Abstract
Somatic hypermutation (SHM) of Ig genes depends upon the deamination of C nucleotides in WRCY (W=A/T, R=A/G, Y=C/T) motifs by activation-induced cytidine deaminase (AICDA). Despite this, a large number of mutations occur in WA motifs that can be accounted for by the activity of polymerase eta (POL η). To determine whether there are AICDA-independent mutations and to characterize the relationship between AICDA- and POL η-mediated mutations, 1,470 H chain and 1,313 kappa and lambda chain rearrangements from three AICDA−/− patients were analyzed. The Ig mutation frequency of all VH genes from AICDA−/− patients was 40-fold less than that of normal donors whereas the mutation frequency of mutated VH sequences from AICDA−/− patients was 6.8-fold less than normal donors. AICDA−/− B cells lack mutations in WRCY/RGYW motifs as well as replacement mutations and mutational targeting in complementarity determining regions. A significantly reduced mutation frequency in WA motifs compared to normal donors and an increased percentage of transitions, which may relate to reduced uracil DNA-glycosylase (UNG) activity, suggest a role for AICDA in regulating POL η and UNG activity. Similar results were observed in VL rearrangements. The residual mutations were predominantly G:C substitutions, indicating that AICDA-independent cytidine deamination was a likely, yet inefficient, mechanism for mutating Ig genes.
Keywords: human, activation induced cytidine deaminase, somatic hypermutation, polymerase eta, uracil-DNA glycosylase
Introduction
Expression of B cell specific AICDA in germinal center (GC) B cells is required for generating SHM of Ig genes and affinity maturation of antibodies during an immune response(1, 2). AICDA deficient B cells can undergo a GC reaction but fail to accumulate mutations in the Ig variable regions following immunization(3). The lack of mutation may contribute to shifts in the heavy and light chain repertoires(4). Although AICDA is critical for SHM, other mechanisms contribute to the overall mutational pattern (5) and some mutations may occur independent of AICDA(1, 3). However, the nature of the mutations in the absence of AICDA and the relationship between AICDA induced mutations and those owing to the activity of other mechanisms, such as error-prone polymerase eta (η), are not known.
AICDA preferentially deaminates Cs in the WRC motif (W=A/T, R=A/G) in both DNA strands(6-9)(N.S.L. and P.E.L., manuscript in preparation) and is directly associated with generating the G/C mutation spectrum. The targeting mechanism of AICDA and certain features of Ig heavy chain gene CDR, such as the presence of overlapping targeted motifs, are associated with favoring CDR as opposed to framework region (FR) mutations and also replacement (R) substitutions that impact the antigen binding site (8, 10) and enable affinity maturation.
The A/T mutation spectrum is associated with the activity of error-prone POL η that preferentially targets A residues in the WA motif. Linkage between AICDA and POL η activity was suggested by a detailed analysis of mutation patterning showing that mutations of POL η-targeted WA motifs were increased when these motifs overlapped AICDA-targeted RGYW motifs.(N.S.L. and P.E.L., manuscript in preparation) This pattern of mutations suggested the AICDA-induced U:G lesions may foster additional mutation in adjacent residues through the recruitment of a WA/TW motif mutator, such as POL η. (N.S.L. and P.E.L., manuscript in preparation) Consistent with this mechanism, POL η deficiency results in a loss of mutations in WA motifs in AICDA-targeted WRCY/RGYW motifs(11). However, since POL η deficiency also results in a loss of WA mutations remote from RGYW/WRCY motifs, it is possible that this error-prone POL might also have an AICDA-independent function.
In order to characterize the mutational pattern that occurs in the absence of AICDA and to explore the relationship between AICDA and POL η in more detail and to characterize the mutational pattern that occurs in the absence of AICDA, we analyzed the H chain mutations from three AICDA deficient patients. We focused on mutations in nonproductive rearrangements that are not influenced by selection to identify mutational mechanisms. There was a significant reduction in the frequency of mutations, but still significantly more than could be anticipated in genes of non-B cells. Most of the mutations could be attributed to non-targeted AICDA-independent cytidine deamination resulting in a loss of targeting of mutations to CDRs. The mutation pattern in AICDA deficiency was biased toward transitions and lacked mutations in POL η-targeted WA motifs, indicating that AICDA is necessary for the recruitment of POL η and also UNG during SHM.
Materials and Methods
Patient samples and cell preparation
Three AICDA deficient patients donated peripheral blood samples after signing informed consent forms in accordance with Institutional Review Board approved protocols. The control samples(12-14) and aicda mutations and immunoglobulin levels for each patient (P4 age 11, P5 age 7 and P6 age 21) have been described (3). Blood mononuclear cells were isolated on Ficoll gradients and stained with mouse IgG1 anti-human CD19 PE (Becton Dickinson), mouse IgG2 anti-human IgD FITC (Pharmingen) or goat anti-human IgD FITC (Caltag), and anti-CD27-PE and isotype controls (Becton Dickinson Pharmingen, San Diego, CA). Patient mononuclear cells were sorted into CD19+ or CD19+CD27− and CD19+CD27+ subsets using a Becton Dickinson FACS Vantage SE DiVa or a MoFlo Cell Sorter (DAKO). Sorted populations were 94−98% pure.
Single cell PCR
Aliquots of approximately 1 cell were diluted in buffer (10mM NaCl, 5mM Tris-HCl pH 8 at 25°C, 0.1% Triton X-100) containing 0.4mg/ml proteinase K (Sigma Chemical Co., St. Louis) and genomic DNA from individual cell lysates was linearly amplified using random 15 mers (Qiagen, Valencia, CA). Aliquots of the preamplification product were used as template for internal and external nested amplification of the transcribed strand using VH3, VH4 and VH1 heavy chain-specific upstream primers and downstream JH-specific primers as described previously(14). The AICDA−/− light chains were analyzed with Vκ1, Vκ2, Vκ3 and Vλ1, Vλ2 and Vλ3 specific primers as described previously (15, 16).
Sequencing Analysis
PCR products were separated by electrophoresis on 1.2% agarose gels, purified (Edge Biosystems, Gaithersburg, MD) and directly sequenced using the ABI Prism Big Dye Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA). PCR products were directly sequenced using on an automated capillary sequencer (ABI Prism 3100 Genetic Analyzer; Applied Biosystems, Foster City, CA).
Database
The analysis included 105 nonproductive and 636 productive rearrangements from normal donors that included CD19+IgM+ B cells from accession numbers X87006-X87082, CD19+IgM+CD5+ and CD19+IgM+CD5− B cells from accession numbers Z80363-Z80770(12-14), IgD+CD27+ B cells from accession numbers EF542547-EF542687 and IgD−CD27+ B cells from accession numbers EF542688-EF542796. The AICDA deficient analysis included 320 nonproductive and 1150 productive CD19+CD27− and CD19+CD27+ VHDJH rearrangements with accession numbers (EU237493-EU238970), 294 nonproductive and 482 productive kappa rearrangements, and 142 nonproductive and 395 productive lambda rearrangements to be submitted to Gen Bank.
