Abstract
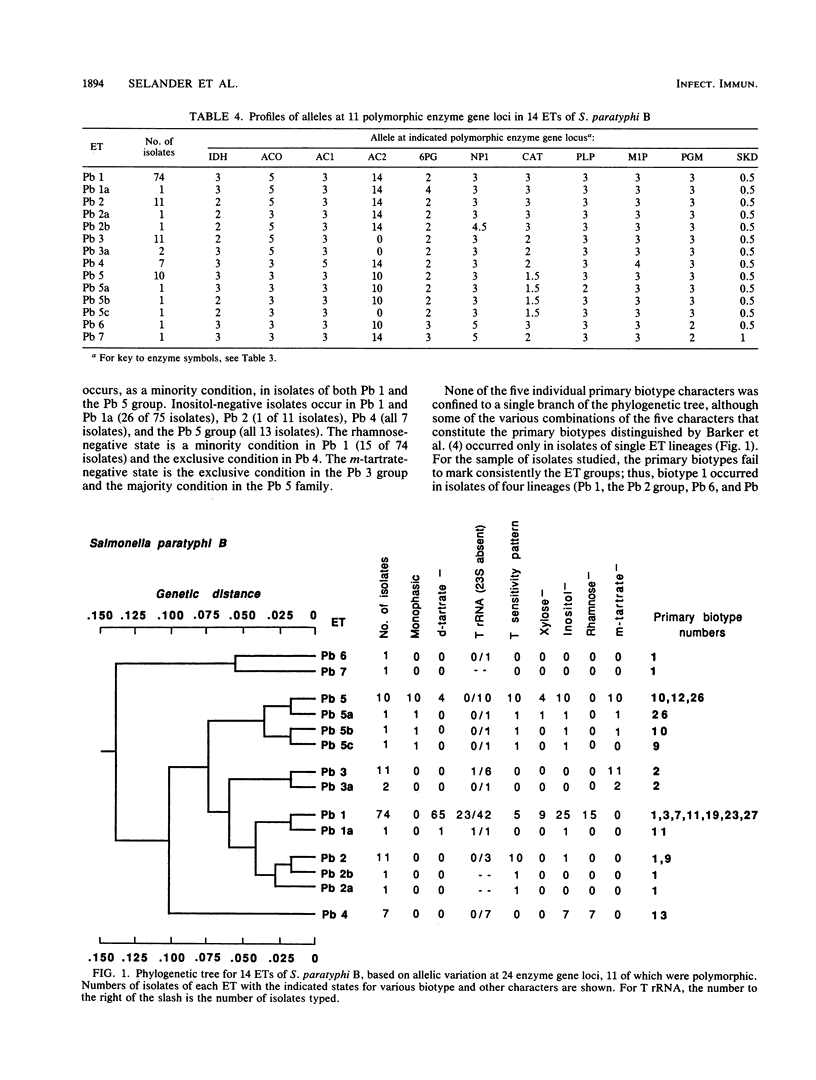
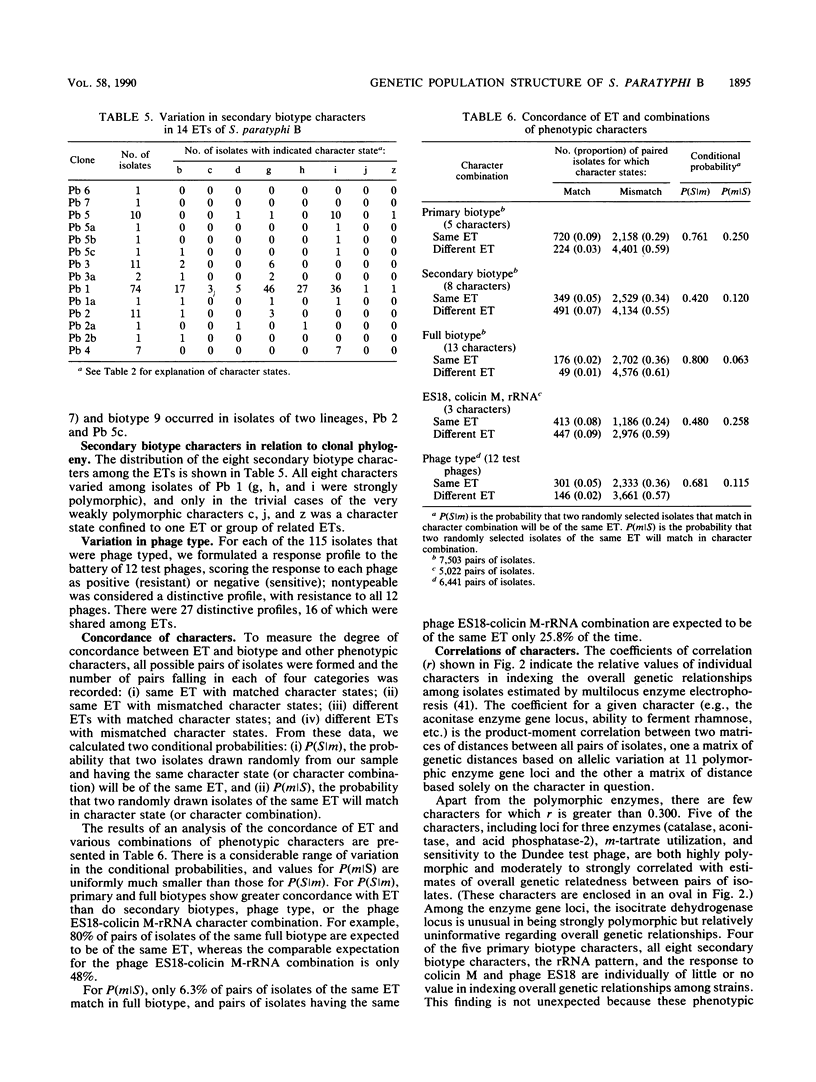
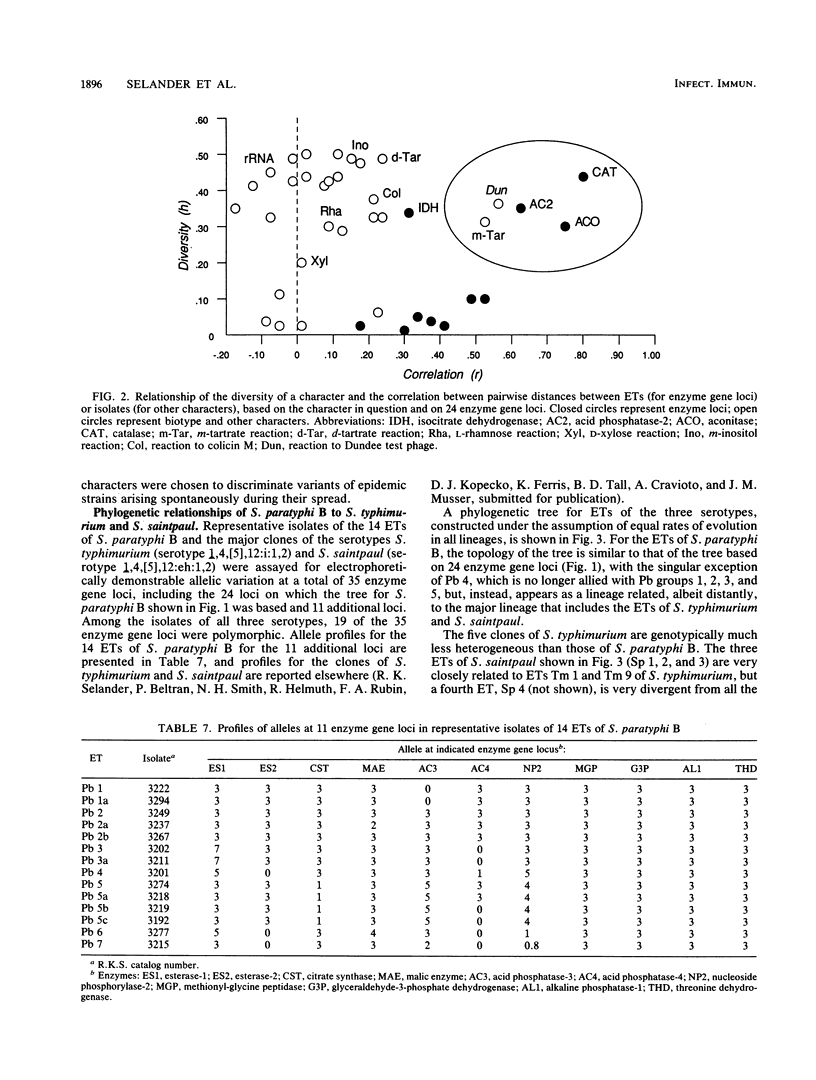
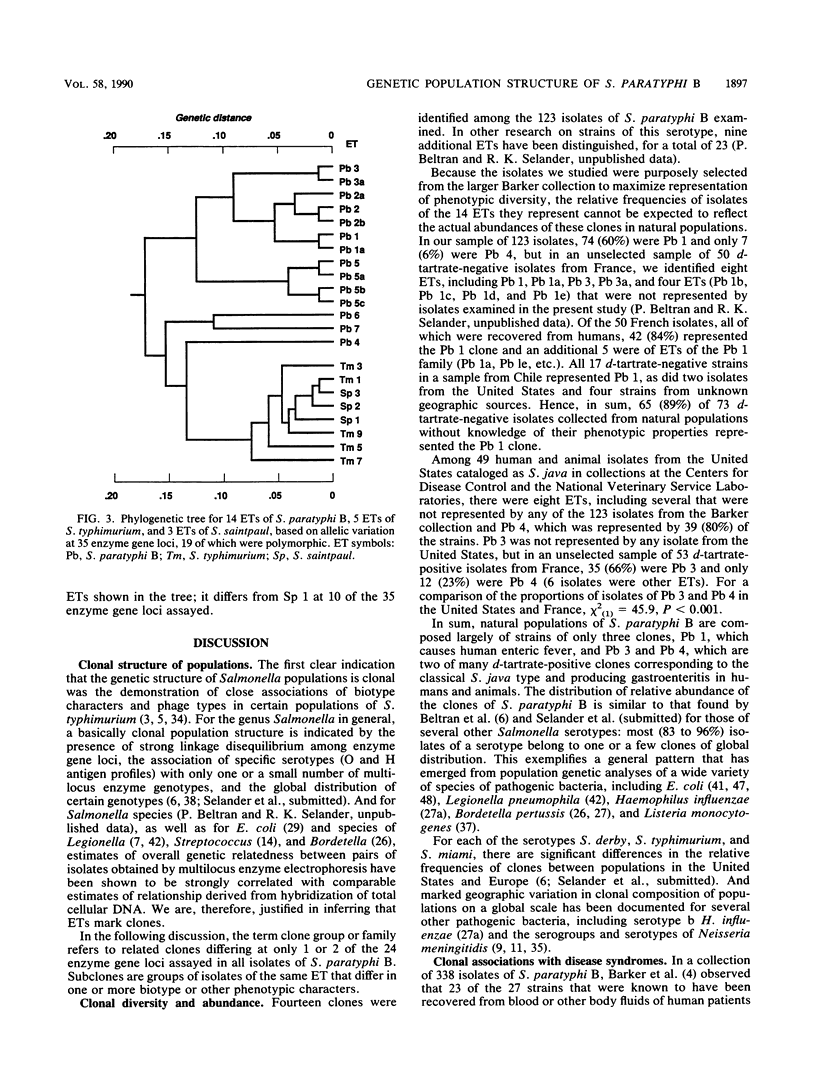
Genetic diversity and relationships among 123 strains of Salmonella paratyphi B (serotype 1,4,[5],12:b:[1,2]) were estimated from an assessment of electrophoretically demonstrable allelic variation at 24 chromosomal enzyme gene loci. Fourteen electrophoretic types, marking clones, were distinguished, the phylogeny of the clonal lineages was reconstructed, and biotype and other phenotypic characters were mapped onto this structure. Most d-tartrate-negative strains are members of an abundant, globally distributed clone (Pb 1) that is polymorphic for many biotype characters (including d-tartrate utilization), bacteriophage type, rRNA pattern, and colicin M and phage ES18 sensitivity. This clone is largely responsible for S. paratyphi B enteric fever in humans. In contrast, d-tartrate-positive strains (formerly known as S. java) occurred in all seven of the clonal lineages identified by population genetic analysis, although most d-tartrate-positive isolates belong to only two clones (Pb 3 and Pb 4), which vary in frequency geographically. Monophasic strains represent four closely related clones forming a distinctive phylogenetic lineage. The Kauffmann hypothesis of convergence in serotype among distantly related cell lineages through recombination (via phage transduction or other means) may account for the considerable genotypic diversity among clones of S. paratyphi B. Pb 4, Pb 6, and Pb 7 are more closely allied with clones of S. typhimurium and S. saintpaul than with other clones of S. paratyphi B. Sensitivity or resistance to colicin M and phage ES18 and the electrophoretic pattern of the rRNA, which were incorporated into a recently proposed scheme for the identification of types of S. paratyphi B, individually or in combination fail to mark clones or other meaningful phylogenetic subdivisions.