Abstract
Many studies reported that oxidative and nitrosative stress might be important for the pathogenesis of Alzheimer’s disease (AD) beginning with arguably the earliest stage of AD, i.e., as mild cognitive impairment (MCI). p53 is a pro-apoptotic protein that plays an important role in neuronal death, a process involved in many neurodegenerative disorders. Moreover, p53 plays a key role in the oxidative stress-dependent apoptosis. We demonstrated previously that p53 levels in brain were significantly higher in MCI and AD IPL compared to control brains. In addition, we showed that in AD IPL, but not in MCI, HNE, a lipid peroxidation product, was significantly bound to p53 protein. In this report, we studied by means of immunoprecipitation analysis, the levels of markers of protein oxidation, 3-nitrotyrosine (3-NT) and protein carbonyls in p53 in a specific region of the cerebral cortex, namely the inferior parietal lobule (IPL), in MCI and AD compared to control brains. The focus of these studies was to measure the oxidation and nitration status of this important pro-apoptotic protein, consistent with hypothesis that oxidative modification of p53 could be involved in the neuronal loss observed in neurodegenerative conditions.
Keywords: mild cognitive impairment (MCI), Alzheimer’s disease (AD), apoptosis, oxidative stress, 3-nytrotyrosine, protein carbonyl, p53
Introduction
Alzheimer disease (AD), the leading cause of dementia, involves regionalized features such as neuronal death, synaptic loss, intracellular neurofibrillary tangles and extracellular amyloid plaques [1]. Although, to date, the mechanism responsible for Alzheimer disease has not yet been identified, several independent hypotheses have been proposed to explain the disease [2–5]. However, none of the hypotheses alone is sufficient to explain the pathological and biochemical alteration in AD. Some of the previous studies showed a role of oxidative stress in development of this neurodegenerative disease [4–9]. Oxidative stress, as well as nitrosative stress, results from an imbalance between oxidants and antioxidants. Oxidants can damage all biological molecules: DNA, RNA, lipid, protein, carbohydrates and antioxidants. In AD brain, the antioxidant levels were found to be decreased, whereas the protein oxidation (protein carbonyl and 3-nitrotyrosine), lipid peroxidation, DNA oxidation, advanced glycation end products were found to be increased [4–9]. Also, in mild cognitive impairment (MCI) a transition phase between normal aging and dementia [10], previous studies showed elevated protein oxidation and lipid peroxidation in specific regions of the brain, such as the hippocampus and the inferior parietal lobule (IPL) [11–14]. This strongly supported the thesis that oxidative stress is involved in the progression of AD from an early phase.
Reactive oxygen species (ROS) and reactive nitrogen species (RNS) attack proteins, leading to the formation of protein carbonyls and 3-nitrotyrosine (3-NT). The levels of protein carbonyls and 3-NT reflect the level of protein oxidation in a cell. Protein oxidation causes the loss of protein function, cellular dysfunction and, ultimately, cell death [11, 15–17]. Oxidative damage can be measured by the determination of levels of protein carbonyls, tyrosine nitration, and protein adducts of alkenals such as acrolein and 4-hydroxynonenal, which are themselves reactive products of lipid peroxidation. Tyrosine nitration is one specific form of protein oxidation that is associated with Alzheimer's disease (AD) [9, 18–20]. Nitric oxide (NO) reacting with the superoxide anion (O2.−) forms the product, peroxynitrite (ONOO−), known to lead to nitration of tyrosine (3-NT) residues [18, 21]. Nitration of proteins results in the inactivation of several important mammalian proteins such as Mn superoxide dismutase (SOD), Cu/Zn SOD, actin, and tyrosine hydroxylase, and likely interferes with tyrosine phosphorylation-mediated cell signaling, as a result of steric effects [17].
Protein carbonyls (aldehydes and ketones, PCO) can arise from direct oxidation of aminoacid side chains (His, Pro, Arg, Lys, and Thr, etc.), by oxidative cleavage of proteins via the α-amidation pathway, or Michael addition reactions of α-, β-unsaturated aldehydes, such as 4-hydroxy-2-nonenal (HNE), malondialdehyde and 2-propenal (acrolein), derived from lipid peroxidation [17]. Elevated levels of PCO are generally associated not only with oxidative stress, but also with the disease-resident protein dysfunction [22]. By using a redox proteomics approach, many proteins involved in energy production, pH regulation and mitochondrial functions were found carbonylated and nitrated in AD inferior parietal lobule [9, 15, 23–25]. In addition, experiments demonstrated other targets of oxidation in different brain regions, and also that oxidatively modified proteins are prone to inactivation [24].
