Abstract
Epstein Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV) are found together in approximately 80 percent of primary effusion lymphomas (PELs), but their contribution to these cancers is unclear (1). We found that dominant-negative derivatives of EBNA1 inhibited EBV-positive PEL cells from forming colonies. Those rare PEL cells that proliferated after expression of the dominant-negative derivatives usually expressed these derivatives at low or undetectable levels and continued to maintain their EBV genomes. Those proliferating cells expressing higher levels of the derivatives, expressed mutant derivatives that could not bind DNA. These findings indicate that EBV is required to sustain proliferation as measured by colony-formation of dually infected PEL cells. The dominant-negative derivatives of EBNA1 had no effect on the colony-forming ability of five, EBV-negative, KSHV-negative hematopoietic cell lines. Surprisingly, they did inhibit the colony forming ability of EBV-negative, KSHV-positive PEL cells. The small fraction of cells that continued to proliferate expressed only mutants of the EBNA1 derivatives that could no longer bind DNA. These findings indicate that the site-specific DNA-binding activity of EBNA1 or its derivatives when expressed efficiently in EBV-negative, KSHV-positive PEL cells inhibits their colony-formation possibly through their binding to the KSHV genome.
Keywords: EBV, KSHV, primary effusion lymphoma, EBNA1, colony-formation
Introduction
Primary effusion lymphomas (PELs) are rare and represent a distinct class of B-cell, non-Hodgkin's Lymphomas for which there is no effective therapy and survival is predicted to be less than one year post-diagnosis. All PELs are infected with KSHV and most with EBV, but they consistently lack the c-myc gene rearrangements typically found in EBV-positive Burkitt's Lymphoma (2). PELs frequently occur in AIDs patients supporting a correlation between immunodeficiency, susceptibility to viral infection, and subsequent tumor development (3-5). It is not clear though how each of the viruses found in dually-infected PELs (EBV+, KSHV+) contributes either to the initiation or maintenance of these tumors.
We have found that EBV is necessary for the proliferation of dually-infected PEL cells under limiting dilution as measured by colony-formation assays. We forced the loss of EBV from these cells by inhibiting EBNA1, the viral protein on which EBV's plasmid replication depends, with dominant-negative derivatives of EBNA1. The majority of dually-infected cells in which EBNA1 was inhibited failed to grow to form colonies under limiting dilution. Those that did grow were found not to have their EBNA1 inhibited and to retain EBV. The dominant-negative derivatives of EBNA1 used to inhibit it were either expressed inefficiently or mutated and non-functional in the minority of cells that did grow.
We also tested the derivatives of EBNA1 in five EBV-negative, KSHV-negative hematopoietic cell lines and three EBV-negative KSHV-positive PEL lines. While these EBNA1 derivatives had no effect on colony formation by the virus-negative hematopoietic lines, they did inhibit that of the EBV-negative, KSHV-positive PEL cells. We have found that EBNA1 binds multiple sites in KSHV's genome (Dresang and Sugden, unpublished) and hypothesize that high levels of DNA-binding derivatives of EBNA1 affect functions of the KSHV genome. We again found that the EBV-negative, KSHV-positive PELs that did grow to form colonies either expressed the derivatives of EBNA1 inefficiently or expressed non-functional mutants of them.
Materials and Methods
Cell lines
The EBV−, KSHV+ PELs, BC-3, BCP-1, and BCBL-1 and the EBV+, KSHV+ PELs, JSC-1 and BC-2, were cultured in RPMI 1640, 10% FBS, 200 units/ml of penicillin, and 200 μg/ml of streptomycin. The EBV−, KSHV− hematopoietic cell lines K562, Jurkat, MOLT-4, and BJAB as well as the EBV-transformed 721 lymphoid clones were cultured in the same media as the PELs. 293T cells used for retroviral production express the SV40 T-antigen and were cultured in DMEM and 10% calf serum. Human fibroblasts (HF) used in colony-formation assays were maintained in DMEM and 10% FBS.
Retroviral production
Retroviruses were produced, concentrated, and titered as described (6).
Retroviral infections
A multiplicity of infection (MOI) equal to 10 − 15 infectious units per cell as titered on BJAB cells was used wherever PEL or other hematopoietic cells were infected. Infections were performed at a concentration of 5 × 105 cells per ml in R10F/50 mM Hepes (pKa = 7.55), placed in 5 ml snap-cap tubes, and rocked 4°C for 1 − 2 hours. Cells were then transferred to 15 ml Falcon tubes, brought to 4 ml with R10F, and spun at 1000 rpm, 4°C for 10 minutes. After infection, cells were washed with 4 ml 1 × PBS then resuspended in 10 ml R10F, transferred to a 10 cm plate, and incubated for 48 to 72 hours.
Western Blotting
Cell lysates were run on 10% polyacrylamide gels and EBNA1 was detected using an alkaline-phosphatase-conjugated rat monoclonal antibody.
Detecting EBV and KSHV by fluorescence in situ hybridization (FISH)
Cells were prepared as described and hybridized with probes that were generated as described (7). Briefly, the DNA probes for the detection of KSHV plasmids were generated by nick-translation using alkali-labile digoxigenin-11-dUTP. The DNA template was derived from cosmid Z8 (8). The DNA probe for the detection of EBV plasmids was derived from the BamH W repeated region and generated by nick-translation using biotinylated-11-dUTP.
