers and Lichten, 2001a; Hunter and Kleckner, 2001; Borner et al., 2004; Bishop and Zickler, 2004). Molecular events leading to noncrossovers are less well defined but appear to involve a synthesis-dependent strand-annealing (SDSA) mechanism (McMahill et al., 2007). In its simplest form, SDSA proposes that one DSB-end invades a homolog to form a transient D-loop, which primes DNA synthesis; the nascent strand is then displaced and anneals to complementary sequences on the second DSB-end (Nassif et al., 1994; Paques and Haber, 1999). The failure to detect discrete JM intermediates specific to the noncrossover pathway suggests that they are more labile and/or transient than JMs along the crossover pathway.
One criteria used to show that JMs identified in vivo were dHJs is that they comprised primarily parental DNA strands and crossover strands only arose following resolution (Schwacha and Kleckner, 1995; Szostak et al., 1983). This feature contradicted a basic tenet of the original Holliday model: that a single HJ (sHJ), comprising both parental and crossover strands, is the central intermediate in HR (Holliday, 1964). However, recent evidence suggests that sHJs are the predominant intermediate during meiosis in fission yeast, Schizosaccharomyces pombe, and may also be a significant intermediate in budding yeast (Cromie et al., 2006; Oh et al., 2007).
We recently identified a third JM type, the multi-chromatid JM (mcJM), which comprises three or four interconnected chromatids and is thought to arise when the two ends of a DSB independently invade different chromatids and/or when DSB-ends sequentially invade multiple templates (Oh et al., 2007). mcJMs are aberrant intermediates that cause unregulated crossing-over and their formation is limited by the RecQ DNA helicase, Sgs1. Like its human ortholog, the Bloom’s Syndrome helicase (BLM), Sgs1 is thought to disassemble joint molecules by unwinding D-loops and/or by “dissolving” dHJs as part of a ternary complex together with the type-I topoisomerase, Top3, and a specificity factor, Rmi1 (Chang et al., 2005; Mullen et al., 2005; van Brabant et al., 2000; Wu et al., 2006; Wu and Hickson, 2003). Recent studies indicate that the Sgs1 complex works in coordination with meiosis-specific JM-stabilizing factors, such as the MutS complex Msh4-Msh5, to effect the orderly formation of two-chromatid dHJs, and thus single exchanges, at designated crossover sites (Oh et al., 2007).
Unambiguous identification of the enzyme(s) that resolve dHJs into crossovers remains an outstanding challenge for the HR field. However, the accumulation of JMs observed in S. cerevisiae mutants lacking the meiosis-specific transcription factor, Ndt80, and the polo-like kinase, Cdc5 (whose expression is positively regulated by Ndt80), suggest that a resolvase is activated via Cdc5-mediated phosphorylation during meiosis (Allers and Lichten, 2001a; Clyne et al., 2003). In fission yeast, meiotic crossing-over is almost entirely dependent on the XPF-family endonuclease, Mus81-Eme1, and mus81 mutants accumulate sHJ intermediates (Boddy et al., 2001; Cromie et al., 2006; Smith et al., 2003). In contrast, only ~25% of crossovers in budding yeast are dependent on the analogous Mus81-Mms4 enzyme. Despite this modest crossover defect, however, S. cerevisiae mus81/mms4 mutants have severe sporulation defects and spore viability is ≤50% (Interthal and Heyer, 2000; Kaliraman et al., 2001; Mullen et al., 2001; de Los Santos et al., 2003; de los Santos et al., 2001; this study). In vitro, the Mus81-Eme1 and Mus81-Mms4 enzymes efficiently cleave a number of branched structures, such as nicked-HJs, D-loops and 3’-flaps but, under standard enzymological conditions, cleavage of intact HJs by recombinant Mus81-Eme1 and Mus81-Mms4 is relatively negligible (Ehmsen and Heyer, 2007; Fricke et al., 2005; Gaskell et al., 2007; Heyer, 2004; Hollingsworth and Brill, 2004; Taylor and McGowan, 2008; Whitby, 2006). Thus, the in vivo substrate(s) of Mus81-Eme1 and Mus81-Mms4 still remains uncertain and controversial.
In mitotically dividing cells, a role of Mus81-Mms4 in processing recombination intermediates was inferred from the synthetic lethality observed when mus81/mms4 mutations are combined with mutations in the Sgs1 complex. Moreover, this lethality is suppressed by mutations that prevent the early steps of HR, e.g. deletion of the strand-exchange protein Rad51. It has been suggested that Mus81-Mms4 and the Sgs1 complex catalyze alternative ways to process either the same or readily interconverted JMs and in the absence of both proteins unresolved JMs cause a lethal cellular event (Bastin-Shanower et al., 2003; Fabre et al., 2002).
To identify the cause of sgs1 mus81/mms4 lethality and the intermediate(s) processed by Sgs1 and Mus81-Mms4, we have utilized conditional mutations to inactivate both SGS1 and MMS4 specifically during meiotic prophase. We find that sgs1 mms4 double mutants undergo recombination-dependent meiotic catastrophe in which chromosome segregation is impeded by unresolved JMs. Unexpectedly, large numbers of both mms4 and sgs1 single mutant cells also experience segregation problems indicating a common defect in these strains. Physical monitoring of JMs suggests a model in which Sgs1 limits the formation of aberrant JMs that are otherwise resolved by Mus81-Mms4. In this way, Sgs1 and Mus81-Mms4 facilitate productive meiotic recombination and chromosome segregation. We also identify a new type of aberrant two-chromatid JM in which one chromatid has undergone crossing-over. These recombinant JMs (rJMs) accumulate in sgs1 mms4 mutants and are likely derived from the partial resolution of mcJMs and/or from secondary strand-invasion events. sHJs and dHJs with relatively short inter-junction distances also accumulate in sgs1 mms4 cells indicating that Sgs1 and Mus81-Mms4 function to prevent and resolve such structures. Together these data provide critical insights into the dynamic nature and regulation of homologous recombination.
Results
To circumvent the vegetative lethality of sgs1 mms4 double mutants, we constructed meiosis-specific conditional alleles by replacing native SGS1 and MMS4 promoters with the CLB2 promoter, which is strongly repressed during meiosis (Lee and Amon, 2003). pCLB2-SGS1 and pCLB2-MMS4 single mutants and pCLB2-SGS1 pCLB2-MMS4 double mutants show normal vegetative growth characteristics and efficient pre-meiotic synchronization (not shown). These three mutant strains were analyzed in parallel together with a wild-type strain.
Recombination-dependent meiotic catastrophe occurs in pCLB2-SGS1 pCLB2-MMS4 mutants
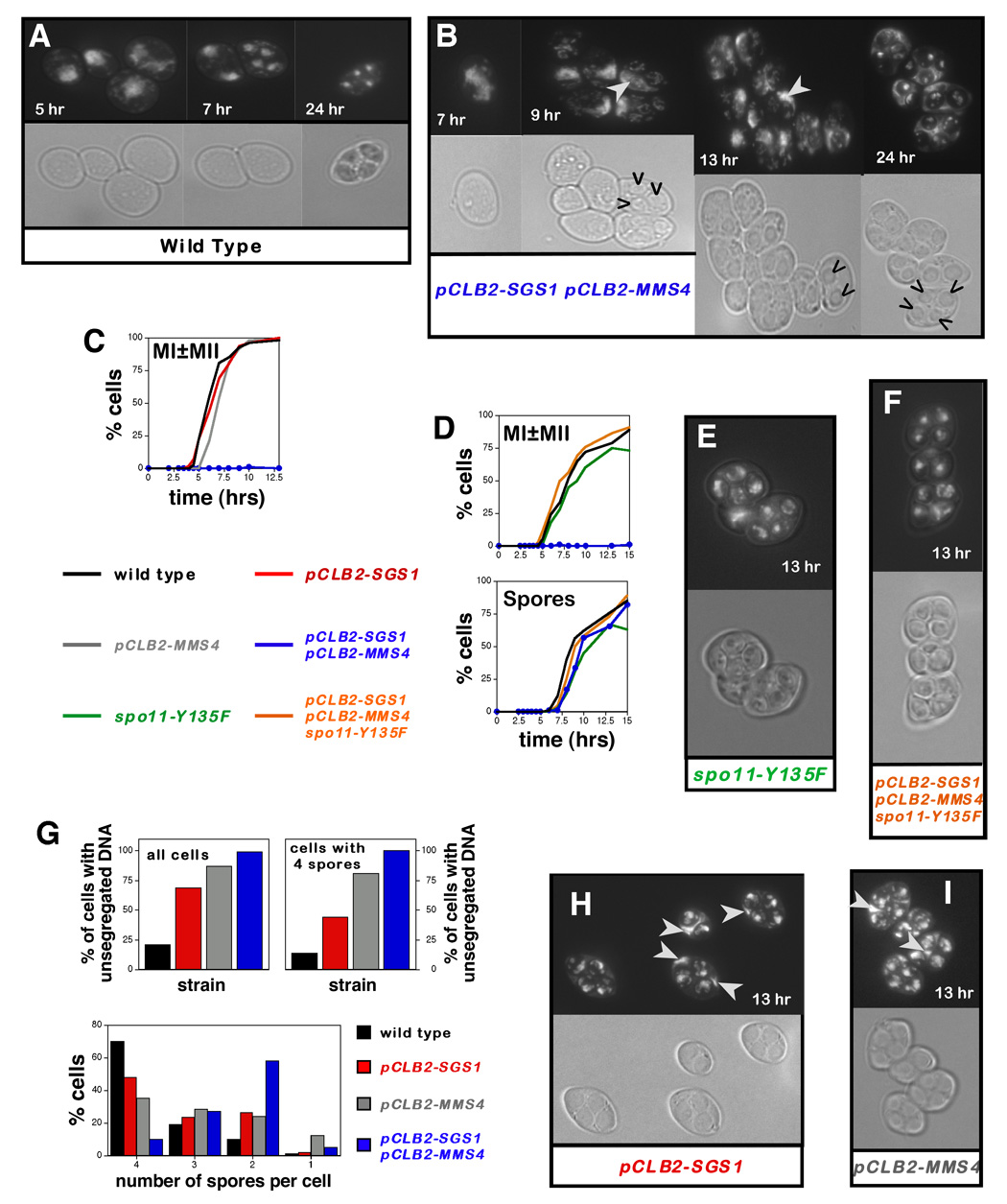
The timing and efficiency of nuclear divisions (MI and MII) and spore formation were monitored microscopically (Figure 1). In wild-type cells, MI begins ~5 hrs after transfer to sporulation media and is rapidly followed by MII (Figure 1A). By ~6 hrs, 50% of cells have completed one or both divisions (MI±MII; Figure 1A and 1C). Spores first appear at 7 hrs and 50% of cells have made spores by 8.5 hrs (Figure 1D). In sharp contrast, normal nuclear divisions completely fail in pCLB2-SGS1 pCLB2-MMS4 double mutants. A number of yeast mutants with defects in meiotic recombination arrest in prophase I, with a single undivided nucleus, and fail to form spores (Hochwagen and Amon, 2006). Unusually, pCLB2-SGS1 pCLB2-MMS4 cells unsuccessfully attempt meiotic divisions and form spores with only a slight delay relative to wild type (Figure 1B and 1D). By 6–7 hrs, elongated and partially separated DAPI-staining structures can be seen (Figure 1B, 7hr panel), but after 8–9 hrs most cells still contain a large unsegregated DNA mass and spores include only very small amounts of DNA (Figure 1B, 9 and 13 hr panels). In addition, pCLB2-SGS1 pCLB2-MMS4 cells typically form fewer than four spores and mature asci are almost never observed (Figure 1B, 24 hr panel; Figure 1G). Moreover, the very rare mature tetrad asci that do form contain only dead spores (not shown). Thus, the sgs1 mms4 mutant combination is lethal in both vegetative and meiotic cells. In an accompanying study, MI and MII spindle elongation cycles are shown to occur normally in sgs1 mus81 double mutants even though nuclear divisions fail in these cells (Jessop and Lichten, in press). Thus, sgs1 mms4/mus81 cells are progressing through meiotic divisions but failing to segregate their chromosomes.