The tumor-suppressor p53 protein plays an important role in cellular response following DNA damage [26]. p53 binds specific DNA sequences and regulates the expression of target genes which encode the proteins that control cell cycle progression or lead cells to apoptosis [27]. Also, p53 might contribute to apoptosis by a mitochondrial pathway [28]. A close connection between NO and p53 may exist because, on the one hand, p53 accumulates in cells following incubation with NO-releasing compounds [29–32]and on the other, p53 mediates transcriptional transrepression of iNOS mRNA expression by a negative feedback loop [31, 32]. Mutated p53 is unable to exert this function. Moreover, high levels of NO can induce a conformational change of wild-type p53 resulting in impairment of its DNA binding activity in vitro [32].
We showed recently that the p53 expression was significantly increased in MCI and AD IPL compared to control samples [33]. In addition, one product of lipid peroxidation, HNE, was found to bind significantly to p53 protein in AD IPL, but not in MCI [33]. The results are consistent with the notion of an involvement of p53, an important regulator of apoptosis, in neurodegenerative conditions, and its special link with oxidative stress.
The prior research on p53 from our laboratory dealt with p53 expression in brain of subjects with AD and MCI. The present work expanded the prior study to examine the oxidation status of p53 protein in these neurodegenerative conditions. Therefore, we performed immunoprecipitation experiments to examine of 3-nitrotyrosine and protein carbonyl levels in p53 protein in MCI and AD inferior parietal lobule compared to control brains. This research tested the hypothesis that p53 is modified by oxidative and nitrosative stress in MCI and AD, suggesting that alteration of p53 pathway could be involved in neuronal death and in the progression of AD.
Materials and Methods
Materials
All chemicals used were purchased from Sigma-Aldrich (St. Louis, MO, USA) with exceptions of nitrocellulose membranes (Bio-Rad, Hercules, CA, USA), electrophoretic transfer system (Trans-blot semi-dry Transfer Cell; Bio-Rad), anti-p53 monoclonal antibody used for immunoprecipitation and Western blotting (Calbiochem, LA Jolla, CA, USA), anti-DNP protein adducts polyclonal antibody (Chemicon International, Temecula, CA, USA).
Patients
Frozen IPL samples from MCI, AD, and age-matched controls were obtained from the University of Kentucky Rapid Autopsy Program of the Alzheimer’s Disease Clinical Center (UK ADC). The diagnosis of probable AD was made according to criteria developed by the National Institute of Neurological and Communicative Disorders and Stroke (NINCDS) and the Alzheimer’s Disease and Related Disorders Association (ADRDA) [34]. All AD patients displayed progressive intellectual decline. Control subjects were without history of dementia or other neurological disorders and underwent annual mental status testing and semi-annual physical and neurological exams as part of the UK ADC normal volunteer longitudinal aging study. In addition, patients had test scores in the normal range (Table II). Samples and demographics used for the AD study were described previously [24]. Additional demographic parameters of control, MCI, and AD patients available from medical records are provided in Table I and Table II.
Table II.
Characteristics of control and AD patients (mean ± SD)
| Demographic Variables | Control Subjects | AD Subjects |
|---|---|---|
| Number of subjects | 5 | 5 |
| Gender (male/female) | 3/2 | 3/2 |
| Age at death (yrs) | 87.0 ± 3.94 | 85.8 ± 6.02 |
| Postmortem interval (h) | 2.9 ± 0.70 | 3.4 ± 1.4 |
| MMSE; number of months prior to death test taken | 28 ± 0.8; 6.6 ± 1.4 | 15.7 ± 2.6; 19.7 ± 1.0 |
| APOE genotype, if known (N) | 3/3 (3) 3/4 (2) | ND |
AD, Alzheimer’s Disease; MMSE, Mini-Mental State Examination; APOE, apolipoprotein E; ND, not determined; N, number of individuals; SD, standard deviation; COPD, chronic obstructive pulmonary disease [adapted from [11].
Table I.
Characteristics of control and MCI patients (mean ± SD)
| Demographic Variables | Control Subjects | MCI Subjects |
|---|---|---|
| Number of subjects | 7 | 7 |
| Gender (male/female) | 3/4 | 3/4 |
| Postmortem Interval (h) | 2.87 ± 1.14 | 3.125 ± 1.033 |
| Brain Weight (g) | 1260 ± 120 | 1120 ± 61 |
| Braak Stage | I–II | III–V |
MCI, Mild Cognitive Impairment.
Sample preparation
The brain tissues (IPL) from control, MCI and AD were homogenized in ice-cold isolation buffer containing 10 mM HEPES buffer, 137 mM NaCl, 4.6 mM KCl, 1.1 mM KH2PO4, and 0.6 mM MgSO4, as well as proteinase inhibitors leupeptin (0.5 mg/mL), pepstatin (0.7 mg/mL). Homogenates were centrifuged at 14,000 × g for 10 min to remove debris. The supernatant was extracted to determine the total protein concentration by BCA method (Pierce, Rockford, IL, USA).