Mutation analysis
Nonproductive and productive VHDJH rearrangements were determined by using the JOINSOLVER® sequence and mutation analysis program(17). Enumeration of nucleotides used an algorithm that takes into consideration the finding that some base pair positions in the germline genes reside within more than one motif and/or within ambiguous motifs (ie, AGCT is an RGYW and WRCY motif; TA is a WA and TW motif). This approach avoided a subjective bias in motif nucleotide designation, but also was the source of non-integer values.
Seven new polymorphisms within codons 25−94 in 6 VH genes were excluded from the mutation analysis.(N.S.L. and P.E.L., manuscript in preparation) The following 12 substitutions in light chain genes (IMGT numbering for codons in brackets) appeared to be new polymorphisms and were excluded from the mutation analysis: IGKV1−16*01 [75], IGKV1D-17*01 [26], IGKV1D-17*01 [52], IGKV1−5*03 [57], IGKV1−9*01 [86], IGKV1−9*01 [90], IGKV1−8*01 [77], IGKV1−27*01 [94], IGKV1−12*01 [24], IGLV l3−21*01 [108] and IGLV 6−57*01 [49].
Statistical Analysis
The χ2 test and the p< 0.05 limit were used to determine statistically significant differences. In Table 3 and Figure 2A, the ratio of nucleotide mutational targeting inside the motif vs mutation of the nucleotide outside the motif was used to compare differences between AICDA and normal donors.
Table III.
Target nucleotide mutation frequency (%) in hypermutable motifs in nonproductive VH rearrangements
| Mutated Base | Motif | AICDA−/− | Normal Donor |
|---|---|---|---|
| G |
inside RGYW | 2/2374† | 94/937* |
| 0.08% | 10.0% | ||
| all other G |
18/13016† | 99/6093 | |
| 0.14% |
1.6% |
||
| C |
inside WRCY | 0/2140† | 62/832* |
| 0% | 7.5% | ||
| all other C |
7/13596† | 75/5685 | |
| 0.05% |
1.3% |
||
| A |
inside WA | 4.5/8273† | 115.9/2473* |
| 0.05% | 4.7% | ||
| all other A |
6.5/8564† | 79.1/3615 | |
| 0.08% |
2.2% |
||
| T | inside TW | 1.5/5180† | 56.5/1550* |
| 0.03% | 3.6% | ||
| all other T | 3.5/8288† | 45.5/3714 | |
| 0.04% | 1.2% |
The targeted nucleotide refers to the base in the underlined position (or two positions in WA where W=A or T). The mutation frequency of the targeted nucleotide inside a motif was calculated from the number of target nucleotide mutations divided by the total number of the motif (or total number of A in WA motifs). To calculate the mutation frequency outside the motif, the total number of base mutations in the sequence minus the number of targeted nucleotide mutations was divided by the total number of the base in the sequence minus the number of target nucleotides inside the motif. The extent of targeting was determined by calculating the fold difference between the mutation frequency inside a motif vs all other positions.
Significant (p<0.01) difference between the nucleotide mutation frequency inside a motif vs all other positions
significant (p<.0001) difference between AICDA−/− and normals.
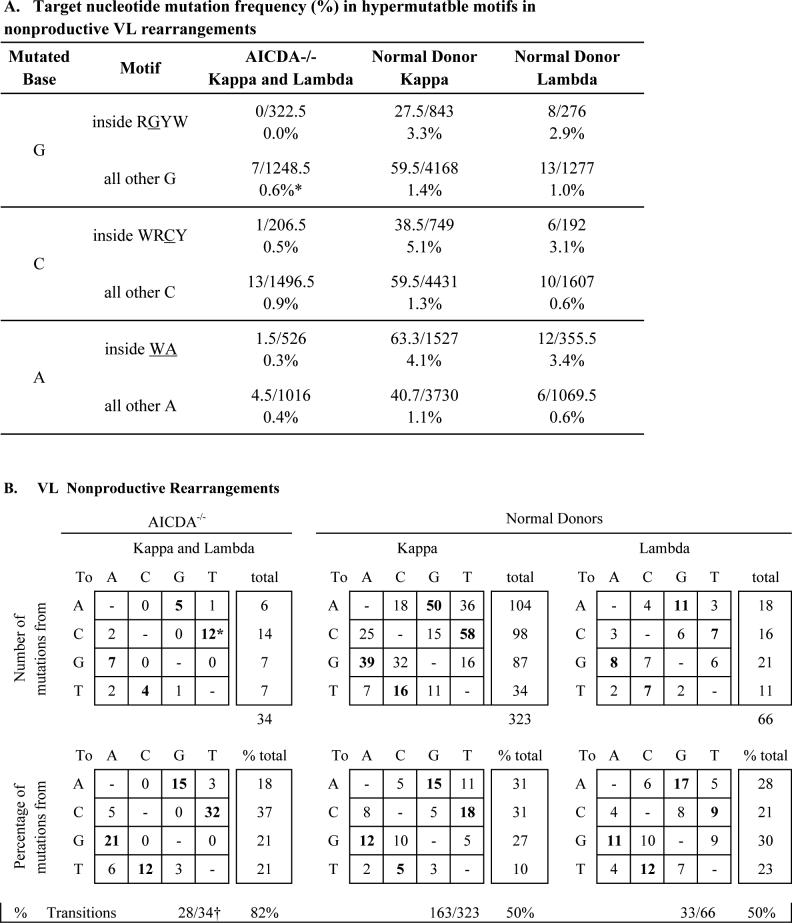
Figure 2. The nucleotide mutation pattern in nonproductive light chain rearrangements in AICDA−/− B cells.
The light chain data is collected from AICDA−/− patients P4 and P5 and consists of 18 mutations from 294 nonproductive kappa rearrangements and 16 mutations from 142 nonproductive lambda rearrangements. (A) Individual nucleotide substitutions in the light chain repertoires are presented in which the percentage of all mutations (lower panels) are scored after correction for base composition of the target sequence. (B) The mutation frequency of a specific nucleotide inside vs outside a hypermutable motif was calculated as shown in Table 3. *Significant (p=.02) difference between AICDA−/− and normal donors; †significant (p<.0001) difference between transitions and transversions and significant (p=.0008) difference in transitions between AICDA−/− and normal donor.