Figure 1. Recombination-Dependent Meiotic Catastrophe in pCLB2-SGS1 pCLB2-MMS4 Cells.
(A, B) DAPI-fluorescence and bright-field images of cells from wild type and pCLB2-SGS1 pCLB2-MMS4 cultures sampled at the indicated times. Carets indicate spores.
(C) The timing and efficiency of meiotic divisions in parallel cultures of wild type, pCLB2-SGS1, pCLB2-MMS4 and pCLB2-SGS1 pCLB2-MMS4 strains. MI±MII is cells that have completed one or both meiotic divisions.
(D) Analysis of meiotic divisions (top graph) and sporulation (bottom graph) in parallel cultures of wild type, spo11-Y135F, pCLB2-SGS1 pCLB2-MMS4 and spo11-Y135F pCLB2-SGS1 pCLB2-MMS4 strains. “Spores” indicates cells that contain at least one spore.
(E) DAPI-fluorescence and bright-field images of spo11-Y135F cells.
(F) DAPI-fluorescence and bright-field images of spo11-Y135F pCLB2-SGS1 pCLB2-MMS4 cells.
(G) Upper graphs: quantitation of cells with DNA outside of the spores (“unsegregated DNA”) in 13 hr samples. The percentage of cells with unsegregated DNA is shown for all cells and for cells containing four spores.
Lower graphs: percentage of cells containing 4, 3, 2, 1, or 0 spores for wild type, pCLB2-SGS1, pCLB2-MMS4 and pCLB2-SGS1 pCLB2-MMS4 in 24 hr samples.
(H) DAPI-fluorescence and bright-field images of pCLB2-SGS1 cells.
(I) DAPI-fluorescence and bright-field micrographs of pCLB2-MMS4 cells.
White arrows highlight unsegregated DNA masses.
In vegetative cells, sgs1 mms4/mus81 lethality is suppressed when HR is blocked prior to the strand-exchange step (Bastin-Shanower et al., 2003; Fabre et al., 2002). To determine whether the meiotic catastrophe of pCLB-SGS1 pCLB-MMS4 cells is also recombination dependent, meiotic DSB formation was prevented using a catalytically inactive SPO11 allele, spo11-Y135F (Figure 1D, 1E and 1F) (Bergerat et al., 1997). In spo11-Y135F single mutants, meiotic divisions and sporulation occur efficiently, but spores are dead due to random homolog segregation at MI. spo11-Y135F suppresses the catastrophic meiosis of pCLB-SGS1 pCLB-MMS4 cells: nuclear divisions occur efficiently and mature asci are formed, as in spo11-Y135F single mutants (Figure 1D, 1E and 1F). We conclude that pCLB-SGS1 pCLB-MMS4 mutants undergo recombination-dependent catastrophic meiosis.
Segregation defects are common in pCLB-SGS1 and pCLB-MMS4 single mutants
At first glance, both pCLB2-SGS1 and pCLB2-MMS4 single mutants appear to complete divisions and sporulate efficiently (Figure 1C). However, closer inspection reveals that significant numbers of asci from both strains contain DNA that has not been packaged into spores, although the amount of unpackaged DNA is generally much less than that in the pCLB2-SGS1 pCLB2-MMS4 double mutant (Figure 1G, 1H and 1I). 69% of sporulated pCLB2-SGS1 cells (containing at least one spore) have DNA outside of the spores compared to less than 21% of wild-type cells (Figure 1G and 1H; P<0.05). In pCLB2-MMS4 cells, this defect is seen in 87% of cells (Figure 1G and 1I). Qualitatively, the amount of unpackaged DNA negatively correlates with the number of spores formed, and accordingly the fractions of cells with <4 spores is increased in pCLB2-SGS1 and pCLB2-MMS4 cells (Figure 1G). Even amongst cells with 4 spores, however, unpackaged DNA is seen in 44% of pCLB2-SGS1 cells and 81% of pCLB2-MMS4 cells (Figure 1G). We infer that defective chromosome segregation is common to pCLB2-SGS1, pCLB2-MMS4 and pCLB2-SGS1 pCLB2-MMS4 strains, suggesting a common underlying defect.
Crossing-over is reduced in pCLB2-MMS4 and pCLB2-SGS1 pCLB2-MMS4 cells
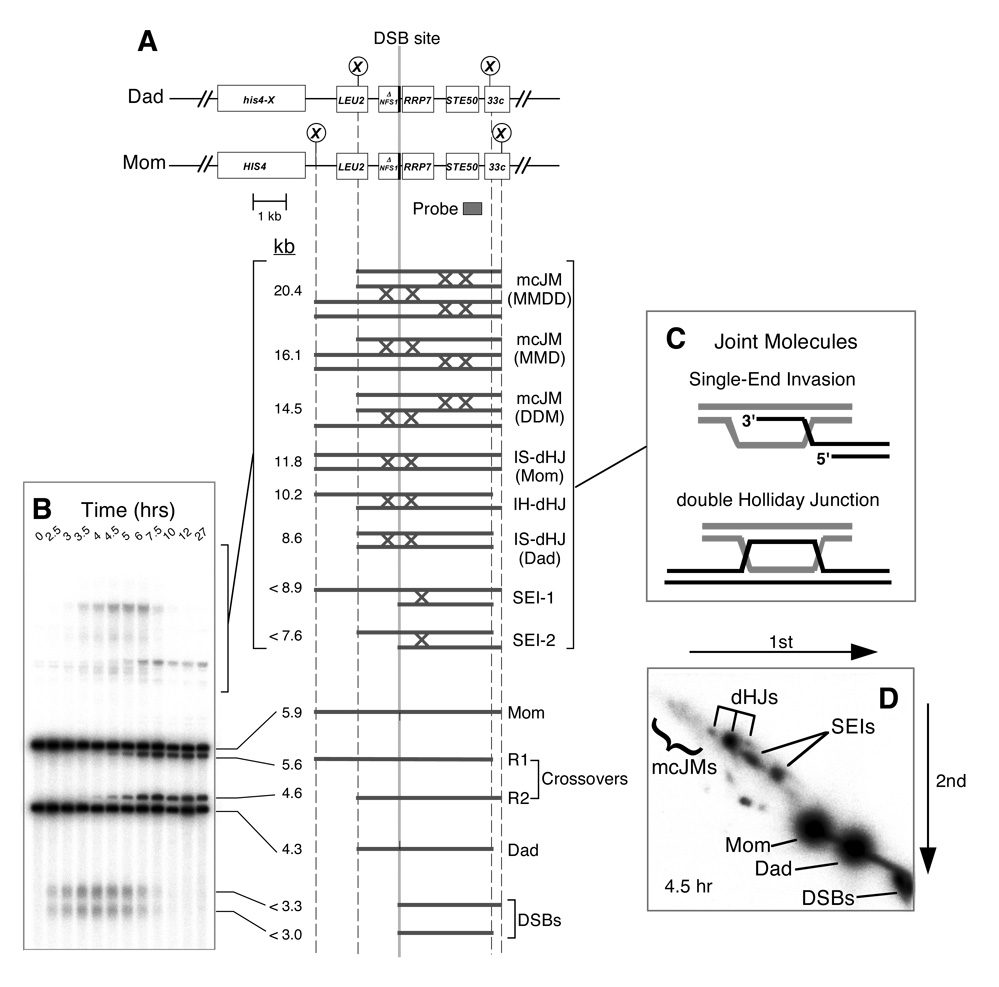
To determine the molecular defect that impedes segregation, DNA events of meiotic recombination were monitored using the HIS4LEU2 physical assay system (Figure 2; Cao et al., 1990; Hunter and Kleckner, 2001; Schwacha and Kleckner, 1995). DNA events are monitored over time in synchronized meiotic cultures. Cell samples are first treated with psoralen to produce DNA interstrand crosslinks, which stabilize JM intermediates. Species of interest are then detected by gel electrophoresis and Southern blot hybridization (Figure 2). XhoI polymorphisms between parental “Mom” and “Dad” homologs produce diagnostic restriction fragments for parental and recombinant chromosomes, DSBs and JMs. Each hybridizing signal is quantified using a Phosphorimager. DSBs and crossovers are quantified from one-dimensional gels (Figure 2B). Native/native two-dimensional gels reveal the branched structure of JMs and are used to individually quantitate SEIs, dHJs and mcJMs (Bell and Byers, 1983; Hunter and Kleckner, 2001). Figure 3 shows data for the parallel analysis of wild-type, pCLB2- SGS1, pCLB2-MMS4 and pCLB2-SGS1 pCLB2-MMS4 cultures and is described below. Very similar data for a second set of time-course experiments are presented in Supplemental Figure S1.