Immunoprecipitations
For immunoprecipitation experiments, 150 µg of protein extracts were resuspended in 500 µl RIPA buffer (10mM Tris, pH 7.6; 140mM NaCl; 0.5% NP40 including protease inhibitors) and then incubated with 1 µg of monoclonal conformation-specific antibody against p53 protein (wild type specific - PAb11) at 4°C overnight. Immunocomplexes were collected by using protein A/G suspension for 2 h at 4°C and washed five times with immunoprecipitation buffer. Immunoprecipitated p53 was recovered by resuspending the pellets in loading buffer, and protein was detected by Western blotting.
Western blotting analysis
For immunoblotting analysis proteins immunoprecipited (30 µl) were electrophoresed through a 10% polyacrylamide gel and transferred to nitrocellulose paper (Bio-Rad Trans-blot semi-dry Transfer Cell) at 45 mA for 2 h. The membranes were blocked for 1 h at room temperature with blocking solution in 5% non-fat dried milk in phosphate-buffered saline containing 0.01% (w/v) sodium azide and 0.2% (v/v) Tween 20 (PBST) at 4°C for 1 h. The membranes were then incubated for 2 h at room temperature with primary antibodies: Anti-nitrotyrosine polyclonal antibody (3-NT), diluted 1:100 in wash blot and anti-DNP protein adducts polyclonal antibody (1:100). After three washes for 5 min with wash blot, the membranes were incubated for 1 h at room temperature with IgG alkaline phosphatase polyclonal secondary antibody diluted 1:2000 in wash blot and developed using 5-bromo-4-chloro-3-indolyl-phosphate/nitroblue tetrazolium (BCIP/NBT) color developing reagent. Blots were dried and scanned with Adobe Photoshop and quantitated with Scion Image (PC version of Macintosh-compatible NIH Image) software.
Post derivatization of protein
Samples were post-derivatized with DNPH on the membrane and probed with anti-DNPH antibody to identify the oxidized proteins. The nitrocellulose membranes where equilibrated in solution A (20% (v/v) methanol:80%(v/v) wash blot buffer) for 5 min, followed by incubation of membranes in 2N HCl for 5 min. The proteins on blots were then derivatized in solution B (0.5 mM DNPH in 2N HCl) for exactly 10 min as described by Conrad et al. [35]. The membranes were washed three times in 2N HCl for 5 min each and then five times with 50% methanol and two times with wash blot each for 5 min. The 2,4-dinitrophenylhydrazone (DNP) adducts of the carbonyls of the brain proteins were detected immunochemically as described above.
Statistics
The results are presented as means ± SD. Statistical analysis was performed using two-tailed Student’s t-test. A value of P < 0.05 was considered statistically significant.
Results
Protein carbonyl levels in p53 protein in MCI and AD IPL
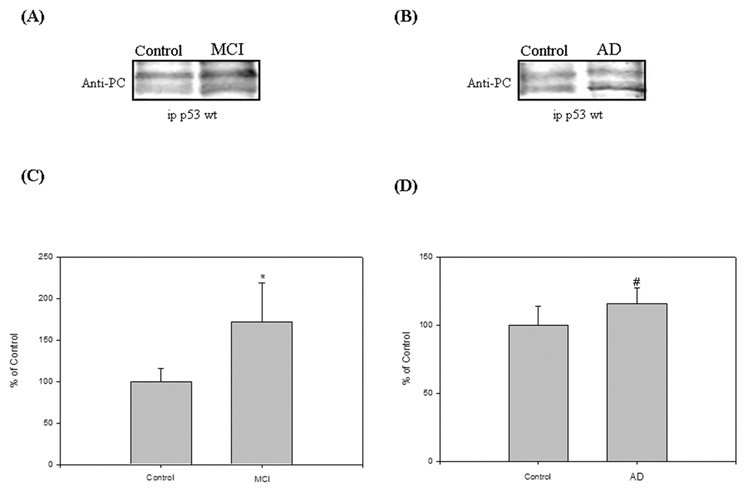
Previous results from our laboratory demonstrated elevated protein carbonylation in specific brain regions of subjects with MCI and AD compared to controls [14, 23–25]. Therefore, to examine whether the p53 protein from MCI and AD IPL were carbonylated compared to age-matched control brains, we performed immunoprecitation and Western blotting analysis. In order to avoid the recognition of the heavy and light chain of immunoglobulins of the antibody in the immunoprecipitation by the second antibody used for Western blotting analysis, we used a mouse monoclonal antibody for immunoprecipitation and a rabbit polyclonal antibody for Western blotting. Protein extracts from controls, MCI, and AD IPL were immunoprecipitated with a conformational specific antibody against p53 (wild type specific - PAb11) and then subjected to Western blotting analysis. To measure the levels of protein carbonyls in p53, the protein on the membrane was derivatized with DNPH and subsequently probed with anti-protein-DNP hydrazone antibody. As shown in Fig. 1, p53 was found to exhibit a significant increase in protein carbonylation by about 72% (Fig. 1C; *P<0.02) and 32% (Fig. 1D; #P<0.05), respectively, in both MCI and AD IPL compared to controls.