Calculation of the PCR error rate
The PCR error of this technique was initially demonstrated to be 1×10−4 by amplifying an unmutated germline Ig sequence 96 times using the same amplification and sequencing conditions employed here(18). The low PCR error rate was confirmed by amplifying ckit genes from B cells (0.8×10−4, 6/72,261) (19). Finally, unrearranged VH family gene segments were amplified and sequenced from CD19− peripheral blood cells. Analysis of 96 VH4 segments from CD19 negative cells demonstrated a PCR error rate of 0.76 × 10−4 (2/25,986)
Results
Decreased mutation frequency of B cells from AICDA−/− patients
B cell phenotyping and mutation analysis confirmed previous reports (3) that AICDA deficient patients have a normal percentage of CD19+CD27+ B cells but unlike normal donors in which CD27+ expression correlates with a mutated memory B cell population, the AICDA−/− CD27+ B cells are infrequently mutated. In this study, 20−28% of the peripheral blood B cells from AICDA−/− patients were CD27+ (data not shown). Both subsets of CD19+CD27+ and CD19+CD27− B cells contained mutated sequences. However, there was no significant difference in the percentage of mutated sequences or the mutation frequency between the CD27− and CD27+ subsets. Since CD27 expression did not correlate with mutation in AICDA−/− B cells and CD27− human memory cells have been identified,(20) the subsets were combined for the mutation analysis. The percentages of mutated VHDJH sequences in AICDA−/− nonproductive (NP) and productive (P) repertoires (12% and 9%, respectively) were significantly (p<.0001) less than normal donors (71−75%) (Table I). The overall mutation frequency was 40-fold less than normals, whereas the mutation frequency of mutated sequences was only 6.8-fold less than normals. Most of the mutated nonproductive AICDA−/− sequences had a unique, single nucleotide substitution, except four sequences that contained two mutations. In the mutated productive repertoire, 100/104 (96%) rearrangements had 1−2 mutations, 3 sequences had 3 mutations and one sequence had 4 mutations. These mutations clearly were not polymorphisms and occurred at approximately a 10 fold higher frequency than the PCR error for this method (6 mutations/75,182bp; 8×10−5) (19) or 2/25,986 (0.76 × 10−4) determined for VH4 sequences in non-B cells.
Table I.
Mutation frequency of VH segment of AICDA deficient B cells
| AICDA−/− |
Normal Donors |
|||
|---|---|---|---|---|
| NP | P | NP | P | |
| Total number of Rearrangements | 320 | 1150 | 105 | 636 |
| Total number of Mutations | 43 | 124 | 627 | 4864 |
| Percentage of Sequences Mutated | 39/320* | 104/1150* | 76/105 | 479/636 |
| 12% | 9% | 71% | 75% | |
| Mutation Range | 1−2 | 1−4 | 1−33 | 1−38 |
| Mutation Frequency Overall | 43/64081* | 124/230514* | 627/22693 | 4864/152977 |
| 0.07% | 0.05% | 2.8% | 3.2% | |
| Mutation Frequency of Mutated | 43/7799* | 124/19938* | 627/15149 | 4864/113434 |
| Rearrangements | 0.6% | 0.6% | 4.1% | 4.3% |
NP = nonproductive rearrangements, P = productive rearrangements. The AICDA−/− data includes CD19+CD27− and CD19+CD27+ sequences from three patients. The control data comes from previous reports of 4 individuals and three different experiments that collected IgM+, IgM+CD5+, IgM+CD5−, IgD+CD27− and IgD−CD27+ B cells which represent normal unmutated and mutated populations in peripheral blood. The mutation frequencies were calculated from the number of mutations/total number of bases analyzed or from the total number of bases analyzed from mutated sequences only.
Significant (p<.0001) difference between AICDA−/− and normal donors.
Ig hypermutable motifs are not mutated in AICDA−/− B cells
Because mutations do occur in Ig genes in the absence of AICDA, we next characterized the frequency of mutation in Ig hypermutable motifs, particularly those in the nonproductive rearrangements that are not influenced by positive and negative selection. An increased focus of mutations in any of the four bases constituting WRCY (W=A/T, R=A/G, Y=C/T) and the inverse complementary motif, RGYW, are the hallmarks of AICDA-dependent somatic hypermutation (SHM) (21, 22). In the nonproductive repertoire, the percentage of mutations that occurred in WRCY motifs was significantly (p=0.004) reduced compared to normals (3% vs 16%) (Table II). Similarly, the frequency of mutations in RGYW (10%) was no greater than that expected from random chance (12.5%) and significantly less than in normal controls (24%, p=0.03). The frequency of mutations in either motif (13%) was also significantly (p= 0.01) less than in normal B cells (40%). These motifs become hot spots for hypermutation primarily because of AICDA targeting C in WRCY on both strands of DNA (7, 9, 23). In the AICDA deficient B cells, the hypermutable nucleotides (C in WRCY and G in RGYW), which represents an AICDA attack on the opposite strand) were not targeted for mutation more frequently than mutation of C or G residues anywhere else in the heavy chain variable segment (Table III), consistent with a complete loss of AICDA activity in these subjects. By comparison, there was nearly 4-fold more targeting of these specific nucleotides inside WRCY/RGYW motifs in normal B cells. A similar loss of G/C mutational targeting was noted in the AICDA−/− L chain rearrangements (Fig. 2A). These results indicate that the preferential targeting of the C in WRCY (and G in RGYW) is completely dependent on AICDA activity and provides the opportunity to examine the AICDA dependence of other known components of the SHM machinery.
Table II.
Percentage of mutations in RGYW/WRCY and WA/TW motifs.
| AICDA−/− |
Normal Donors |
|||
|---|---|---|---|---|
| NP | P | NP | P | |
| WRCY | 1.5/43* | 13.8/124† | 98.1/627† | 756.7/4864† |
| 3% | 11% | 16% | 16% | |
| RGYW | 4.5/43* | 30.3/124 | 152.4/627 | 1285.9/4864 |
| 10% | 24% | 24% | 26% | |
| WA | 4.5/43* | 25.5/124‡ | 140.4/627‡ | 888.5/4864‡ |
| 10% | 21% | 22% | 18% | |
| TW | 1.5/43 | 9/124 | 73.7/627 | 490.5/4864 |
| 4% | 7% | 12% | 10% | |
The enumeration of nucleotides was adjusted for nucleotides that occupy a position in more than one motif or ambiguous motifs. This calculation therefore, can assign a fraction of a mutation for a given motif.
Significant (p≤.05) difference between AICDA−/− and ND
significant (p<.008) difference between WRCY and RGYW
significant (p<.009) difference between WA and TW.