Figure 2. Physical Assay System for Monitoring Recombination.
(A) Map of the HIS4LEU2 locus showing diagnostic restriction sites and position of the probe. DNA species detected following Southern hybridization are shown below. SEI-1 and SEI-2 are the two major SEI species detected with Probe 4 (Hunter and Kleckner, 2001). Lollipops indicate restriction sites: X, XhoI.
(B) Image of one-dimensional (1D) Southern analysis showing DNA species detailed in (A).
(C) Predicted structures of SEI and dHJ joint molecule intermediates.
(D) Image of a native/native 2D analysis. Species detailed in (A) are highlighted.
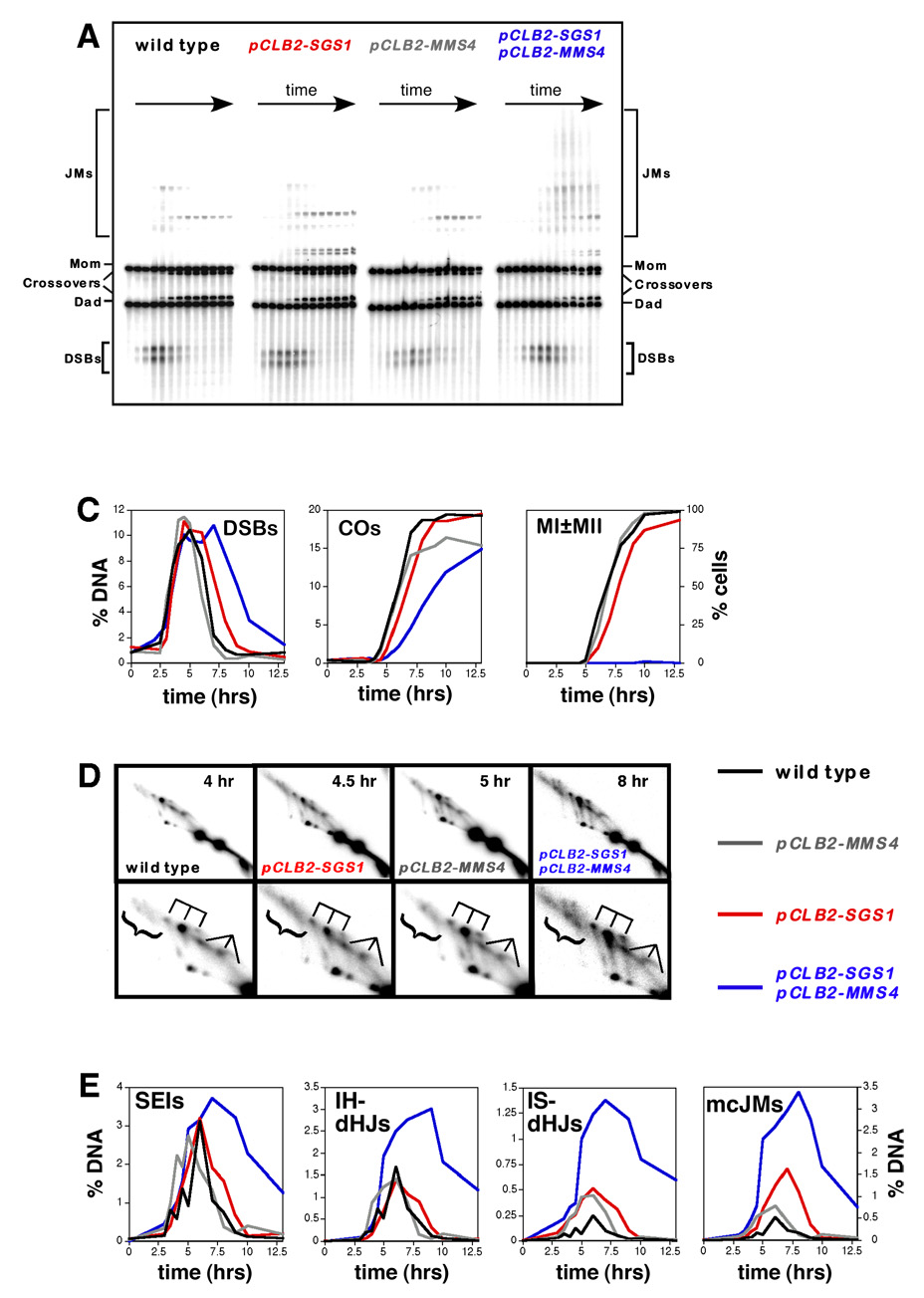
Figure 3. Physical Analysis of Recombination in Wild-Type, pCLB2-SGS1, pCLB2-MMS4 and pCLB2-SGS1 pCLB2-MMS4 Cells.
(A) Representative images of 1D Southern analysis.
(B) Quantitative analysis of DSBs, crossovers (COs) and meiotic divisions (MI±MII). “% DNA” is percent of total hybridizing DNA.
(C) 2D analysis of JMs. For each strain a representative 2D panel is shown together with a blowup of the JM region, below. dHJ species are highlighted by a trident; SEIs are indicated by a fork; mcJMs are indicated by a bracket.
(D) Quantitative analysis of JM formation.
DSBs
In wild-type cells, DSBs are first detected 2.5 hrs after transfer to sporulation medium, peak at ~4–5 hrs at ~10% of hybridizing DNA and disappear by 7 hrs (Figure 3A and 3B). DSB timing and levels are similar in pCLB2-SGS1, pCLB2-MMS4 and pCLB2-SGS1 pCLB2-MMS4 cells (Figure 3B), but a significant subset of the DSBs in pCLB2-SGS1 pCLB2-MMS4 cells is repaired with a delay of 2–3 hrs.
Crossovers
In wild type, crossovers plateau at ~20% of chromosomes after 8 hrs (Figure 3A and 3B). The timing and level of crossovers in pCLB2-SGS1 cells are similar to those in wild-type cells, consistent with previous studies that utilized both the pCLB2-SGS1 conditional allele and the sgs1-ΔC795 truncation mutation (Jessop et al., 2006; Mullen et al., 2000; Oh et al., 2007). In pCLB2-MMS4 cells, the maximum crossover level (15%) reaches ~75% of the wild-type level (Figure 3A and 3B and Supplemental Figure S1). Previous analysis of an mms4Δ null mutation reported that crossing-over at HIS4LEU2 reached only ~56% of wild-type levels (de Los Santos et al., 2003). The discrepancy may be explained by a difference in the efficiency of meiotic divisions in pCLB2-MMS4 (97% efficient) and mms4Δ (~75% efficient) strains. We suggest that cells that fail to complete meiotic divisions in mms4Δ cultures also fail to initiate or complete recombination. When this is taken into account, the difference between crossover levels in pCLB2-MMS4 and mms4Δ strains becomes negligible. Crossovers in the pCLB2-SGS1 pCLB2-MMS4 double mutant show a general delay of ~2.0 hrs relative to wild type but, like the pCLB2-MMS4 single mutant, reach ~75% of wild-type levels (Figure 3A, 3B and Supplemental Figure S1). In Supplemental Figure S2, we show that noncrossover products also form at high levels in pCLB2-SGS1, pCLB2-MMS4 and pCLB2-SGS1 pCLB2-MMS4 strains. A similar pattern of product formation is observed for pCLB2-SGS1 mus81Δ cells (Jessop and Lichten, in press).
pCLB2-SGS1 pCLB2-MMS4 mutants accumulate high levels of joint molecules
Four types of JM were monitored using 2D gels: SEIs, interhomolog dHJs (IH-dHJs), intersister JMs (IS-JMs; presumed to be dHJs) and mcJMs (Figure 2 and Figure 3; Supplemental Figure S1 shows analysis of a second set of time-course experiments). Consistent with our previous study of the sgs1-ΔC795 mutant (Oh et al., 2007), pCLB2-SGS1 cells form high levels of IS-JMs and mcJMs (Figure 3C and 3D). pCLB2-MMS4 cells also appear to form slightly higher than normal levels of IS-JMs and mcJMs and JM disappearance is generally delayed (Figure 3D and Supplemental Figure S3). Strikingly, the pCLB2-SGS1 pCLB2-MMS4 double mutant forms very high levels of all JM types, which also persist for several hours (Figure 3C and 3D; Supplemental Figure S1). The ratio of JM types is very similar to that of pCLB2-SGS1 cells, with high fractions of IS-JMs and mcJMs. JM levels are increased by 1.3- to 6-fold relative to wild type and peak between 7–8 hrs, that is, when these cells are attempting to segregate their chromosomes. The logical conclusion is that unresolved JMs physically impede chromosome segregation in pCLB2-SGS1 pCLB2-MMS4 mutants. The same conclusion is made in an accompanying study (Jessop and Lichten, in press). Late persistence of JMs in the pCLB2-MMS4 single mutant suggests a similar, though much less severe, defect may also occur in this mutant (Supplemental Figure S3). Moreover, in both pCLB2-SGS1 and sgs1ΔC795 mutants, JM resolution is also slightly delayed and low levels of JMs are detected at very late times, although this defect appears to be less severe than in pCLB2-MMS4 cells (Figure 3D; Supplemental Figure S3; Oh et al., 2007). Thus, unresolved JMs may also account for segregation defects detected in pCLB2-SGS1 cells.