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson E. S., Ward L. R., De Saxe M. J., Old D. C., Barker R., Duguid J. P. Correlation of phaga type, biotype and source in strains of Salmonella typhimurium. J Hyg (Lond) 1978 Oct;81(2):203–217. doi: 10.1017/s0022172400025031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker R. M., Kearney G. M., Nicholson P., Blair A. L., Porter R. C., Crichton P. B. Types of Salmonella paratyphi B and their phylogenetic significance. J Med Microbiol. 1988 Aug;26(4):285–293. doi: 10.1099/00222615-26-4-285. [DOI] [PubMed] [Google Scholar]
- Barker R., Old D. C., Sharp J. C. Phage type/biotype groups of Salmonella typhimurium in Scotland 1974-6: variation during spread of epidemic clones. J Hyg (Lond) 1980 Feb;84(1):115–125. doi: 10.1017/s0022172400026607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beltran P., Musser J. M., Helmuth R., Farmer J. J., 3rd, Frerichs W. M., Wachsmuth I. K., Ferris K., McWhorter A. C., Wells J. G., Cravioto A. Toward a population genetic analysis of Salmonella: genetic diversity and relationships among strains of serotypes S. choleraesuis, S. derby, S. dublin, S. enteritidis, S. heidelberg, S. infantis, S. newport, and S. typhimurium. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7753–7757. doi: 10.1073/pnas.85.20.7753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner D. J., Steigerwalt A. G., Epple P., Bibb W. F., McKinney R. M., Starnes R. W., Colville J. M., Selander R. K., Edelstein P. H., Moss C. W. Legionella pneumophila serogroup Lansing 3 isolated from a patient with fatal pneumonia, and descriptions of L. pneumophila subsp. pneumophila subsp. nov., L. pneumophila subsp. fraseri subsp. nov., and L. pneumophila subsp. pascullei subsp. nov. J Clin Microbiol. 1988 Sep;26(9):1695–1703. doi: 10.1128/jcm.26.9.1695-1703.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgin A. B., Parodos K., Lane D. J., Pace N. R. The excision of intervening sequences from Salmonella 23S ribosomal RNA. Cell. 1990 Feb 9;60(3):405–414. doi: 10.1016/0092-8674(90)90592-3. [DOI] [PubMed] [Google Scholar]