Loss of polymerase eta (POL η) mutational targeting in AICDA−/− B cells
There is substantial evidence that error-prone POL η plays a significant role in SHM (11, 24, 25). Analysis of Ig gene mutational patterns from individuals deficient in POL η has indicated that this polymerase specifically targets the A in WA motifs,(11, 26) and preferentially targets the TA dinucleotide (N.S.L. and P.E.L., manuscript in preparation). More recently, evidence of WA mutations overlapping RGYW motifs suggested that POL η may be recruited to the site of the AICDA-induced lesion during mismatch repair (MMR) in normal human B cells. In support of the human data, transgenic mice with SHM substrates acquired the capacity to acquire A/T mutations only when the substrate contained an AICDA-targeted cytosine(27). To examine the role of POL η in the absence of AICDA, we examined mutations in WA motifs in the H chain genes of the AICDA−/− B cells. WA motif mutations (10%) were no more frequent than that expected from random mutation (12.5%) and were significantly less frequent than WA motif mutations found in normal B cells (Table II). Notably, mutations of the hypermutable A in WA motifs were no more frequent than A mutations elsewhere in the variable segment, indicating that POL η activity was greatly reduced (Table III). Similar results were noted with L chain rearrangements (Fig. 2A).
Despite reduced targeting in all hypermutable motifs, mutations in RGYW motifs appeared to be positively selected as indicated by the increased mutation frequency in the VH productive compared to nonproductive repertoires (Table II). A similar trend was observed with the other motifs, but significant differences were not found. Therefore, some form of selection is intact, although this is not evident in normals in which a large number of mutations have been analyzed, presumably because large numbers of mutations of Ig genes may disable the Ig molecule and contribute to B cell loss.
Mutations in AICDA−/− B cells are not associated with a new consensus mutation motif
Although the known hypermutable motifs were not targeted in AICDA−/− B cells, there was the possibility that another motif was targeted. To address this question, the Ig sequences were divided into oligomers each of which contained a mutated nucleotide from the AICDA−/− B cells in the context of five nucleotides flanking both sides of the mutation. The sequences were interrogated for shared motifs by using the web based Genio/logo program (http://www.biogenio.com/logo/logo.cgi). This procedure failed to identify a motif within which residual mutations occurred (data not shown). Other potential patterns of mutational activity were investigated, such as UV–induced dipyrimidine substitutions, or oxidative damage generating mutations of guanine dinucleotides, but no pattern with a mutation frequency greater than normal was found (data not shown). We concluded that the major mutation mechanism in AICDA−/− was a random event involving individual Ig gene nucleotides.
Mutational targeting of CDRs is decreased in AICDA deficiency
In normal nonproductive VH rearrangements, the frequency of mutation in the CDRs in VH genes is twofold greater than the frequency of mutations in the FRs after normalization for the number of nucleotides analyzed in each region (Table IV) (8). By contrast, in the AICDA deficient VH rearrangements the preferential mutation targeting of CDRs was lost. These data indicate that AICDA activity is essential for targeting of mutations to CDRs. Enrichment of mutations is normally observed in CDR regions in the productive repertoire when productive and nonproductive VH rearrangements are compared (Table IV). However, in the AICDA−/− rearrangements an increased frequency of CDR mutations and a decrease in FR mutations was noted in the productive repertoire and as a result, the ratio of mutations in CDRs versus FRs was normalized, presumably because of positive selection of CDR mutations and negative selection of FR mutations.
Table IV.
The effect of AICDA deficiency on amino acid and nucleotide substitutions in CDRs and FRs of heavy chain rearrangements.
| AICDA−/− |
Normal Donors |
|||
|---|---|---|---|---|
| NP | P | NP | P | |
| CDR R:S | 3/1† | 41/2* | 170/32† | 1383/391 |
| 3.0 | 20.5 | 5.3 | 3.5 | |
| FR R:S | 24/15* | 54/27 | 329/96† | 1940/1150 |
| 1.6 | 2.0 | 3.4 | 1.7 | |
| CDR Mutation Frequency | 4/11393*† | 43/40699* | 198/4826† | 1741/29250 |
| 3.5 × 10−4 | 1 × 10−3 | 4.1 × 10−2 | 6.0 × 10−2 | |
| FR Mutation Frequency | 39/52688*† | 81/189815* | 426/20244† | 3092/120907 |
| 7.4 × 10−4 | 0.4 × 10−3 | 2.1 × 10−2 | 2.6 × 10−2 | |
| Mutation Frequency CDR:FR | 3.5:7.4*† | 1:0.4 | 4.1:2.1 | 6.0:2.6 |
| 0.5 | 2.5 | 2.0 | 2.3 | |
The FR:CDR mutation frequency ratio was normalized to correct for the number of nucleotides analyzed in FRs and CDRs. The analysis excludes the CDR3 and the first 25 codons of FR1.
Significant (p=.03) difference between AICDA−/− and normal donors
significant (p<.007) difference between NP and P rearrangements.
The ratio of replacement:silent mutations (R/S) is 5.3 in normal donor nonproductive CDRs compared to the 3.4 in FRs (Table IV). By comparison, the R/S ratio anticipated by random chance is 4.5 in CDRs and 3.2 in FRs because of codon bias. In normal donors, the R/S ratios of both the CDRs and the FRs in the nonproductive repertoire were 1.5 to 2-fold greater than those of the productive rearrangements, suggesting that large numbers of R mutation in both regions result in the loss of cells, as previously reported(8). In the absence of AICDA, R substitutions in the nonproductive repertoire were less common in both CDRs and FRs than in normal rearrangements. Notably, R substitutions appeared to be positively selected when occurring in CDRs and not eliminated when occurring in FRs, as evidenced by comparison of R/S ratios in nonproductive and productive rearrangements. Presumably, the very low mutational activity in AICDA−/− B cells introduces fewer deleterious mutations so that elimination of rearrangements with R mutations is not greater than the increase resulting from positive selection.