Figure 1.
(A) & (B) The oxidation status of p53 was studied by immunoprecipitation analysis in Alzheimer’s disease (AD), mild cognitive impairment (MCI) and control IPL. Equal amounts of protein (150 µg/lane) were immunoprecipitated by anti-p53 antibody, and immunoprecipitates were analyzed for protein carbonyl immunoreactivity by Western blotting. ‘A’ is a representative blot of data obtained from 7 control and MCI samples, and ‘B’ is representative blot of data obtained from 5 control and AD samples, respectively.
(C) & (D) Graphical analysis of MCI and AD band intensities, respectively. The respective control values were set to 100%, to which experimental values were compared. Data are shown in arbitrary units on the ordinate axis as mean ±S.D. MCI, *<0.02; AD, #P<0.05.
3-nitrotyrosine levels in p53 protein in AD in MCI IPL
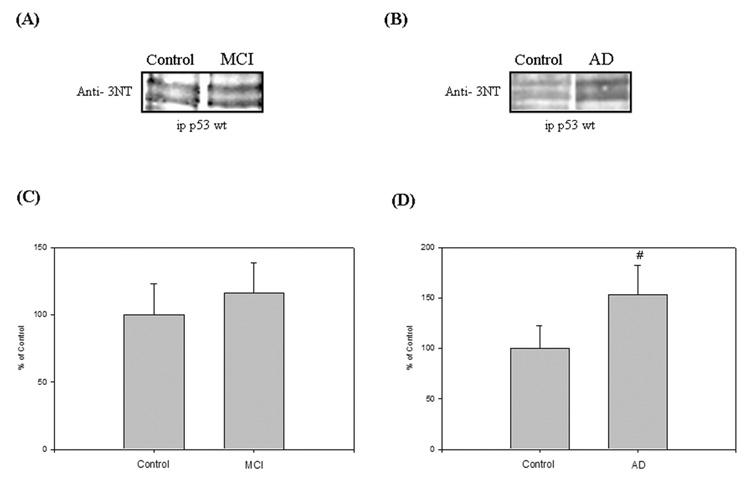
In the same way as protein carbonyls, we studied another oxidative stress marker in p53 protein, the levels of 3-nitrotyrosine. Previous observations from our laboratory showed an increased of protein nitration in specific cerebral regions of MCI and AD subjects compared to controls [9, 12, 13]. Consequently, we performed an immunoprecipitation experiment to check whether tyrosine residues in p53 protein were nitrated in MCI and AD IPL compared to controls. As shown in Fig. 2, p53 was found to exhibit a significant increase in protein nitration by about 58% (Fig. 2D; #P<0.007) in AD IPL compared to controls, but the level of 3-NT was not statistically different from controls in MCI IPL.
Figure 2.
(A) & (B) Nitration status of p53 was studied by immunoprecipitation analysis in Alzheimer’s disease (AD), mild cognitive impairment (MCI) and control IPL. Equal amounts of protein (150 µg/lane) were immunoprecipitated by anti-p53 antibody, and immunoprecipitates were analyzed for 3-nitrotyrosine immunoreactivity by Western blotting. ‘A’ is a representative blot of data obtained from 7 control and MCI samples, and ‘B’ is representative blot of data obtained from 5 control and AD samples, respectively.
(C) & (D) Graphical analysis of MCI and AD band intensities, respectively. The respective control values were set to 100%, to which experimental values were compared. Data are shown in arbitrary units on the ordinate axis as mean ±S.D. AD, #P<0.007.
Discussion
Many studies reported that oxidative and nitrosative stress are early events in the progression of AD and involved in neurodegeneration [9, 25]. Oxidative stress could also stimulate additional damage via the overexpression of inducible (i) and neuronal (n) specific NO synthase (NOS: iNOS and nNOS) leading to increased levels of NO. NO and O2.− react at diffusion controlled rates to produce peroxynitrite, an extremely strong oxidant that affects lipids, DNA, carbohydrates and proteins (particularly the amino acids cysteine, methionine, tryptophan, phenylalanine and especially tyrosine) and, consequently, an increase of oxidative damage [36–38]. Peroxynitrite can nitrate tyrosine [39] at the 3-position, that, by steric effects, could prevent the phosphorylation of the OH moiety on tyrosine residue. Therefore, 3-NT can cause the loss of protein functionality and potentially lead to cell death [36, 40]. Peroxynitrite can also avidly react with thiols to form nitrosothiols, affecting the function of proteins [39]. Nitration of proteins may lead to their irreversible damage [36–38, 40] and also affect the energy status of neurons by inactivating key enzymes [41]. These oxidative alterations not only decrease or eliminate the normal functions of these macromolecules [22], but may also activate an inflammatory response in AD brain.