- CHERRY W. B., DAVIS B. R., EDWARDS P. R. Observations on the types and typing of Salmonella paratyphi B cultures in the United States. Am J Public Health Nations Health. 1953 Oct;43(10):1280–1286. doi: 10.2105/ajph.43.10.1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caugant D. A., Mocca L. F., Frasch C. E., Frøholm L. O., Zollinger W. D., Selander R. K. Genetic structure of Neisseria meningitidis populations in relation to serogroup, serotype, and outer membrane protein pattern. J Bacteriol. 1987 Jun;169(6):2781–2792. doi: 10.1128/jb.169.6.2781-2792.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe B. A., Wall R. A., Kusecek B., Neumann B., Olyhoek T., Abdillahi H., Hassan-King M., Greenwood B. M., Poolman J. T., Achtman M. Clonal and variable properties of Neisseria meningitidis isolated from cases and carriers during and after an epidemic in The Gambia, West Africa. J Infect Dis. 1989 Apr;159(4):686–700. doi: 10.1093/infdis/159.4.686. [DOI] [PubMed] [Google Scholar]
- Duguid J. P., Anderson E. S., Alfredsson G. A., Barker R., Old D. C. A new biotyping scheme for Salmonella typhimurium and its phylogenetic significance. J Med Microbiol. 1975 Feb;8(1):149–166. doi: 10.1099/00222615-8-1-149. [DOI] [PubMed] [Google Scholar]
- Gilmour M. N., Whittam T. S., Kilian M., Selander R. K. Genetic relationships among the oral streptococci. J Bacteriol. 1987 Nov;169(11):5247–5257. doi: 10.1128/jb.169.11.5247-5257.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham A. C., Stocker B. A. Genetics of sensitivity of Salmonella species to colicin M and bacteriophages T5, T1, and ES18. J Bacteriol. 1977 Jun;130(3):1214–1223. doi: 10.1128/jb.130.3.1214-1223.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- KAUFFMANN F. On the transduction of serological properties in the Salmonella group. Acta Pathol Microbiol Scand. 1953;33(4):409–420. doi: 10.1111/j.1699-0463.1953.tb01537.x. [DOI] [PubMed] [Google Scholar]
- KAUFFMANN F. Zur Differential diagnose und Pathogenität von Salmonella java und Salmonella paratyphi B. Z Hyg Infektionskr. 1955;141(6):546–550. [PubMed] [Google Scholar]
- LEDERBERG J., EDWARDS P. R. Sero-typic recombination in Salmonella. J Immunol. 1953 Oct;71(4):232–240. [PubMed] [Google Scholar]