Mutation of individual nucleotides in AICDA deficiency
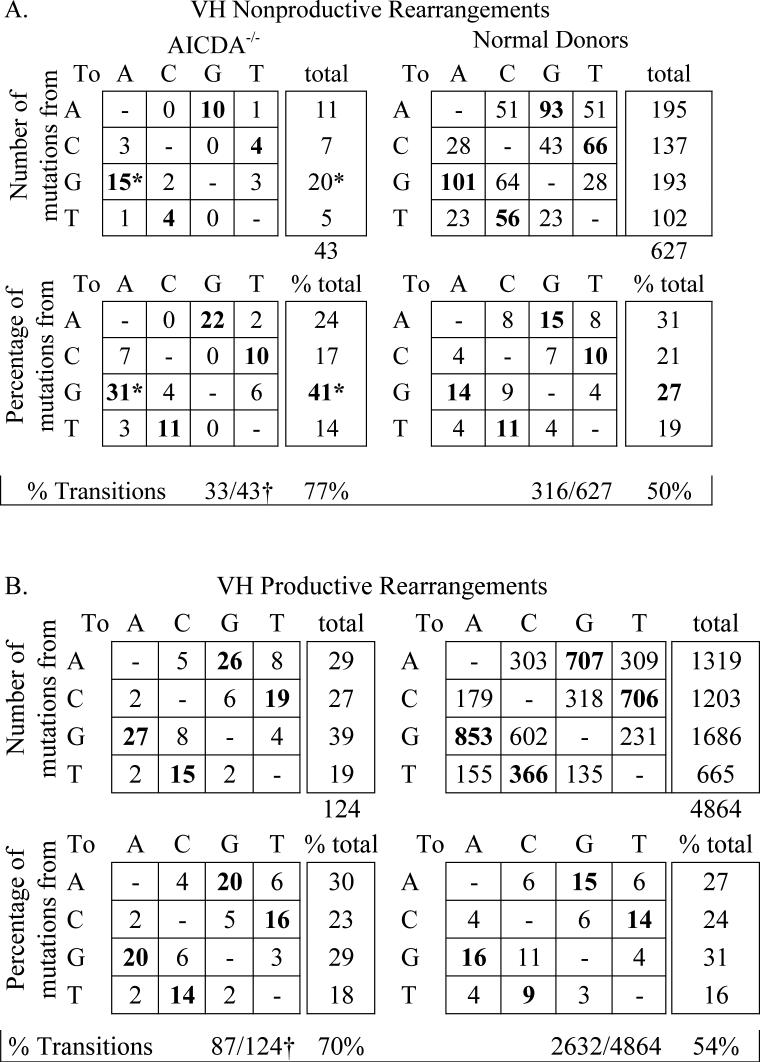
In the nonproductive AICDA deficient repertoire, G residues were the most frequently mutated (41%), followed by A (24%), C (17%) and T residues (14%) compared to A > G > C > T in normal donors (Figure 1). Similar to normals, a bias favoring A mutations more than T mutations was present in AICDA−/− B cells. Although POL η targets A residues more than T, the absence of targeting in WA motifs suggested that another polymerase, such as POL ζ, POLθ, or POLι, eacḥ of which is expressed in human tonsilar B cells, may be contributing to the residual A:T mutations in AICDA−/− B cells(28, 29).
Figure 1. The nucleotide mutation pattern in productive and nonproductive heavy chain rearrangements in AICDA−/− B cells.
The individual nucleotide substitutions detected in the variable segment of (A) nonproductive VHDJH and (B) productive VHDJH rearrangements in peripheral blood B cells from 4 normal donors and 3 AICDA−/− patients are presented. The percentages of all mutations (lower panels in each group) are scored after correction for base composition of the target sequence. Transitions appear in bold face type. *Significant (p=0.03) difference between AICDA−/− and normal donors; †significant (p<0.0001) difference between transitions and transversions and significant (p<0.0008) difference in transitions between AICDA−/− and normal donor.
Of the few mutations in AICDA−/− VH genes, the percentage of C mutation was similar to normal (AICDA−/−, 17% vs normal, 21%,; p=0.39) whereas the percentage of G mutations was significantly increased in AICDA−/− compared to normal (AICDA−/−, 41% vs normal, 27%; p=0.03). This suggested that a major source of mutation in AICDA-deficient cells was a G:C focused mutational mechanism (Figure 1). AICDA-independent cytidine deamination is the most common form of DNA damage in non-B cells and could contribute to the mutation pattern in AICDA−/− B cells(30). Presumably, this could target both the transcribed and nontranscribed strands with a bias toward the latter with little or no involvement of POL η. By comparing the mutation frequency of AICDA−/− B cells to our estimated mutation frequency of non-B cells, it appears that AICDA-independent C deamination in B cells is subjected to less of the normal repair mechanisms operative in non-B cells.
AICDA-independent mutations are biased toward transitions
The individual nucleotide substitutions in the AICDA−/− nonproductive rearrangements were biased toward transitions (77%), compared to normal donors in which transitions and transversions occur at comparable frequencies (Figure 1). The most evident reason for this mutation shift was the significant (p=0.01) increase in G > A transitions and to a lesser extent A > G transitions. Similarly, mutations in nonproductive AICDA-deficient light chain rearrangements were skewed significantly toward transitions (Figure 2B). One of the mechanisms known to contribute to a loss of transversions is UNG deficiency(31). Even though UNG expression is increased in tonsillar GC cells, UNG may be less active or not accessible when AICDA is not available. A similar phenomenon was noted when mutations in X-HIgM B cells which are also characterized by less functional AICDA activity were analyzed. These data suggest that AICDA plays a role not only in the initiation and targeting of SHM, but also the utilization of UNG and POL η that are involved in processing the AICDA-induced lesion.
Discussion
The current study demonstrated that a small number of mutations can develop in B cell Ig genes of individuals who totally lack expression of AICDA. However, in the absence of AICDA, the mutational frequency is markedly reduced and there is a loss of RGYW/WRCY mutations as well as of mutations in POL η targeted WA motifs. In addition, UNG-dependent transversions were markedly reduced. These results suggest that AICDA induces a process that involves POL η and UNG to induce the normal spectrum of SHM. The loss of AICDA results in the absence of a mutational focus on CDRs and a bias toward R mutations in CDRs and, therefore, little ability of SHM to contribute to affinity maturation of the antibody response.
Because the measured mutational frequency was relatively low, it was essential to be certain that PCR error did not confound interpretation of the data. PCR amplification of genomic DNA from individual cells with direct sequencing of the PCR products used here has been documented to have a very low PCR error rate (0.8−1.0 × 10−4). To confirm this, we sequenced unrearranged Ig VH gene segments from CD19− non-B cells and confirmed the very low PCR error rate (0.76 × 10−4). Thus few of the detected base pair changes could be accounted for by PCR error. Moreover, since the majority of the data related to the absence of expected mutational targeting (G in RGYW, C in WRCY, A in WA), it was unlikely that these few base pair changes potentially introduced by the PCR amplification affected the interpretation of the results.
Importantly, the Ig mutation frequency of AICDA−/− B cells was greater than that expected from DNA instability in non-B cells. We directly measured the mutational frequency of unrearranged VH4 genes in non-B cells and determined that it was no more than the PCR error rate. Thus, the mutation frequency of AICDA−/− B cells was nearly 10 fold that of Ig genes in non-B cells. Other approaches have been employed to estimate the mutation frequency of eukaryotic cells which is generally reported to be ≤10−10/base pair/cell division(32) compared to 1 × 10−3/base pair/cell division for Ig genes in B cells(33). Assuming a DNA length of 240 base pairs, equivalent to that of VH gene sequences, and that the cells undergo 25 cell divisions, which would yield a predicted Ig gene mutational frequency of 2.5% (similar to that measured for normal B cells), the AICDA deficient B cells have as much as a 2.8 × 105-fold increase in mutational frequency compared to that of DNA in non-B cells. This estimate does not take into account the increased mutability of Ig genes within B cells, but it clearly is consistent with the current data that the frequency of mutations of Ig genes is significantly greater in AICDA−/− B cells than that of non-Ig genes in B cells or Ig genes in non-B cells. Whether this reflects an increased likelihood of introducing base pair changes or an altered tendency for error prone repair is currently unknown.