Increased levels of protein carbonyls and protein-bound HNE were reported in IPL and the hippocampus of subjects with MCI compared to that of controls [11, 14], suggesting the build up of oxidative stress [11, 14, 42]. A recent study reported the excess protein carbonylation (protein oxidation) of alpha-enolase, glutamine synthetase, pyruvate kinase M2 and peptidyl–prolyl cis/trans isomerase 1 (Pin1) in hippocampus of subjects with amnestic MCI using a redox proteomics approach [14].
In an earlier study, we showed the levels of p53 were elevated in brain from subjects with AD and MCI and that p53 was modified by covalent binding of the lipid peroxidation prodect HNE [33]. In the current paper, we expanded this prior study to show that p53, a pro-apoptotic protein, is a target for oxidative and nitrosative stress in these neurodegenerative conditions. By immunoprecipitation analysis, the oxidation and nitration status in pro-apoptotic p53 was studied in MCI and AD IPL compared to age-matched control IPL. The wild type isoform of p53 protein was immunoprecipitated and then subjected to Western blot analysis to investigate the levels of 3-nitrotyrosine and protein carbonyls. The results reported in this current study suggest that wild type p53 had significantly increased levels of 3-nitrotyrosine and protein carbonyl in AD IPL compared to controls, while MCI showed a significant increase of protein carbonyl levels, but not 3-nitrotyrosine. We have previously reported that a highly toxic product from lipid peroxidation, 4-hydroxy-2-nonenal (HNE), bound p53 in AD IPL compared to controls, but not in MCI IPL. Taken together, these data support the notion of oxidative and nitrosative stress in AD and MCI, and in particular of an important protein involved in crucial cellular processes. Although the effects on p53 conformation and DNA-binding activity remain to be determined, our results perhaps are consistent with the concept that stress conditions may play a role in the alteration of p53 protein function.
Some scientists have shown that treatment of tumor cells with high concentrations of NO can result in tyrosine nitration and mutant conformation of wild-type p53 with inactivation of functionality [32, 43, 44]. Because eight of the nine tyrosines in the p53 molecule are located in the critical DNA binding region, it is possible that covalent modification of these residues by p53 could result in a mutant p53 conformation or directly interfere with DNA binding residues. Further, it is been suggested that tyrosine residues next to glutamate in an amino acid sequence could be easily nitrated in neurofilaments [45]. In human p53 protein, there are three tyrosines with match these characteristics, i.e., Y 205, Y 220 and Y 327. These tyrosine-glutamate sequences are localized in the central core domain as well as in the tetramerization domain of p53. Peroxynitrite could also inhibit wild-type p53 protein function through other mechanisms. Oxidizing agents are known to modify both conformation and sequence-specific DNA binding of p53 in vitro, and peroxynitrite causes zinc to be released from the zinc-thiolate center of zinc finger transcription factors [46, 47]. Zinc binding and redox regulation are, at least in part, distinct determinants of the binding of p53 to DNA. Moreover, researchers have shown that p53 is subject to more than one level of conformational modulation through oxidation-reduction of cysteines at or near the p53-DNA interface [46]. Thus, oxidation of these critical p53 cysteines by peroxynitrite could have a dramatic effects on p53 function.
In summary, we showed for the first time, that wild-type p53 protein, an important molecule involved in fundamental cellular process such as apoptosis, the cell cycle, and DNA repair, is modified by oxidative and nitrosative stress, particularly in particular in advance stage of AD. One important consequence likely would be a conformational change and dysregulation of p53 transcriptional activity and downstream pathways. Current investigations in our laboratory are underway to determine if the oxidative modifications of wild type p53 in brain of subjects with AD and early stage MCI result in change of p53 conformation and functional activity. These observations may provide definitive support for the notion that oxidative and nitrosative stress are involved in neuronal death in neurodegenerative conditions like AD. Since new therapeutic strategies are designated to modulate protein oxidation and lipid peroxidation early in the course of disease, p53 could be a new therapeutic target to possibly prevent or to slow neuronal loss in MCI and AD, and possibly other neurodegenerative disorders as well.