- Le Minor L., Véron M., Popoff M. Proposition pour une nomenclature des Salmonella. Ann Microbiol (Paris) 1982 Sep-Oct;133(2):245–254. [PubMed] [Google Scholar]
- Morgenroth A., Duguid J. P. Demonstration of different mutational sites controlling rhamnose fermentation in FIRN and non-FIRN rha-strains of Salmonella typhimurium: an essay in bacterial archaeology. Genet Res. 1968 Apr;11(2):151–169. doi: 10.1017/s0016672300011320. [DOI] [PubMed] [Google Scholar]
- Musser J. M., Bemis D. A., Ishikawa H., Selander R. K. Clonal diversity and host distribution in Bordetella bronchiseptica. J Bacteriol. 1987 Jun;169(6):2793–2803. doi: 10.1128/jb.169.6.2793-2803.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser J. M., Hewlett E. L., Peppler M. S., Selander R. K. Genetic diversity and relationships in populations of Bordetella spp. J Bacteriol. 1986 Apr;166(1):230–237. doi: 10.1128/jb.166.1.230-237.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musser J. M., Kroll J. S., Granoff D. M., Moxon E. R., Brodeur B. R., Campos J., Dabernat H., Frederiksen W., Hamel J., Hammond G. Global genetic structure and molecular epidemiology of encapsulated Haemophilus influenzae. Rev Infect Dis. 1990 Jan-Feb;12(1):75–111. doi: 10.1093/clinids/12.1.75. [DOI] [PubMed] [Google Scholar]
- Ochman H., Whittam T. S., Caugant D. A., Selander R. K. Enzyme polymorphism and genetic population structure in Escherichia coli and Shigella. J Gen Microbiol. 1983 Sep;129(9):2715–2726. doi: 10.1099/00221287-129-9-2715. [DOI] [PubMed] [Google Scholar]
- Old D. C., Dawes P. F., Barker R. M. Transduction of inositol-fermenting ability demonstrating phylogenetic relationships among strains of Salmonella typhimurium. Genet Res. 1980 Apr;35(2):215–224. doi: 10.1017/s0016672300014063. [DOI] [PubMed] [Google Scholar]
- Old D. C., Duguid J. P. Transduction of fimbriation demonstrating common ancestry in FIRN strains of Salmonella typhimurium. J Gen Microbiol. 1979 Jun;112(2):251–259. doi: 10.1099/00221287-112-2-251. [DOI] [PubMed] [Google Scholar]