During the GC reaction, expression of AICDA and multiple enzymes involved in DNA repair and translesion synthesis is uniquely altered resulting in high frequency mutation with specific complex patterns targeted to Ig genes. A greater understanding of how individual components contribute to SHM in humans has resulted from the analysis of the Ig gene mutational pattern in B cells from individuals with a deficiency in one of the enzymes involved in SHM or the GC reaction itself. In normal B cells, WA mutations occur both when motifs overlap RGYW/WRCY tetrads, as well as when they are distant(11). This pattern has made it difficult to distinguish whether the underlying mutation process targeting WA motifs was related to AICDA(34). The loss of POL η activity in xeroderma pigmentosa variant (XPV) resulted in a reduction in WA mutations regardless of their position inside or outside RGYW/WRCY motifs, indicated that WA mutations in RGYW/WRCY were related to POL η targeting and not to the direct activity of AICDA(11, 35). Thus the loss of POL η targeting in WA motifs in AICDA−/− Ig genes may reflect the observation that pol η is constitutively expressed in B cells and actually downregulated in GCs that are present and accentuated in size in subjects with AICDA deficiency(3). Based upon the current finding that there was a marked reduction of WA targeting in AICDA−/− B cells, it is likely that AICDA may directly or indirectly recruit POL η into a mutation complex. This conclusion is consistent with recent findings that the induction of A/T mutations in a transgenic mutation substrate required a potentially AICDA targeted G/C site(27). However, the current data extend this finding to implicate POL η as the error prone polymerase targeting WA motifs in an AICDA-dependent manner. These findings also support the data from X-HIgM studies suggesting a linkage between POL η targeting WA within the AICDA-induced lesion. Notably, however, the current data indicate that AICDA is also responsible for recruiting POL η activity to sites remote from the AICDA-targeted lesion, possibly through long patch repair(36).
The AICDA-deficient B cells demonstrated a marked skewing toward transitions whereas normal B cells had equal numbers of transitions and transversions. A similar skewing toward transitions was attributed to the absence of UNG in a chicken cell line,(31) mouse,(37) and human B cells(38). Importantly, these results suggest a role for AICDA in recruiting UNG2 to sites of AICDA-dependent uracil generation, perhaps by forming a multiprotein structure with proliferating cell nuclear antigen and replication protein A.
The lack of targeting to WRCY/RGYW motifs and the decrease in CDR mutations as well as R substitutions in the CDRs provide support for the proposal that two intrinsic features of Ig genes may have co-evolved with AICDA to focus the mutation mechanism on the antigen-binding site. First, WRCY/RGYW motifs are more concentrated in CDRs than in the FRs of the VH genes(8). This may bias the mutational machinery initiated by AICDA to induce mutations preferentially into CDRs, as the current data clearly show that in the absence of AICDA, the bias for replacement mutations to localize into CDRs was markedly reduced. The second feature of Ig genes noted in normal B cells to influence mutational patterning is the biased use of codons that favor replacement substitutions. This was highlighted by the increase in the predicted R/S ratio in CDRs (5.2) compared to FRs (3.4) in the current analysis. It is notable that positive selection of R mutations in the CDRs was apparent with the low mutation frequency of AICDA−/− B cells, but absent from the normal mutated B cells that have a markedly higher mutation frequency compared to AICDA−/− mutated B cells. This may be attributed to the increased loss of B cells that accumulate large numbers of mutations either because the R mutations in the CDRs introduce autoreactivity or R mutations in the FRs alter the structural integrity of molecule(8). The very low mutation frequency in AICDA−/− B cells may make it less likely that deleterious mutations are introduced and therefore, more probable that positive selection of mutations can be observed.
The residual mutations in AICDA−/− Ig genes did not appear to be targeted by a new mutator to an identifiable motif. Moreover, the mutations did not exhibit a pattern of targeting within G-G or Y-Y dinucleotides compatible with oxidative damage to DNA. A likely source of the predominant G/C nucleotide mutations was spontaneous cytidine deamination that is common to all cells but apparently occurs more frequently in Ig genes possibly because of the general down regulation of conventional high fidelity repair during a GC reaction. Spontaneous cytidine deamination occurs without any sequence specificity(30) but is strongly dependent on DNA structure. The rate of deamination is 2 orders of magnitude greater for single-stranded DNA than for double-stranded DNA because of cytosine accessibility during replication or transcription(39). Although it is possible that the high rate of B cell proliferation during GC reactions(33) or the level of Ig gene transcription makes spontaneous cytidine deamination more likely in B cells, there is no evidence that this is the case. Alternatively, B cells may be more likely to employ error prone repair mechanisms to correct mutations and thereby be more likely to accumulate G mutations. The nature of such error prone polymerase is currently unknown but the targeting to G/C nucleotides indicates that POL η is not involved, leaving POL θ or POL ζ as likely candidates. In mice it appears that all A/T mutations are attributed to POL η recruitment by MSH2(40). However, A/T mutations remain in the pol η and msh2 single knock-outs when AICDA and UNG are available, suggesting that MSH2 may be capable of recruiting other enzymes if POL η is absent or in the absence of MSH2, error–prone enzymes other than POL η may leave a mild A/T footprint. Thus, the mechanism that accounts for the other 40% of mutations in AICDA deficient B cells remains unknown.
Overall, the mutations that occur in the absence of AICDA are likely to have little impact on the conventional antigen-binding site or the nonvariable FR regions that superantigens bind(41). In spite of the positive selection of mutated B cells that occurs in AICDA−/− subjects, the data suggest the process may not be sufficiently robust to allow B cells to undergo affinity maturation. The lack of affinity maturation and class switching is reflected in the absence of anti-tetanus IgG antibodies in AICDA−/− patients after immunization (3) although detailed affinity measurements of these antibodies has not been carried out.
In conclusion, the absence of AICDA results in a loss of targeting to known AICDA and POL η hypermutable motifs and decreased mutation (and R substitutions) in the CDRs. The current study provides evidence that AICDA not only initiates SHM but is also involved in the recruitment of POL η and possibly, UNG. The mutation pattern in AICDA−/− B cells suggests the G:C mutation spectrum is most likely introduced by AICDA-independent cytidine deamination on both DNA strands and a low level A:T mutation spectrum that is not associated with POL η. Finally, because of the loss of CDR targeting, somatic mutation in AICDA−/− B cells provides an inefficient mechanism for altering the antigen-binding site.