pCLB2-SGS1 and pCLB2-MMS4 mutations cause early defects in JM formation and late defects in JM resolution
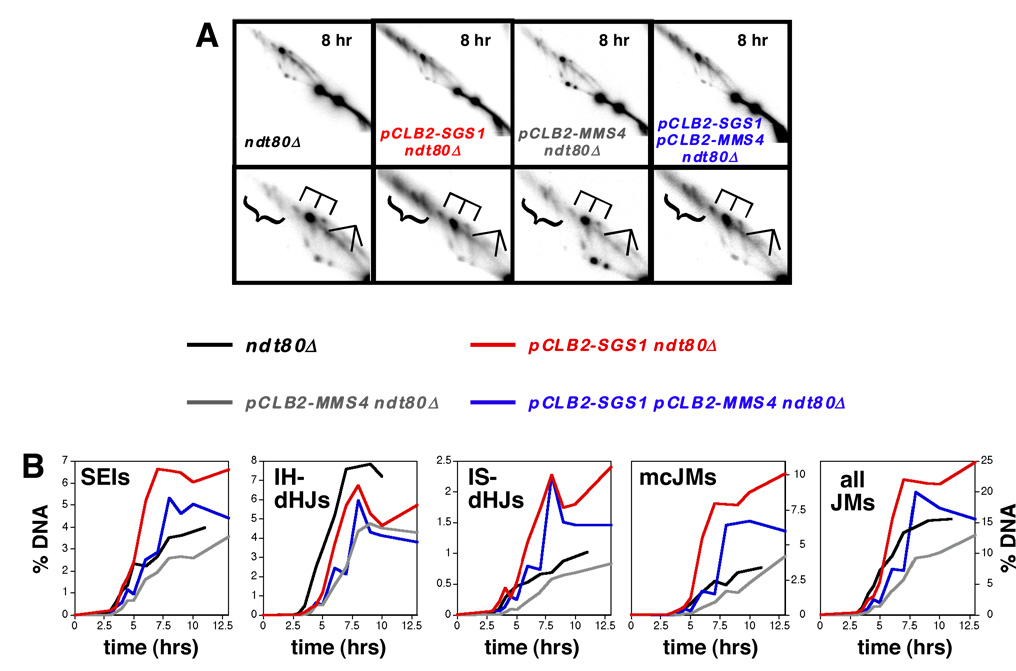
In theory, the high levels of JMs detected in pCLB2-SGS1 pCLB2-MMS4 cells could result from a defect in JM resolution or simply from greater JM formation. To distinguish these possibilities, we measured JM levels in the ndt80Δ background in which unresolved JMs accumulate (Allers and Lichten, 2001a). Mirroring the situation in pCLB2-SGS1 single mutants, pCLB2-SGS1 ndt80Δ cells accumulate higher than normal levels of IS-JMs and mcJMs, but IH-dHJs are slightly reduced relative to wild type (Figure 4A and 4B). These data are consistent with our previous analysis of the sgs1ΔC795 ndt80Δ mutant (Oh et al., 2007). As also noted previously, SEI-like signals accumulate in ndt80Δ cells and these species accumulate to ~1.5-fold higher levels in pCLB2-SGS1 ndt80Δ cells. Below we show that these SEI-like signals contain novel JM species that are distinct from SEIs formed at early times. Overall, ~37% more chromosomes are engaged in JMs in pCLB2-SGS1 ndt80Δ cells than in the ndt80Δ single mutant. These data underscore our proposal that Sgs1 functions to limit unproductive JM formation during meiotic prophase (Oh et al., 2007).
Figure 4. Physical Analysis of Recombination In The ndt80Δ Background.
(A) Representative images from 2D analysis of ndt80Δ, pCLB2-SGS1 ndt80Δ, pCLB2-MMS4 ndt80Δ and pCLB2-SGS1 pCLB2-MMS4 ndt80Δ strains. Lower panels show blowups of the JM regions.
(B) Quantitative analysis of JM formation.
In pCLB2-MMS4 ndt80Δ cells, total JM levels are ~17% lower than in the ndt80Δ single mutant, due primarily to a ~40% decrease in IH-dHJ levels (Figure 4A and 4B), i.e. Mms4 appears to specifically promote formation of IH-dHJs, as inferred previously (de Los Santos et al., 2003). This phenotype likely accounts for the reduced crossing-over in pCLB-MMS4 cells.
In the pCLB2-SGS1 pCLB2-MMS4 ndt80Δ triple mutant, the JM spectrum is most similar to that of pCLB2-SGS1 ndt80Δ cells, with higher than normal levels of SEIs, IS-JMs and mcJMs, but decreased IH-dHJs (Figure 4A and 4B). Notably, total JM levels in pCLB2-SGS1 pCLB2-MMS4 ndt80Δ cells are not higher than in pCLB2-SGS1 ndt80Δ, and are only slightly higher than in the ndt80Δ single mutant (Figure 4B).
Overall, we infer that Sgs1 and Mms4 have distinct roles in JM formation prior to the Ndt80-dependent late-prophase transition. However, segregation defects detected in pCLB2-SGS1, pCLB2-MMS4 and pCLB2-SGS1 pCLB2-MMS4 cells appear to reflect post-Ndt80 defects in JM resolution.
sHJs and dHJs with short inter-junction distances accumulate at late times in pCLB2- SGS1 pCLB2-MMS4 cells
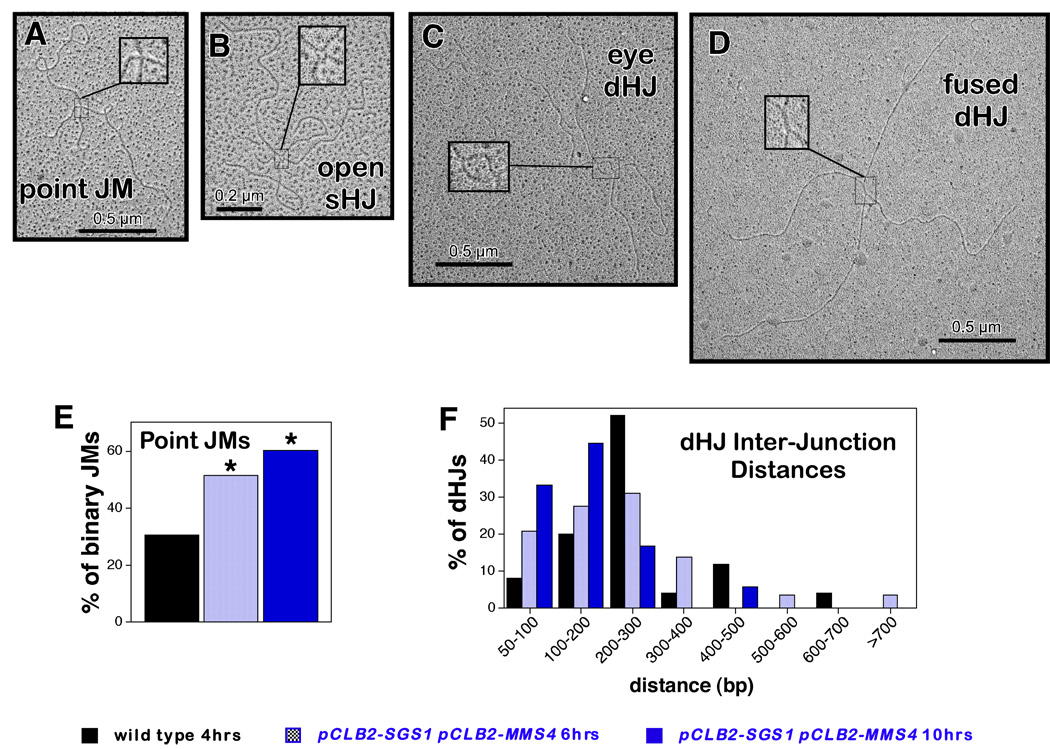
S. pombe mus81 mutants accumulate primarily single-HJ (sHJ) intermediates and undergo meiotic catastrophe analogous to S. cerevisiae pCLB2-SGS1 pCLB2-MMS4 double mutants (Boddy et al., 2001; Cromie et al., 2006). This similarity prompted us to ask whether S. cerevisiae pCLB2-SGS1 pCLB2-MMS4 mutants also accumulate sHJs or other unusual structures. To this end, JMs were purified from 2D gels and examined by electron microscopy (Cromie et al., 2006; Oh et al., 2007). Four types of JM were identified by EM: Y structures, mcJMs, two-chromatid JMs with “eye” junctions or “fused” junctions (presumed dHJs), and two-chromatid JMs connected at a single point (Figure 5A, 5C, 5D and data not shown; Oh et al., 2007). The latter “point JMs” are potentially sHJs, but could also be nicked-HJs, hemicatenanes or dHJs with less than ~50 bp of heteroduplex between the two HJs. However, under partially denaturing conditions, point junctions occasionally adopt an open center configuration (Figure 5B) unambiguously identifying them as sHJs; note that this analysis cannot rule out the presence of a nick adjacent to the junction point. Random sampling of EM grids reveals significantly higher frequencies of point junctions in pCLB2-SGS1 pCLB2-MMS4 samples when compared to wild type (Figure 5E). As shown previously, point junctions are the minority JM type (14/46 two-chromatid JMs) in wild-type cells sampled at 4 hrs, when JM levels peak. In contrast, point junctions comprised ~51% (36/70) of two-chromatid JMs in pCLB2-SGS1 pCLB2-MMS4 cells sampled at 6 hrs, just before JMs reach their peak level. This level rose to ~60% (38/63) in a 10 hr pCLB2-SGS1 pCLB2-MMS4 sample, when JMs levels are decreasing; moreover, 4/14 point junctions from the same sample showed open centers when prepared under partially denaturing conditions, identifying them as sHJs, (not shown).