- Old D. C., Mortlock R. P. Phylogenetic relationships between different D-xylose biogroups in wild-type Salmonella typhimurium strains and a suggested evolutionary pathway. J Appl Bacteriol. 1979 Aug;47(1):167–174. doi: 10.1111/j.1365-2672.1979.tb01181.x. [DOI] [PubMed] [Google Scholar]
- Old D. C. Phylogeny of strains of Salmonella typhimurium. Microbiol Sci. 1984 Jun;1(3):69–72. [PubMed] [Google Scholar]
- Olyhoek T., Crowe B. A., Achtman M. Clonal population structure of Neisseria meningitidis serogroup A isolated from epidemics and pandemics between 1915 and 1983. Rev Infect Dis. 1987 Jul-Aug;9(4):665–692. doi: 10.1093/clinids/9.4.665. [DOI] [PubMed] [Google Scholar]
- Piffaretti J. C., Kressebuch H., Aeschbacher M., Bille J., Bannerman E., Musser J. M., Selander R. K., Rocourt J. Genetic characterization of clones of the bacterium Listeria monocytogenes causing epidemic disease. Proc Natl Acad Sci U S A. 1989 May;86(10):3818–3822. doi: 10.1073/pnas.86.10.3818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves M. W., Evins G. M., Heiba A. A., Plikaytis B. D., Farmer J. J., 3rd Clonal nature of Salmonella typhi and its genetic relatedness to other salmonellae as shown by multilocus enzyme electrophoresis, and proposal of Salmonella bongori comb. nov. J Clin Microbiol. 1989 Feb;27(2):313–320. doi: 10.1128/jcm.27.2.313-320.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selander R. K., Caugant D. A., Ochman H., Musser J. M., Gilmour M. N., Whittam T. S. Methods of multilocus enzyme electrophoresis for bacterial population genetics and systematics. Appl Environ Microbiol. 1986 May;51(5):873–884. doi: 10.1128/aem.51.5.873-884.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selander R. K., McKinney R. M., Whittam T. S., Bibb W. F., Brenner D. J., Nolte F. S., Pattison P. E. Genetic structure of populations of Legionella pneumophila. J Bacteriol. 1985 Sep;163(3):1021–1037. doi: 10.1128/jb.163.3.1021-1037.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith N. H., Beltran P., Selander R. K. Recombination of Salmonella phase 1 flagellin genes generates new serovars. J Bacteriol. 