Acknowledgments
We thank Igor Rogozin (National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, Bethesda, MD) for comments and discussions. The authors are grateful for the cooperation of the patients, their families and normal donors for their participation in the study and thank Jim Simone, Flow Cytometry Section, Office of Science and Technology, NIAMS, for cell sorting.
This work was supported in part by the NIH, NIAMS Intramural Research Program and grants from the l'Institut National de la Santé et de la Recherche Médicale (INSERM), l'Association de la Recherche Contre le Cancer (ARC), the European Community n° PL 006411 (EUROPOLICY-PID) – 6th Programme Cadre de Recherche Développment (PCRD), the Association Nationale pour la Recherche (ANR), INCa (Institut National du Cancer) and a grant from The Fondazione C. Golgi and the Associazione Immunodeficienze Primitive, Brescia, Italy.
Nonstandard abbreviations
- AICDA
activation induced cytidine deaminase
- FR
framework region
- MMR
mismatch repair
- POL η
polymerase eta
- R
replacement
- S
silent
- SHM
somatic hypermutation
Footnotes
The submitted manuscript which is cited in the text is titled “Analysis of somatic hypermutation in X-HIgM syndrome reveals specific deficiencies in mutational targeting” by Nancy S. Longo, Patricia L. Lugar, Sule Yavuz, Wen Zhang, Peter H. L. Krijger, Daniel E. Russ, Amrie C. Grammer and Peter E. Lipsky, manuscript in preparation.
Disclosures: The authors declare no competing financial interests.
References
- 1.Muramatsu M, Kinoshita K, Fagarasan S, Yamada S, Shinkai Y, Honjo T. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000;102:553–563. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 2.Milstein C. Diversity and the genesis of high affinity antibodies. Biochem. Soc. Trans. 1987;15:779–787. doi: 10.1042/bst0150779. [DOI] [PubMed] [Google Scholar]
- 3.Revy P, Muto T, Levy Y, Geissmann F, Plebani A, Sanal O, Catalan N, Forveille M, Dufourcq-Labelouse R, Gennery A, Tezcan I, Ersoy F, Kayserili H, Ugazio AG, Brousse N, Muramatsu M, Notarangelo LD, Kinoshita K, Honjo T, Fischer A, Durandy A. Activation-induced cytidine deaminase (AID) deficiency causes the autosomal recessive form of the Hyper-IgM syndrome (HIGM2). Cell. 2000;102:565–575. doi: 10.1016/s0092-8674(00)00079-9. [DOI] [PubMed] [Google Scholar]
- 4.Meffre E, Catalan N, Seltz F, Fischer A, Nussenzweig MC, Durandy A. Somatic hypermutation shapes the antibody repertoire of memory B cells in humans. J. Exp. Med. 2001;194:375–378. doi: 10.1084/jem.194.3.375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barreto VM, Ramiro AR, Nussenzweig MC. Activation-induced deaminase: controversies and open questions. Trends Immunol. 2005;26:90–96. doi: 10.1016/j.it.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 6.Yu K, Huang FT, Lieber MR. DNA substrate length and surrounding sequence affect the activation-induced deaminase activity at cytidine. J. Biol. Chem. 2004;279:6496–6500. doi: 10.1074/jbc.M311616200. [DOI] [PubMed] [Google Scholar]
- 7.Shen HM, Storb U. Activation-induced cytidine deaminase (AID) can target both DNA strands when the DNA is supercoiled. Proc. Natl. Acad. Sci. U. S. A. 2004;101:12997–13002. doi: 10.1073/pnas.0404974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dörner T, Brezinschek HP, Brezinschek RI, Foster SJ, Domiati-Saad R, Lipsky PE. Analysis of the frequency and pattern of somatic mutations within nonproductively rearranged human variable heavy chain genes. J. Immunol. 1997;158:2779–2789. [PubMed] [Google Scholar]
- 9.Xue K, Rada C, Neuberger MS. The in vivo pattern of AID targeting to immunoglobulin switch regions deduced from mutation spectra in msh2−/− ung−/− mice. J. Exp. Med. 2006;203:2085–2094. doi: 10.1084/jem.20061067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milstein C, Neuberger MS, Staden R. Both DNA strands of antibody genes are hypermutation targets. Proc. Natl. Acad. Sci. U. S. A. 1998;95:8791–8794. doi: 10.1073/pnas.95.15.8791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yavuz S, Yavuz AS, Kraemer KH, Lipsky PE. The role of polymerase eta in somatic hypermutation determined by analysis of mutations in a patient with xeroderma pigmentosum variant. J. Immunol. 2002;169:3825–3830. doi: 10.4049/jimmunol.169.7.3825. [DOI] [PubMed] [Google Scholar]
- 12.Brezinschek HP, Brezinschek RI, Lipsky PE. Analysis of the heavy chain repertoire of human peripheral B cells using single-cell polymerase chain reaction. J. Immunol. 1995;155:190–202. [PubMed] [Google Scholar]
- 13.Dörner T, Brezinschek HP, Foster SJ, Brezinschek RI, Farner NL, Lipsky PE. Comparable impact of mutational and selective influences in shaping the expressed repertoire of peripheral IgM+/CD5− and IgM+/CD5+ B cells. Eur. J. Immunol. 1998;28:657–668. doi: 10.1002/(SICI)1521-4141(199802)28:02<657::AID-IMMU657>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 14.Brezinschek HP, Foster SJ, Brezinschek RI, Dörner T, Domiati-Saad R, Lipsky PE. Analysis of the human VH gene repertoire. Differential effects of selection and somatic hypermutation on human peripheral CD5(+)/IgM+ and CD5(-)/IgM+ B cells. J. Clin. Invest. 1997;99:2488–2501. doi: 10.1172/JCI119433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Monson NL, Dorner T, Lipsky PE. Targeting and selection of mutations in human Vlambda rearrangements. Eur. J. Immunol. 2000;30:1597–1605. doi: 10.1002/1521-4141(200006)30:6<1597::AID-IMMU1597>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 16.Foster SJ, Brezinschek HP, Brezinschek RI, Lipsky PE. Molecular mechanisms and selective influences that shape the kappa gene repertoire of IgM+ B cells. J. Clin. Invest. 1997;99:1614–1627. doi: 10.1172/JCI119324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Souto-Carneiro MM, Longo NS, Russ DE, Sun HW, Lipsky PE. Characterization of the human Ig heavy chain antigen binding complementarity determining region 3 using a newly developed software algorithm, JOINSOLVER. J. Immunol. 2004;172:6790–6802. doi: 10.4049/jimmunol.172.11.6790. [DOI] [PubMed] [Google Scholar]