Acknowledgements
This research was supported in part by NIH grants to D.A.B. [AG-05119, AG10836; AG-029839].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Katzman R, Saitoh T. Advances in Alzheimer's disease. Faseb J. 1991;5:278–286. [PubMed] [Google Scholar]
- 2.Ho GJ, Drego R, Hakimian E, Masliah E. Mechanisms of cell signaling and inflammation in Alzheimer's disease. Curr Drug Targets Inflamm Allergy. 2005;4:247–256. doi: 10.2174/1568010053586237. [DOI] [PubMed] [Google Scholar]
- 3.Hynd MR, Scott HL, Dodd PR. Glutamate-mediated excitotoxicity and neurodegeneration in Alzheimer's disease. Neurochem Int. 2004;45:583–595. doi: 10.1016/j.neuint.2004.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Markesbery WR. Oxidative stress hypothesis in Alzheimer's disease. Free Radic Biol Med. 1997;23:134–147. doi: 10.1016/s0891-5849(96)00629-6. [DOI] [PubMed] [Google Scholar]
- 5.Butterfield DA, Drake J, Pocernich C, Castegna A. Evidence of oxidative damage in Alzheimer's disease brain: central role for amyloid beta-peptide. Trends Mol Med. 2001;7:548–554. doi: 10.1016/s1471-4914(01)02173-6. [DOI] [PubMed] [Google Scholar]
- 6.Butterfield DA, Lauderback CM. Lipid peroxidation and protein oxidation in Alzheimer's disease brain: potential causes and consequences involving amyloid beta-peptide-associated free radical oxidative stress. Free Radic Biol Med. 2002;32:1050–1060. doi: 10.1016/s0891-5849(02)00794-3. [DOI] [PubMed] [Google Scholar]
- 7.Smith MA, Richey PL, Taneda S, Kutty RK, Sayre LM, Monnier VM, Perry G. Advanced Maillard reaction end products, free radicals, and protein oxidation in Alzheimer's disease. Ann N Y Acad Sci. 1994;738:447–454. doi: 10.1111/j.1749-6632.1994.tb21836.x. [DOI] [PubMed] [Google Scholar]
- 8.Lovell MA, Xie C, Markesbery WR. Acrolein is increased in Alzheimer's disease brain and is toxic to primary hippocampal cultures. Neurobiol Aging. 2001;22:187–194. doi: 10.1016/s0197-4580(00)00235-9. [DOI] [PubMed] [Google Scholar]
- 9.Castegna A, Thongboonkerd V, Klein JB, Lynn B, Markesbery WR, Butterfield DA. Proteomic identification of nitrated proteins in Alzheimer's disease brain. J Neurochem. 2003;85:1394–1401. doi: 10.1046/j.1471-4159.2003.01786.x. [DOI] [PubMed] [Google Scholar]
- 10.Petersen RC. Mild cognitive impairment clinical trials. Nat Rev Drug Discov. 2003;2:646–653. doi: 10.1038/nrd1155. [DOI] [PubMed] [Google Scholar]
- 11.Butterfield DA, Reed T, Perluigi M, De Marco C, Coccia R, Cini C, Sultana R. Elevated protein-bound levels of the lipid peroxidation product, 4-hydroxy-2-nonenal, in brain from persons with mild cognitive impairment. Neurosci Lett. 2006;397:170–173. doi: 10.1016/j.neulet.2005.12.017. [DOI] [PubMed] [Google Scholar]
- 12.Butterfield DA, Reed TT, Perluigi M, De Marco C, Coccia R, Keller JN, Markesbery WR, Sultana R. Elevated levels of 3-nitrotyrosine in brain from subjects with amnestic mild cognitive impairment: implications for the role of nitration in the progression of Alzheimer's disease. Brain Res. 2007;1148:243–248. doi: 10.1016/j.brainres.2007.02.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sultana R, Reed T, Perluigi M, Coccia R, Pierce WM, Butterfield DA. Proteomic identification of nitrated brain proteins in amnestic mild cognitive impairment: a regional study. J Cell Mol Med. 2007;11:839–851. doi: 10.1111/j.1582-4934.2007.00065.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Butterfield DA, Poon HF, St Clair D, Keller JN, Pierce WM, Klein JB, Markesbery WR. Redox proteomics identification of oxidatively modified hippocampal proteins in mild cognitive impairment: insights into the development of Alzheimer's disease. Neurobiol Dis. 2006;22:223–232. doi: 10.1016/j.nbd.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 15.Castegna A, Aksenov M, Aksenova M, Thongboonkerd V, Klein JB, Pierce WM, Booze R, Markesbery WR, Butterfield DA. Proteomic identification of oxidatively modified proteins in Alzheimer's disease brain. Part I: creatine kinase BB, glutamine synthase, and ubiquitin carboxy-terminal hydrolase L-1. Free Radic Biol Med. 2002;33:562–571. doi: 10.1016/s0891-5849(02)00914-0. [DOI] [PubMed] [Google Scholar]