1990 May;172(5):2209–2216. doi: 10.1128/jb.172.5.2209-2216.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith N. H., Crichton P. B., Old D. C., Higgins C. F. Ribosomal-RNA patterns of Escherichia coli, Salmonella typhimurium and related Enterobacteriaceae. J Med Microbiol. 1988 Jul;26(3):223–228. doi: 10.1099/00222615-26-3-223. [DOI] [PubMed] [Google Scholar]
- Smith N. H., Selander R. K. Sequence invariance of the antigen-coding central region of the phase 1 flagellar filament gene (fliC) among strains of Salmonella typhimurium. J Bacteriol. 1990 Feb;172(2):603–609. doi: 10.1128/jb.172.2.603-609.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Sakazaki R., Kuramochi S., Yoshizaki E. [On propriety of distinguishing Salmonella java from Salmonella paratyphi-B]. Kansenshogaku Zasshi. 1982 Nov;56(11):1025–1031. doi: 10.11150/kansenshogakuzasshi1970.56.1025. [DOI] [PubMed] [Google Scholar]
- Vieu J. F., Binette H., Leherissey M. Salmonella paratyphi B d-tartrate positif (var. java): lysotypie de 1200 souches isolées en France (1975-1985). Zentralbl Bakteriol Mikrobiol Hyg A. 1988 May;268(3):424–432. doi: 10.1016/s0176-6724(88)80027-0. [DOI] [PubMed] [Google Scholar]
- Whittam T. S., Wilson R. A. Genetic relationships among pathogenic Escherichia coli of serogroup O157. Infect Immun. 1988 Sep;56(9):2467–2473. doi: 10.1128/iai.56.9.2467-2473.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittam T. S., Wilson R. A. Genetic relationships among pathogenic strains of avian Escherichia coli. Infect Immun. 1988 Sep;56(9):2458–2466. doi: 10.1128/iai.56.9.2458-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler M. E. Ribosomal ribonucleic acid isolated from Salmonella typhimurium: absence of the intact 23S species. J Bacteriol. 1979 Sep;139(3):842–849. doi: 10.1128/jb.139.3.842-849.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ZINDER N. D., LEDERBERG J. Genetic exchange in Salmonella. J Bacteriol. 1952 Nov;64(5):679–699. doi: 10.1128/jb.64.5.679-699.1952. [DOI] [PMC free article] [PubMed] [Google Scholar]