- 18.Farner NL, Dörner T, Lipsky PE. Molecular mechanisms and selection influence the generation of the human V lambda J lambda repertoire. J. Immunol. 1999;162:2137–2145. [PubMed] [Google Scholar]
- 19.Yavuz AS, Lipsky PE, Yavuz S, Metcalfe DD, Akin C. Evidence for the involvement of a hematopoietic progenitor cell in systemic mastocytosis from single-cell analysis of mutations in the c-kit gene. Blood. 2002;100:661–665. doi: 10.1182/blood-2002-01-0203. [DOI] [PubMed] [Google Scholar]
- 20.Wei C, Anolik J, Cappione A, Zheng B, Pugh-Bernard A, Brooks J, Lee EH, Milner EC, Sanz I. A new population of cells lacking expression of CD27 represents a notable component of the B cell memory compartment in systemic lupus erythematosus. J. Immunol. 2007;178:6624–6633. doi: 10.4049/jimmunol.178.10.6624. [DOI] [PubMed] [Google Scholar]
- 21.Rogozin IB, Kolchanov NA. Somatic hypermutagenesis in immunoglobulin genes. II. Influence of neighbouring base sequences on mutagenesis. Biochim. Biophys. Acta. 1992;1171:11–18. doi: 10.1016/0167-4781(92)90134-l. [DOI] [PubMed] [Google Scholar]
- 22.Dörner T, Foster SJ, Brezinschek HP, Lipsky PE. Analysis of the targeting of the hypermutational machinery and the impact of subsequent selection on the distribution of nucleotide changes in human VHDJH rearrangements. Immunol. Rev. 1998;162:161–171. doi: 10.1111/j.1600-065x.1998.tb01439.x. [DOI] [PubMed] [Google Scholar]
- 23.Dörner T, Foster SJ, Farner NL, Lipsky PE. Somatic hypermutation of human immunoglobulin heavy chain genes: targeting of RGYW motifs on both DNA strands. Eur. J. Immunol. 1998;28:3384–3396. doi: 10.1002/(SICI)1521-4141(199810)28:10<3384::AID-IMMU3384>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 24.Mayorov VI, Rogozin IB, Adkison LR, Gearhart PJ. DNA polymerase eta contributes to strand bias of mutations of A versus T in immunoglobulin genes. J. Immunol. 2005;174:7781–7786. doi: 10.4049/jimmunol.174.12.7781. [DOI] [PubMed] [Google Scholar]
- 25.Wilson TM, Vaisman A, Martomo SA, Sullivan P, Lan L, Hanaoka F, Yasui A, Woodgate R, Gearhart PJ. MSH2-MSH6 stimulates DNA polymerase eta, suggesting a role for A:T mutations in antibody genes. J. Exp. Med. 2005;201:637–645. doi: 10.1084/jem.20042066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rogozin IB, Pavlov YI, Bebenek K, Matsuda T, Kunkel TA. Somatic mutation hotspots correlate with DNA polymerase eta error spectrum. Nat. Immunol. 2001;2:530–536. doi: 10.1038/88732. [DOI] [PubMed] [Google Scholar]
- 27.Unniraman S, Schatz DG. Strand-biased spreading of mutations during somatic hypermutation. Science. 2007;317:1227–1230. doi: 10.1126/science.1145065. [DOI] [PubMed] [Google Scholar]
- 28.Diaz M, Lawrence C. An update on the role of translesion synthesis DNA polymerases in Ig hypermutation. Trends Immunol. 2005;26:215–220. doi: 10.1016/j.it.2005.02.008. [DOI] [PubMed] [Google Scholar]
- 29.Masuda K, Ouchida R, Hikida M, Kurosaki T, Yokoi M, Masutani C, Seki M, Wood RD, Hanaoka F, W. J. O. DNA polymerases eta and theta function in the same genetic pathway to generate mutations at A/T during somatic hypermutation of Ig genes. J. Biol. Chem. 2007;282:17387–17394. doi: 10.1074/jbc.M611849200. [DOI] [PubMed] [Google Scholar]
- 30.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 31.Di Noia J, Neuberger MS. Altering the pathway of immunoglobulin hypermutation by inhibiting uracil-DNA glycosylase. Nature. 2002;419:43–48. doi: 10.1038/nature00981. [DOI] [PubMed] [Google Scholar]
- 32.Kunkel TA, Bebenek K. DNA replication fidelity. Annu. Rev. Biochem. 2000;69:497–529. doi: 10.1146/annurev.biochem.69.1.497. [DOI] [PubMed] [Google Scholar]
- 33.McKean D, Huppi K, Bell M, Staudt L, Gerhard W, Weigert M. Generation of antibody diversity in the immune response of BALB/c mice to influenza virus hemagglutinin. Proc. Natl. Acad. Sci. USA. 1984;81:3180–3184. doi: 10.1073/pnas.81.10.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dörner T, Lipsky PE. Smaller role for pol eta? Nat. Immunol. 2001;2:982–984. doi: 10.1038/ni1101-982. [DOI] [PubMed] [Google Scholar]
- 35.Zeng X, Negrete GA, Kasmer C, Yang WW, Gearhart PJ. Absence of DNA polymerase eta reveals targeting of C mutations on the nontranscribed strand in immunoglobulin switch regions. J. Exp. Med. 2004;199:917–924. doi: 10.1084/jem.20032022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annu. Rev. Biochem. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 37.Rada C, Williams GT, Nilsen H, Barnes DE, Lindahl T, Neuberger MS. Immunoglobulin isotype switching is inhibited and somatic hypermutation perturbed in UNG-deficient mice. Curr. Biol. 2002;12:1748–1755. doi: 10.1016/s0960-9822(02)01215-0. [DOI] [PubMed] [Google Scholar]
- 38.Imai K, Slupphaug G, Lee WI, Revy P, Nonoyama S, Catalan N, Yel L, Forveille M, Kavli B, Krokan HE, Ochs HD, Fischer A, Durandy A. Human uracil-DNA glycosylase deficiency associated with profoundly impaired immunoglobulin class-switch recombination. Nat. Immunol. 2003;4:1023–1028. doi: 10.1038/ni974. [DOI] [PubMed] [Google Scholar]
- 39.Frederico LA, Kunkel TA, Shaw BR. A sensitive genetic assay for the detection of cytosine deamination: determination of rate constants and the activation energy. Biochem. 1990;29:2532–2537. doi: 10.1021/bi00462a015. [DOI] [PubMed] [Google Scholar]
- 40.Delbos F, Aoufouchi S, Faili A, Weill JC, Reynaud CA. DNA polymerase {eta} is the sole contributor of A/T modifications during immunoglobulin gene hypermutation in the mouse. J. Exp. Med. 2007;204:17–23. doi: 10.1084/jem.20062131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Silverman GJ. B cell superantigens: possible roles in immunodeficiency and autoimmunity. Semin. Immunol. 1998;10:43–55. doi: 10.1006/smim.1997.0104. [DOI] [PubMed] [Google Scholar]