Figure 5. Analysis of JM Structures by Electron Microscopy.
(A–D) JM structures identified by EM. Insets show magnified images of each junction structure.
(E) Percentage of two-chromatid (binary) JMs that are Point JMs in wild-type and pCLB2-SGS1 pCLB2-MMS4 samples. Asterisks indicate that the distribution of Point JMs and dHJs is different to the wild-type distribution by χ2 analysis (P≤0.05).
(F) Distributions of inter-junction distances amongst dHJs in wild-type and pCLB2-SGS1 pCLB2-MMS4 JM samples. Inter-junction distances were measured and converted to base pairs as described previously (Cromie et al., 2006). JMs were then sorted into 100 bp bins and the distributions for the different samples compared by G-test analysis. The distribution of inter-junction distances is different between dHJs from the 10 hr pCLB2-SGS1 pCLB2-MMS4 sample (n=18 dHJs) and dHJs from the wild-type sample (n=25; P≤0.05).
In addition, amongst dHJ structures (eye and fused dHJs, Figure 5C and 5D), the mean distance (± standard error) between the two junction points was smaller in the 10 hr pCLB2-SGS1 pCLB2-MMS4 sample when compared to wild type (155±25 bp versus 250±25 bp; the distributions of inter-junction distances are compared in Figure 5F). Thus, pCLB2-SGS1 pCLB2-MMS4 cells accumulate sHJs and closely-spaced dHJs suggesting that such structures are prevented form forming and/or resolved by Sgs1 and Mus81-Mms4.
JMs with recombinant arms are also enriched at late times in pCLB2-SGS1 pCLB2-MMS4 cells
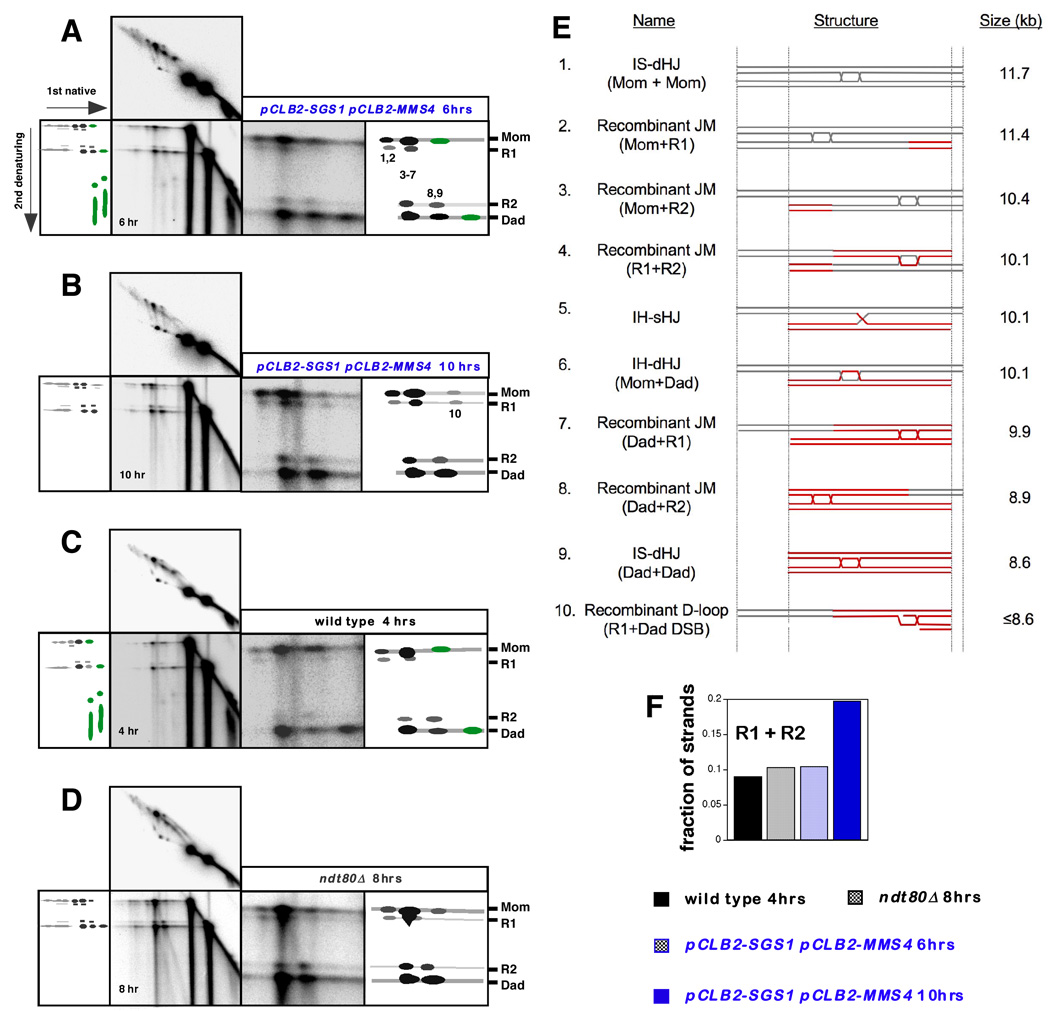
To confirm the inference that sHJs accumulate in pCLB2-SGS1 pCLB2-MMS4 cells, the strand composition of JMs formed at HIS4LEU2 was analyzed using native/denaturing 2D gels, in which psoralen crosslinks are removed prior to running a second dimension gel under denaturing conditions (Figure 6). Interhomolog dHJs contain two HJs and thus the component strands are all parental length. In contrast, sHJs are comprised of both parental and crossover length strands (Cromie et al., 2006; Schwacha and Kleckner, 1995; see Introduction). Interhomolog-JMs in wild-type S. cerevisiae cells were previously shown to contain ~90% parental strands, consistent with their assignment as dHJs (Schwacha and Kleckner, 1995). We were surprised, therefore, to detect prominent crossover-length strands in multiple JM species from wild-type, ndt80Δ and pCLB2-SGS1 pCLB2-MMS4 cells (Figure 6A–D). Notably, JMs that migrate at a position overlapping, but slightly larger than, that of Dad-Dad IS-JMs contain only the shorter of the two crossover strands (species 8,9 in Figure 6A). Oppositely, JMs predicted to be slightly smaller than Mom-Mom IS-JMs contain only the longer crossover strand (species 1,2). Moreover, the reciprocal crossover strands detected at the position of IH-dHJs are slightly offset from the major parental signals and from one another (species 3–7). The presence of interhomolog sHJs does not fully explain this pattern and the existence of two additional JM species can be inferred: one slightly smaller than the IH-dHJs/IH-sHJs containing the longer of the two recombinant strands; and one slightly larger containing the smaller recombinant strand.
Figure 6. Identification and Analysis of Recombinant Joint Molecules.
(A–D) “Pullapart” analysis of the component strands of JMs. In each case, a full image of the native/denaturing 2D blot is shown together with a blowup of the region of interest, to the right. The corresponding native/native 2D panel is shown above to orient native JM species with their component strands. Interpretative cartoons show the positions of the species of interest. Numbers 1–10 correspond to the species detailed in (E) and indicate JM migration positions in the first dimension. Species colored green were previously identified as the component strands of SEIs (Hunter and Kleckner, 2001).
(A) Analysis of a 6 hr sample from the pCLB2-SGS1 pCLB2-MMS4 time-course experiment shown in Figure S1.
(B) Analysis of a 10 hr sample from a pCLB2-SGS1 pCLB2-MMS4 time-course experiment shown in Figure S1. Note the absence of SEI component strands.
(C) Analysis of a 4 hr sample from the wild-type time-course experiment shown in Figure 3.
(D) Analysis of an 8 hr sample from the ndt80Δ time-course experiment shown in Figure 4.
(E) Predicted structures, sizes and DNA strand composition of the joint molecule species analyzed in panels A through D. The fully recombinant dHJ (#4) is the same size as an IH-dHJ (#6) and an IH-sHJ (#5). The recombinant D-loop structure (#10) reconciles the size and strand composition of corresponding JM species highlighted in panel B.
(F) Quantitation of the fraction of crossover-length strands in the JMs analyzed in panels A through D.
Structures that reconcile the size and strand composition of detected JMs are shown in Figure 6E. These include novel recombinant-JM (rJM) structures comprising two chromatids, one or both of which has undergone crossing-over (Figure 6E; species 2, 3, 7 and 8). Such aberrant intersister JMs could arise when the interhomolog-dHJs of a three- and four-chromatid mcJMs are resolved as crossovers, but the intersister-junction (which could be sHJ or dHJ) remains unresolved (see Supplemental Figure S4). Alternatively, an rJM would arise if an IS-JM formed as a secondary event after interhomolog crossing-over had already occurred. Consistent with the latter possibility, we also detect a distinct rJM species (species 10; Figure 6B) that is smaller than the smallest two-chromatid JM (the Dad+Dad intersister-JM). This could be the result of a secondary strand-invasion by a single DSB-end into a crossover duplex to form a “recombinant D-loop” as shown in Figure 6E (species 10). It seems likely that additional, less discrete D-loop species (either recombinant or non-recombinant) also form at late times in pCLB2-SGS1 pCLB-MMS4 and ndt80Δ cells resulting in the smeared SEI-like signals detected in these cells (see the native/native 2D panels of Figure 6B and 6D).