- 16.Sultana R, Butterfield DA. Oxidatively modified GST and MRP1 in Alzheimer's disease brain: implications for accumulation of reactive lipid peroxidation products. Neurochem Res. 2004;29:2215–2220. doi: 10.1007/s11064-004-7028-0. [DOI] [PubMed] [Google Scholar]
- 17.Butterfield DA, Stadman ER. Protein oxidation processes in aging brain. Adv Cell Aging Gerontol. 1997;2:161–191. [Google Scholar]
- 18.Gow AJ, Duran D, Malcolm S, Ischiropoulos H. Effects of peroxynitrite-induced protein modifications on tyrosine phosphorylation and degradation. FEBS Lett. 1996;385:63–66. doi: 10.1016/0014-5793(96)00347-x. [DOI] [PubMed] [Google Scholar]
- 19.Smith MA, Richey Harris PL, Sayre LM, Beckman JS, Perry G. Widespread peroxynitrite-mediated damage in Alzheimer's disease. J Neurosci. 1997;17:2653–2657. doi: 10.1523/JNEUROSCI.17-08-02653.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sultana R, Poon HF, Cai J, Pierce WM, Merchant M, Klein JB, Markesbery WR, Butterfield DA. Identification of nitrated proteins in Alzheimer's disease brain using a redox proteomics approach. Neurobiol Dis. 2006;22:76–87. doi: 10.1016/j.nbd.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 21.Butterfield DA, Kanski J. Brain protein oxidation in age-related neurodegenerative disorders that are associated with aggregated proteins. Mech Ageing Dev. 2001;122:945–962. doi: 10.1016/s0047-6374(01)00249-4. [DOI] [PubMed] [Google Scholar]
- 22.Hensley K, Hall N, Subramaniam R, Cole P, Harris M, Aksenov M, Aksenova M, Gabbita SP, Wu JF, Carney JM, et al. Brain regional correspondence between Alzheimer's disease histopathology and biomarkers of protein oxidation. J Neurochem. 1995;65:2146–2156. doi: 10.1046/j.1471-4159.1995.65052146.x. [DOI] [PubMed] [Google Scholar]
- 23.Castegna A, Aksenov M, Thongboonkerd V, Klein JB, Pierce WM, Booze R, Markesbery WR, Butterfield DA. Proteomic identification of oxidatively modified proteins in Alzheimer's disease brain. Part II: dihydropyrimidinase-related protein 2, alpha-enolase and heat shock cognate 71. J Neurochem. 2002;82:1524–1532. doi: 10.1046/j.1471-4159.2002.01103.x. [DOI] [PubMed] [Google Scholar]
- 24.Sultana R, Boyd-Kimball D, Poon HF, Cai J, Pierce WM, Klein JB, Merchant M, Markesbery WR, Butterfield DA. Redox proteomics identification of oxidized proteins in Alzheimer's disease hippocampus and cerebellum: an approach to understand pathological and biochemical alterations in AD. Neurobiol Aging. 2006;27:1564–1576. doi: 10.1016/j.neurobiolaging.2005.09.021. [DOI] [PubMed] [Google Scholar]
- 25.Butterfield DA, Reed T, Newman SF, Sultana R. Roles of amyloid beta-peptide-associated oxidative stress and brain protein modifications in the pathogenesis of Alzheimer's disease and mild cognitive impairment. Free Radic Biol Med. 2007;43:658–677. doi: 10.1016/j.freeradbiomed.2007.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levine AJ. p53, the cellular gatekeeper for growth and division. Cell. 1997;88:323–331. doi: 10.1016/s0092-8674(00)81871-1. [DOI] [PubMed] [Google Scholar]
- 27.Almog N, Rotter V. Involvement of p53 in cell differentiation and development. Biochim Biophys Acta. 1997;1333:F1–F27. doi: 10.1016/s0304-419x(97)00012-7. [DOI] [PubMed] [Google Scholar]
- 28.Marchenko ND, Zaika A, Moll UM. Death signal-induced localization of p53 protein to mitochondria. A potential role in apoptotic signaling. J Biol Chem. 2000;275:16202–16212. doi: 10.1074/jbc.275.21.16202. [DOI] [PubMed] [Google Scholar]
- 29.Messmer UK, Brune B. Nitric oxide-induced apoptosis: p53-dependent and p53-independent signalling pathways. Biochem J. 1996;319(Pt 1):299–305. doi: 10.1042/bj3190299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Messmer UK, Ankarcrona M, Nicotera P, Brune B. p53 expression in nitric oxide-induced apoptosis. FEBS Lett. 1994;355:23–26. doi: 10.1016/0014-5793(94)01161-3. [DOI] [PubMed] [Google Scholar]