Overall, crossover strands account for ~10% of all JM strands in wild-type and ndt80Δ cells sampled at a time when JMs are at their peak levels (4 hr and 8 hr respectively) (Figure 6F). Given that rJMs are predicted to contain equal numbers of recombinant and parental strands (Figure 6E), we infer that ~20% of all JMs at the HIS4LEU2 locus are rJMs. Thus, JM signals previously interpreted to contain only nonrecombinant IS-JMs and IH-dHJs also contain significant levels of rJMs. In a pCLB2-SGS1 pCLB2-MMS4 sample taken at 6 hrs, when JMs are still accumulating, rJMs also account for ~20% of all JMs. However, at 10 hrs when JMs are turning over, rJMs represent ~40% of total JMs. This enrichment of rJMs at late times in pCLB2-SGS1 pCLB-MMS4 cells is consistent with the proposals that they arise as a consequence of mcJM resolution and/or as secondary intermediates subsequent to crossover formation (above). We can also infer that Sgs1 and Mus81-Mms4 facilitate their resolution.
Discussion
Delayed resolution of JMs impedes meiotic chromosome segregation
The logical interpretation of our data is that JMs that persist beyond metaphase-I mechanically impede chromosome segregation. This same defect was previously inferred for fission yeast mus81/eme1 mutants (Boddy et al., 2001; Cromie et al., 2006), which accumulate sHJs and undergo meiotic catastrophe analogous to that described for budding yeast sgs1 mms4 and sgs1 mus81 double mutants (this study; Jessop and Lichten, in press).
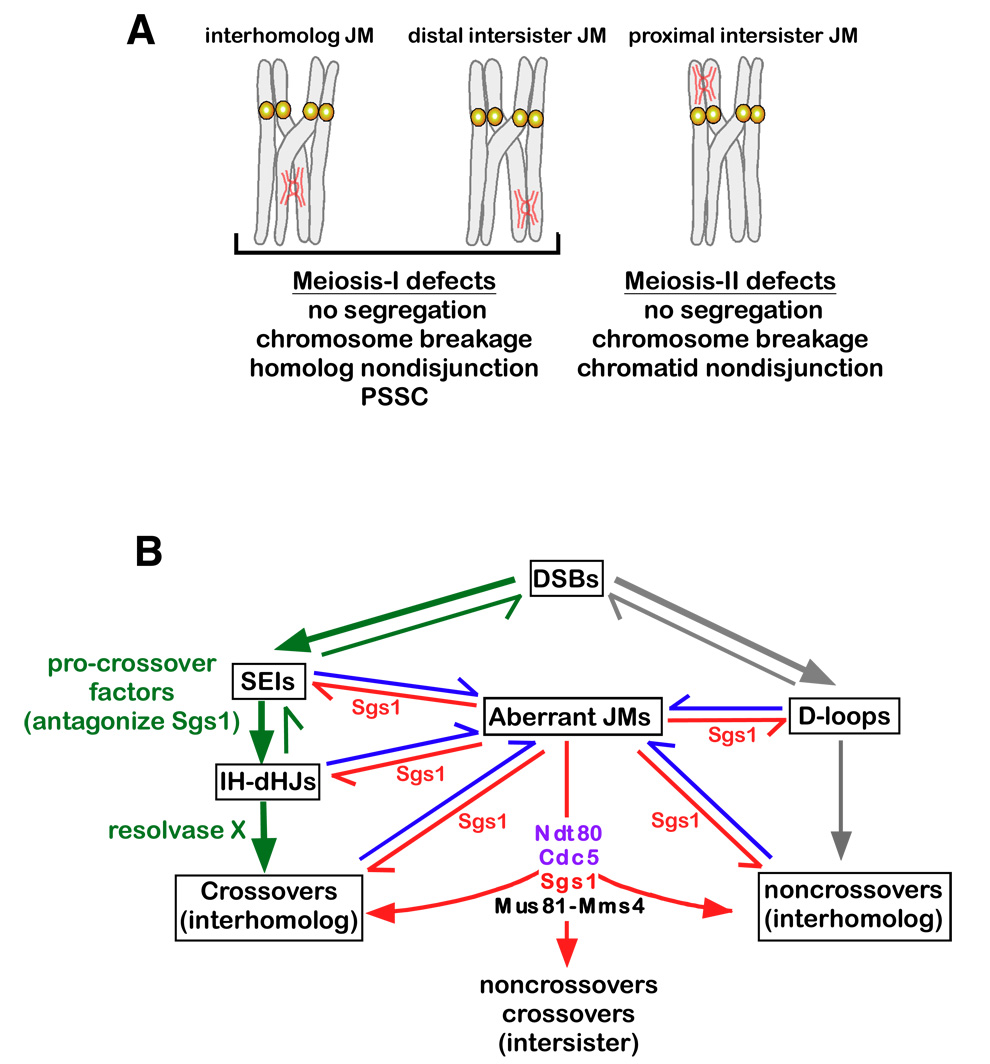
Both IH-JMs and IS-JMs will impede chromosome segregation, but in distinct ways (Figure 7A). An unresolved IH-JM at any location along the chromosomes will hinder homolog disjunction directly. An IS-JM located at a crossover-distal locus will also impede homolog disjunction, but in this case by preventing complete resolution of the chiasma, which normally occurs when arm cohesion is cleaved by separase (Buonomo et al., 2000). Crossover proximal IS-JMs, however, pose no obvious block to MI disjunction, but will impede sister-chromatid disjunction at MII. Unresolved JMs could prevent the segregation of homolog pairs or could lead to mechanical breakage of chromosomes. Alternatively, chromosome missegregation could occur if sister-centromeres separated at MI instead of MII. In fact, Rockmill et al. (2006) showed that such “precocious separation of sister-chromatids” (PSSC) is a major cause of spore inviability in both wild-type cells and sgs1 mutants. Moreover, PSSC is often associated with centromere proximal crossing over, which is proposed to destabilize sister-centromere cohesion (Rockmill et al., 2006). Extending this model, we suggest that unresolved JMs and centromere-proximal crossovers could conspire to provoke PSCC (Figure 7A).
Figure 7. Sgs1 and Mus81-Mms4 Facilitate Interhomolog Recombination and Homolog Segreation by Resolving Aberrant JMs.
(A) Unresolved JMs between homologs and sister-chromatids will hinder chromosome segregation. See text for details. PSSC, precocious segregation of sister-chromatids. “No segregation” means failure to move chromosomes from the metaphase plane.
(B) Model of Sgs1 and Mus81-Mms4 during meiosis. DSBs designated a crossover fate form SEIs and IH-dHJs, which are promoted and stabilized by the ZMM meiotic procrossover factors (green arrows)(Borner et al., 2004; Jessop et al., 2006; Oh et al., 2007). Crossover-designated IH-dHJs are resolved primarily by an unidentified resolvase. Other DSBs proceed to noncrossover products via transient D-loop formation and synthesis-dependent strand-annealing (grey arrows)(McMahill et al., 2007). Formation of aberrant JMs (IS-JMs, mcJMs and rJMs containing sHJs and closely-spaced dHJs) occurs via secondary strand-invasion events (blue arrows), which are antagonized by Sgs1 (red arrows)(Oh et al., 2007). Aberrant JMs that escape Sgs1-mediated disassembly, or that can’t be processed by Sgs1, are resolved by Mus81-Mms4. See text for further details.
The defects described here for meiotic cells could also account for the lethality of sgs1 mms4/mus81 mutants in vegetative cells. Il et al. (2007) recently showed that sgs1 mus81 mutants arrest with a 2n DNA content and a G2 cellular morphology. Whether these cells subsequently undergo mitotic catastrophe has not been reported, however, and it’s possible that these cells undergo regulatory arrest in response to replication problems.
Do meiotic cells sense and respond to unresolved JMs?
Failure to resolve JMs in a timely fashion does not cause arrest of meiotic progression in pCLB2-SGS1, pCLB2-MMS4 or pCLB2-SGS1 pCLB2-MMS4 mutants. This raises the possibility that there is no mechanism to sense unresolved JMs or, more intriguingly, that Sgs1 and/or Mus81-Mms4 are the sensors of such lesions. Notably, meiotic divisions are slightly delayed in both mms4 and sgs1 cultures (Figure 3; Supplemental Figure S1; Oh et al. 2007; de Los Santos et al. 2003). We suggest that cells that progress beyond the Ndt80-dependent transition (exit from the pachytene stage) with persistent JMs are incapable of eliciting a robust regulatory arrest, but may be able to delay the onset of MI to allow more time for JMs to be resolved.
Mus81-Mms4 has both early and late functions in JM metabolism and is essential for normal meiosis in most cells
Cells with unsegregated DNA often contain fewer than four spores and fail to form mature asci. Moreover, even when four spores are present, unsegregated DNA appears to interfere with ascus maturation (data not shown). These phenomena explain the severe sporulation phenotypes previously described for mus81/mms4 cells, which form ~10% mature asci of which only ~10% produce four viable spores (de Los Santos et al., 2003; de los Santos et al., 2001), and indicate that Mus81-Mms4 is required for normal meiosis in most cells.
Mus81-Mms4 appears to be required for the formation or stabilization of a subset of IH-JMs (Figure 4)(de Los Santos et al., 2003). This early function of Mus81-Mms4 likely accounts for the reduced crossing-over in mus81/mms4 mutants. How Mus81-Mms4 facilitates JM formation is unclear. It was previously suggested that Mus81-Mms4 could cleave 3’-flaps that might otherwise hinder dHJ formation (de Los Santos et al., 2003). Alternatively, Mus81-Mms4 could resolve aberrant JMs to liberate DSB-ends that can undergo a second attempt at dHJ formation. Finally, Mus81-Mms4 could somehow antagonize the JM disassembly activity of Sgs1 and/or other resolving enzymes.
Mus81-Mms4 also functions after meiotic prophase to facilitate JM resolution. This conclusion is supported by the finding that induction of MUS81 during mid-prophase in pCLB2-SGS1 mus81Δ cells leads to a marked reduction of JMs (Jessop and Lichten, in press). Putative in vivo substrates for Mus81-Mms4 are the sHJs and closely-spaced dHJs that accumulate in pCLB2-SGS1 pCLB2-MMS4 (Figure 5). However, this inference is not reconciled with the preferred in vitro substrates of recombinant Mus81-Mms4 (see Introduction). It is possible that Mus81-Mms4 activity is modified during meiosis such that it can catalyze HJ resolution directly. Alternatively, Mus81-Mms4 could resolve HJs in coordination with other factors, e.g. endonucleases and/or helicases that could present the enzyme with a preferred substrate such as a nicked-HJ. Another possibility is that Mus81-Mms4 acts indirectly by facilitating a second unidentified HJ resolving enzyme. We propose that the late resolution role of Mus81-Mms4 does not directly contribute to interhomolog crossover levels because it functions primarily to resolve JMs that involve sister-chromatids (IS-JMs, mcJMs and rJMs).
High levels of crossovers and noncrossovers form in pCLB2-SGS1 pCLB2-MMS4 and pCLB2-SGS1 mus81Δ cells (this study; Jessop and Lichten, in press). It is clear, therefore, that meiotic budding yeast cells contain an additional JM-resolving activity that promotes the majority of interhomolog crossing-over. This activity could be equivalent to the “Resolvase X” activity characterized in human cell extracts (Constantinou et al., 2002; Figure 7B).
Sgs1 functions during meiotic prophase to limit the formation of aberrant JMs
The data presented here echo our previous study, which showed that Sgs1 limits the formation of aberrant JMs that arise from secondary strand-invasion events (Oh et al, 2007). We further show that pCLB2-SGS1 cells have a detectable defect in JM resolution and frequently experience chromosome segregation defects. We can infer that sHJs and closely-spaced dHJs result when Sgs1 is absent. Closely-spaced dHJs could reflect the failure to unwind short and normally labile D-loop intermediates and the subsequent capture of a second DSB-end. Similarly, sHJ formation could ensue if the displaced strand of a persistent D-loop intermediate is cleaved as proposed by Cromie et al. (2006). It is currently unclear whether Sgs1 acts primarily by unwinding early strand-exchange intermediates (D-loops) or by “dissolving” dHJs as part of the previously identified Sgs1-Top3-Rmi1 ternary complex (Chang et al., 2005; Mullen et al., 2005; van Brabant et al., 2000; Wu et al., 2006; Wu and Hickson, 2003). Analysis of top3 mutants may shed light on this question.
The functional relationship between the Sgs1 complex and Mus81-Mms4
The distinct phenotypes of pCLB2-SGS1 and pCLB2-MMS4 mutants argue against simple redundancy between the corresponding JM processing activities. Instead, available data argue that Sgs1 limits the formation of aberrant JMs that would subsequently require Mus81-Mms4 for their normal resolution (Figure 7B). This includes the observation that induction of SGS1 during mid-prophase in pCLB2-SGS1 mus81Δ cells prevents further accumulation of JMs (Jessop and Lichten, in press). Aberrant JMs include mcJMs, rJMs and IS-JMs, all of which may include sHJs or closely-spaced dHJs. Within the context of this model, the fact that aberrant JMs are not peculiar to mutant situations, being detected at low levels in wild-type cells (Figure 3–Figure 6), explains why Mus81-Mms4 is essential for meiosis in most cells. When Sgs1 is absent, the high levels of aberrant JMs that result may overload the resolution capabilities of Mus81-Mms4.
We further suggest that disassembly of aberrant JMs by Sgs1 is the preferred resolution mode during meiotic prophase because it always results in a noncrossover outcome thereby limiting unregulated crossing-over (Figure 7B) (Oh et al., 2007). Mus81-Mms4 catalyzed JM resolution may be limited to post-pachytene cells where it would function as an important backup mechanism to resolve aberrant JMs that have escaped Sgs1 or that Sgs1 is incapable of disassembling, e.g. sHJs and nicked-HJs.
Ndt80-dependent and -independent JM resolution
The identification of rJMs and their enrichment at late times in pCLB2-SGS1 pCLB2-MMS4 cells supports our proposal that Sgs1 and Mus81-Mms4 are important for resolving aberrant JMs formed by secondary strand-invasion events (Oh et al., 2007). Since rJMs include crossover-length molecules, equivalent levels of the reciprocal crossover products should accompany their formation (see Supplemental Figure S4). Given that ~20% of JMs in ndt80Δ cells are rJMs it follows that comparable levels of crossover products should also be detected in this background. In fact, crossovers form at ~40% of the wild-type level at the HIS4LEU2 locus in ndt80Δ cells (data not shown)(Xu et al., 1995). This level of crossing-over (~8% of chromosomes) is more than twice the estimated level of rJMs in ndt80Δ cells (~3% of chromosomes). Thus, a substantial fraction of crossover-designated JMs appear to be resolved independently of Ndt80 and Cdc5. It is possible that activation of the resolving enzyme is only partially dependent on phosphorylation by Cdc5. Alternatively, Cdc5-dependent and independent resolving enzymes may be present in the cell. Our observation that accumulated JM levels in pCLB2-SGS1 pCLB2-MMS4 ndt80Δ cells are no higher than in pCLB2-SGS1 ndt80Δ cells indicates that Mus81-Mms4 acts after the Ndt80-dependent transition out of prophase to facilitate JM resolution. This inference raises the possibility that Mus81-Mms4 is activated via Cdc5-mediated phosphorylation.
Experimental Procedures
Yeast strains
Strains are described in Supplementary Table 1). The HIS4LEU2 locus has been described (Cha et al., 2000; Oh et al., 2007). The promoters of SGS1 and MMS4 were replaced with the CLB2 promoter using the pFA6a-KANMX6-pCLB2-3HA cassette as described (Lee and Amon, 2003). The spo11-Y135F and ndt80Δ mutations have been described (Cha et al., 2000; Oh et al., 2007).
Meiotic time courses and DNA physical assays
Meiotic time courses were essentially as described by Goyon and Lichten (1993). DNA physical assays were performed as described (Borner et al., 2004; Hunter and Kleckner, 2001; Schwacha and Kleckner, 1995).
Light Microscopy
To analyze the timing and efficiency of MI, MII and sporulation, cells were fixed in 40% ethanol 0.1M sorbitol, stained with DAPI and ~200 cells were categorized for each time point (Padmore et al., 1991). Unsegregated DNA and the number of spores per cell were analyzed by mounting DAPI-stained cells in anti-fade (Vectashield®, Vector Laboratories, Inc.) and capturing digital images of ~140 cells from the 13hr and 24hr samples of each meiotic culture using a Zeiss AxioPlan II microscope, Hamamatsu ORCA-ER CCD camera and OpenLab software.
Electron Microscopy
DNA was isolated from 2D gels and analyzed by EM as described in Cromie et al. (2006).