- 31.Forrester K, Ambs S, Lupold SE, Kapust RB, Spillare EA, Weinberg WC, Felley-Bosco E, Wang XW, Geller DA, Tzeng E, Billiar TR, Harris CC. Nitric oxide-induced p53 accumulation and regulation of inducible nitric oxide synthase expression by wild-type p53. Proc Natl Acad Sci U S A. 1996;93:2442–2447. doi: 10.1073/pnas.93.6.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calmels S, Hainaut P, Ohshima H. Nitric oxide induces conformational and functional modifications of wild-type p53 tumor suppressor protein. Cancer Res. 1997;57:3365–3369. [PubMed] [Google Scholar]
- 33.Cenini G, Sultana R, Memo M, Butterfield DA. Elevated Levels of Pro-apoptotic p53 and Its Oxidative Modification by the Lipid Peroxidation Product, HNE, in Brain from Subjects with Amnestic Mild Cognitive Impairment and Alzheimer's Disease. J Cell Mol Med. 2007 doi: 10.1111/j.1582-4934.2008.00163.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKhann G, Drachman D, Folstein M, Katzman R, Price D, Stadlan EM. Clinical diagnosis of Alzheimer's disease: report of the NINCDS-ADRDA Work Group under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939–944. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 35.Conrad CC, Talent JM, Malakowsky CA, Gracy RW. Post-Electrophoretic Identification of Oxidized Proteins. Biol Proced Online. 2000;2:39–45. doi: 10.1251/bpo17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koppal T, Drake J, Yatin S, Jordan B, Varadarajan S, Bettenhausen L, Butterfield DA. Peroxynitrite-induced alterations in synaptosomal membrane proteins: insight into oxidative stress in Alzheimer's disease. J Neurochem. 1999;72:310–317. doi: 10.1046/j.1471-4159.1999.0720310.x. [DOI] [PubMed] [Google Scholar]
- 37.Beckman JS. Oxidative damage and tyrosine nitration from peroxynitrite. Chem Res Toxicol. 1996;9:836–844. doi: 10.1021/tx9501445. [DOI] [PubMed] [Google Scholar]
- 38.Perry JM, Zhao Y, Marletta MA. Cu2+ and Zn2+ inhibit nitric-oxide synthase through an interaction with the reductase domain. J Biol Chem. 2000;275:14070–14076. doi: 10.1074/jbc.275.19.14070. [DOI] [PubMed] [Google Scholar]
- 39.Halliwell B. What nitrates tyrosine? Is nitrotyrosine specific as a biomarker of peroxynitrite formation in vivo? EBS Lett. 1997;411:157–160. doi: 10.1016/s0014-5793(97)00469-9. [DOI] [PubMed] [Google Scholar]
- 40.Yamakura F, Taka H, Fujimura T, Murayama K. Inactivation of human manganese-superoxide dismutase by peroxynitrite is caused by exclusive nitration of tyrosine 34 to 3-nitrotyrosine. J Biol Chem. 1998;273:14085–14089. doi: 10.1074/jbc.273.23.14085. [DOI] [PubMed] [Google Scholar]
- 41.Radi R, Rodriguez M, Castro L, Telleri R. Inhibition of mitochondrial electron transport by peroxynitrite. Arch Biochem Biophys. 1994;308:89–95. doi: 10.1006/abbi.1994.1013. [DOI] [PubMed] [Google Scholar]
- 42.Keller JN, Schmitt FA, Scheff SW, Ding Q, Chen Q, Butterfield DA, Markesbery WR. Evidence of increased oxidative damage in subjects with mild cognitive impairment. Neurology. 2005;64:1152–1156. doi: 10.1212/01.WNL.0000156156.13641.BA. [DOI] [PubMed] [Google Scholar]
- 43.Chazotte-Aubert L, Hainaut P, Ohshima H. Nitric oxide nitrates tyrosine residues of tumor-suppressor p53 protein in MCF-7 cells. Biochem Biophys Res Commun. 2000;267:609–613. doi: 10.1006/bbrc.1999.2003. [DOI] [PubMed] [Google Scholar]
- 44.Cobbs CS, Whisenhunt TR, Wesemann DR, Harkins LE, Van Meir EG, Samanta M. Inactivation of wild-type p53 protein function by reactive oxygen and nitrogen species in malignant glioma cells. Cancer Res. 2003;63:8670–8673. [PubMed] [Google Scholar]
- 45.Crow JP, Ye YZ, Strong M, Kirk M, Barnes S, Beckman JS. Superoxide dismutase catalyzes nitration of tyrosines by peroxynitrite in the rod and head domains of neurofilament-L. J Neurochem. 1997;69:1945–1953. doi: 10.1046/j.1471-4159.1997.69051945.x. [DOI] [PubMed] [Google Scholar]
- 46.Hainaut P, Mann K. Zinc binding and redox control of p53 structure and function. Antioxid Redox Signal. 2001;3:611–623. doi: 10.1089/15230860152542961. [DOI] [PubMed] [Google Scholar]
- 47.Crow JP, Beckman JS, McCord JM. Sensitivity of the essential zinc-thiolate moiety of yeast alcohol dehydrogenase to hypochlorite and peroxynitrite. Biochemistry. 1995;34:3544–3552. doi: 10.1021/bi00011a008. [DOI] [PubMed] [Google Scholar]