Supplementary Material
Acknowledgements
We thank Lea Jessop and Michael Lichten for communicating unpublished data, stimulating discussions and helpful suggestions. Steve Brill, Wolf Heyer and Akira Shinohara provided constructive suggestions on the manuscript. This work was supported by grants GM074223 and GM031693 from NIH and CF0406 from the Concern Foundation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allers T, Lichten M. Differential timing and control of noncrossover and crossover recombination during meiosis. Cell. 2001a;106:47–57. doi: 10.1016/s0092-8674(01)00416-0. [DOI] [PubMed] [Google Scholar]
- Allers T, Lichten M. Intermediates of yeast meiotic recombination contain heteroduplex DNA. Mol Cell. 2001b;8:225–231. doi: 10.1016/s1097-2765(01)00280-5. [DOI] [PubMed] [Google Scholar]
- Bastin-Shanower SA, Fricke WM, Mullen JR, Brill SJ. The mechanism of Mus81-Mms4 cleavage site selection distinguishes it from the homologous endonuclease Rad1–Rad10. Mol Cell Biol. 2003;23:3487–3496. doi: 10.1128/MCB.23.10.3487-3496.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell L, Byers B. Separation of branched from linear DNA by two-dimensional gel electrophoresis. Analyt Biochem. 1983;130:527–535. doi: 10.1016/0003-2697(83)90628-0. [DOI] [PubMed] [Google Scholar]
- Bergerat A, de Massy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- Bishop DK, Zickler D. Early decision; meiotic crossover interference prior to stable strand exchange and synapsis. Cell. 2004;117:9–15. doi: 10.1016/s0092-8674(04)00297-1. [DOI] [PubMed] [Google Scholar]
- Boddy MN, Gaillard PH, McDonald WH, Shanahan P, Yates JR, 3rd, Russell P. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- Borner GV, Kleckner N, Hunter N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell. 2004;117:29–45. doi: 10.1016/s0092-8674(04)00292-2. [DOI] [PubMed] [Google Scholar]
- Buonomo SB, Clyne RK, Fuchs J, Loidl J, Uhlmann F, Nasmyth K. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell. 2003;103:387–398. doi: 10.1016/s0092-8674(00)00131-8. [DOI] [PubMed] [Google Scholar]
- Cao L, Alani E, Kleckner N. A pathway for generation and processing of double-strand breaks during meiotic recombination in S. cerevisiae. Cell. 1990;61:1089–1101. doi: 10.1016/0092-8674(90)90072-m. [DOI] [PubMed] [Google Scholar]
- Cha RS, Weiner BM, Keeney S, Dekker J, Kleckner N. Progression of meiotic DNA replication is modulated by interchromosomal interaction proteins, negatively by Spo11p and positively by Rec8p. Genes Dev. 2000;14:493–503. [PMC free article] [PubMed] [Google Scholar]
- Chang M, Bellaoui M, Zhang C, Desai R, Morozov P, Delgado-Cruzata L, Rothstein R, Freyer GA, Boone C, Brown GW. RMI1/NCE4, a suppressor of genome instability, encodes a member of the RecQ helicase/Topo III complex. Embo J. 2005;24:2024–2033. doi: 10.1038/sj.emboj.7600684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyne RK, Katis VL, Jessop L, Benjamin KR, Herskowitz I, Lichten M, Nasmyth K. Polo-like kinase Cdc5 promotes chiasmata formation and cosegregation of sister centromeres at meiosis I. Nat Cell Biol. 2003;5:480–485. doi: 10.1038/ncb977. [DOI] [PubMed] [Google Scholar]
- Constantinou A, Chen XB, McGowan CH, West SC. Holliday junction resolution in human cells: two junction endonucleases with distinct substrate specificities. EMBO J. 2002;21:5577–5585. doi: 10.1093/emboj/cdf554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cromie GA, Hyppa RW, Taylor AF, Zakharyevich K, Hunter N, Smith GR. Single Holliday Junctions Are Intermediates of Meiotic Recombination. Cell. 2006;127:1167–1178. doi: 10.1016/j.cell.2006.09.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Los Santos T, Hunter N, Lee C, Larkin B, Loidl J, Hollingsworth NM. The Mus81/Mms4 endonuclease acts independently of double-holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics. 2003;164:81–94. doi: 10.1093/genetics/164.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de los Santos T, Loidl J, Larkin B, Hollingsworth NM. A role for MMS4 in the processing of recombination intermediates during meiosis in Saccharomyces cerevisiae. Genetics. 2001;159:1511–1525. doi: 10.1093/genetics/159.4.1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehmsen K, Heyer WD. Saccharomyces cerevisiae Mus81-Mms4 is a catalytic, DNA structure-selective endonuclease. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm1152. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre F, Chan A, Heyer WD, Gangloff S. Alternate pathways involving Sgs1/Top3, Mus81/ Mms4, and Srs2 prevent formation of toxic recombination intermediates from single-stranded gaps created by DNA replication. Proc Natl Acad Sci U S A. 2002;99:16887–16892. doi: 10.1073/pnas.252652399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fricke WM, Bastin-Shanower SA, Brill SJ. Substrate specificity of the Saccharomyces cerevisiae Mus81-Mms4 endonuclease. DNA Repair (Amst) 2005;4:243–251. doi: 10.1016/j.dnarep.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Gaskell LJ, Osman F, Gilbert RJ, Whitby MC. Mus81 cleavage of Holliday junctions: a failsafe for processing meiotic recombination intermediates? Embo J. 2007;26:1891–1901. doi: 10.1038/sj.emboj.7601645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyon C, Lichten M. Timing of molecular events in meiosis in Saccharomyces cerevisiae: stable heteroduplex DNA is formed late in meiotic prophase. Mol Cell Biol. 1993;13:373–382. doi: 10.1128/mcb.13.1.373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer WD. Recombination: Holliday junction resolution and crossover formation. Curr Biol. 2004;14:R56–R58. doi: 10.1016/j.cub.2003.12.043. [DOI] [PubMed] [Google Scholar]
- Hochwagen A, Amon A. Checking your breaks: surveillance mechanisms of meiotic recombination. Curr Biol. 2006;16:R217–R228. doi: 10.1016/j.cub.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Holliday R. A mechanism for gene conversion in fungi. Genet. Res. 1964;5:282–304. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- Hollingsworth NM, Brill SJ. The Mus81 solution to resolution: generating meiotic crossovers without Holliday junctions. Genes Dev. 2004;18:117–125. doi: 10.1101/gad.1165904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter N. Meiotic Recombination. In: Aguilera A, Rothstein R, editors. Molecular Genetics of Recombination. Heidelberg: Springer-Verlag; 2006. pp. 381–442. [Google Scholar]
- Hunter N, Kleckner N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell. 2001;106:59–70. doi: 10.1016/s0092-8674(01)00430-5. [DOI] [PubMed] [Google Scholar]
- Interthal H, Heyer WD. MUS81 encodes a novel helix-hairpin-helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol Gen Genet. 2000;263:812–827. doi: 10.1007/s004380000241. [DOI] [PubMed] [Google Scholar]
- Jessop L, Rockmill B, Roeder GS, Lichten M. Meiotic Chromosome Synapsis-Promoting Proteins Antagonize the Anti-Crossover Activity of Sgs1. PLoS Genet. 2006;2 doi: 10.1371/journal.pgen.0020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessop L, Lichten M. Mus81/Mms4 endonuclease and Sgs1 helicase collaborate to ensure proper recombination intermediate metabolism during meiosis. Mol Cell. 2008 doi: 10.1016/j.molcel.2008.05.021. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaliraman V, Mullen JR, Fricke WM, Bastin-Shanower SA, Brill SJ. Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 2001;15:2730–2740. doi: 10.1101/gad.932201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao JP, Oh SD, Shinohara M, Shinohara A, Hunter N. Rad52 promotes post-invasion steps of meiotic double-strand-break repair. Mol Cell. 2008;29:517–524. doi: 10.1016/j.molcel.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee BH, Amon A. Role of Polo-like kinase CDC5 in programming meiosis I chromosome segregation. Science. 2003;300:482–486. doi: 10.1126/science.1081846. [DOI] [PubMed] [Google Scholar]
- Ii M, Ii T, Brill SJ. Mus81 functions in the quality control of replication forks at the rDNA and is involved in the maintenance of rDNA repeat number in Saccharomyces cerevisiae. Mutat Res. 2007;625:1–19. doi: 10.1016/j.mrfmmm.2007.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahill MS, Sham CW, Bishop DK. Synthesis-dependent strand annealing in meiosis. PLoS Biol. 2007;5:e299. doi: 10.1371/journal.pbio.0050299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JR, Kaliraman V, Brill SJ. Bipartite structure of the SGS1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2000;154:1101–1114. doi: 10.1093/genetics/154.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JR, Kaliraman V, Ibrahim SS, Brill SJ. Requirement for three novel protein complexes in the absence of the Sgs1 DNA helicase in Saccharomyces cerevisiae. Genetics. 2001;157:103–118. doi: 10.1093/genetics/157.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullen JR, Nallaseth FS, Lan YQ, Slagle CE, Brill SJ. Yeast Rmi1/Nce4 controls genome stability as a subunit of the Sgs1-Top3 complex. Mol Cell Biol. 2005;25:4476–4487. doi: 10.1128/MCB.25.11.4476-4487.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassif N, Penney J, Pal S, Engels WR, Gloor GB. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol Cell Biol. 1994;14:1613–1625. doi: 10.1128/mcb.14.3.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SD, Lao JP, Hwang PY, Taylor AF, Smith GR, Hunter N. BLM ortholog, Sgs1, prevents aberrant crossing-over by suppressing formation of multichromatid joint molecules. Cell. 2007;130:259–272. doi: 10.1016/j.cell.2007.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmore R, Cao L, Kleckner N. Temporal comparison of recombination and synaptonemal complex formation during meiosis in S. cerevisiae. Cell. 1991;66:1239–1256. doi: 10.1016/0092-8674(91)90046-2. [DOI] [PubMed] [Google Scholar]
- Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockmill B, Voelkel-Meiman K, Roeder GS. Centromere-proximal crossovers are associated with precocious separation of sister chromatids during meiosis in Saccharomyces cerevisiae. Genetics. 2006;174:1745–1754. doi: 10.1534/genetics.106.058933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell. 1995;83:783–791. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- Shinohara A, Shinohara M. Roles of RecA homologues Rad51 and Dmc1 during meiotic recombination. Cytogenet Genome Res. 2004;107:201–207. doi: 10.1159/000080598. [DOI] [PubMed] [Google Scholar]
- Smith GR, Boddy MN, Shanahan P, Russell P. Fission yeast Mus81.Eme1 Holliday junction resolvase is required for meiotic crossing over but not for gene conversion. Genetics. 2003;165:2289–2293. doi: 10.1093/genetics/165.4.2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Taylor ER, McGowan CH. Cleavage mechanism of human Mus81-Eme1 acting on Holliday-junction structures. Proc Natl Acad Sci U S A. 2008;105:3757–3762. doi: 10.1073/pnas.0710291105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Brabant AJ, Ye T, Sanz M, German IJ, Ellis NA, Holloman WK. Binding and melting of D-loops by the Bloom syndrome helicase. Biochemistry. 2000;39:14617–14625. doi: 10.1021/bi0018640. [DOI] [PubMed] [Google Scholar]
- Whitby MC. Hollday junction resolution. In: Aguilera A, Rothstein R, editors. Molecular Genetics of Recombination. Heidelberg: Springer-Verlag; 2006. pp. 169–199. [Google Scholar]
- Wu L, Bachrati CZ, Ou J, Xu C, Yin J, Chang M, Wang W, Li L, Brown GW, Hickson ID. BLAP75/RMI1 promotes the BLM-dependent dissolution of homologous recombination intermediates. Proc Natl Acad Sci U S A. 2006;103:4068–4073. doi: 10.1073/pnas.0508295103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L, Hickson ID. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- Xu L, Ajimura M, Padmore R, Klein C, Kleckner N. NDT80, a meiosis-specific gene required for exit from pachytene in Saccharomyces cerevisiae. Mol Cell Biol. 1995;15:6572–6581. doi: 10.1128/mcb.15.12.6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.