Colony-Formation Assays
Infected cells were sorted based on the levels of fluorescent protein expression by FACs analysis and distributed into the wells of a 96-well plate at dilutions ranging from 1 to 100 cells per well. The efficiencies of colony-formation at all dilutions were then averaged. Each well of the plate also contained approximately 2 × 104 human fibroblast (HF) cells in 200 μl of R10F and were plated no more than 3 days prior to sorting. Wells containing colonies with greater than 1000 cells were scored positive while those containing few to no cells were scored negative 14 − 18 days after sorting. The efficiency of colony-formation was then calculated using a Poisson Distribution.
Results
Inhibiting EBNA1 in EBV+, KSHV+ PEL cells inhibits their growth under limiting dilution as measured by assays of colony-formation
We used retroviral vectors to introduce two derivatives of EBNA1 that function dominant-negatively, DBDmut and deltaUR1, into two EBV-positive, KSHV-positive PEL cell lines to inhibit EBNA1. DBDmut dimerizes, binds DNA site-specifically, and inhibits all known functions of EBNA1 (Figure 1a). DeltaUR1 lacks a 25 amino acid region required for EBNA1's transcriptional activity and inhibits only EBNA1's support of transcription (Figure 1a). Both of these derivatives inhibit colony-formation by EBV-positive normal and Burkitt's lymphoma cells and both inhibit their survival by inducing apoptosis (9).
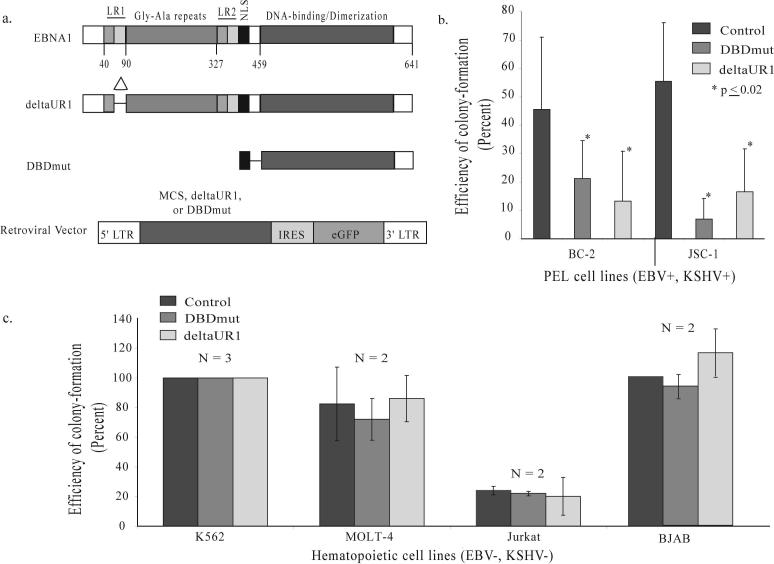
Figure 1. Two derivatives of EBNA1 inhibit EBV+, KSHV+ PEL cells from forming colonies and do not affect EBV−, KSHV− hematopoietic cell lines.
(a) Full-length EBNA1 is 641 amino acids in length, contains two regions (LR1 and LR2) responsible for linking DNA, a nuclear localization sequence (NLS) and a DNA-binding and dimerization domain (15). This carboxy-terminal DNA-binding and dimerization domain is not sufficient to support EBNA1's contributions to DNA replication and acts as a dominant-negative derivative inhibiting all of these functions (9, 15). Numbers indicate the position of amino acids. The deletion within deltaUR1 spans amino acids 65 − 89 (Delta) rendering EBNA1 transcriptionally defective. DBDmut contains only an NLS and the DNA-binding and dimerization domain and is defective in all of EBNA1's known functions. DBDmut, deltaUR1, or a multiple cloning sequence (MCS) as the control was inserted into a retroviral backbone as previously described (9) that co-expresses eGFP. (b) BC-2 and JSC-1 cells were infected with retroviruses expressing DBDmut, deltaUR1, or the control retrovirus containing only the MCS. Infected cells were sorted by FACs 48 hours post-infection, and their efficiency of forming colonies determined 2 weeks post-plating. (c) DBDmut and deltaUR1 do not inhibit EBV(−), KSHV(−) hematopoietic cell lines from forming colonies. Two EBV-negative Burkitt's lymphoma cell lines BJAB and DG75 (analyzed in reference 9); an erythroleukemia cell line (K562); and two T-cell lymphoblastic leukemia lines (MOLT-4 and Jurkat) were tested in assays for colony-formation. These cells were infected with control, DBDmut, or deltaUR1-expressing retroviruses, sorted into 96-well plates and measured for their ability to form colonies 2 weeks post-plating. N, number of independently performed experiments.
The colony-forming ability of BC-2 and JSC-1 cells was significantly inhibited by the expression of either DBDmut or deltaUR1 (Figure 1b). DBDmut-infected BC-2 cells formed colonies at 45% the level of control-infected cells and deltaUR1-infected BC-2 cells at 25% that of control-infected cells (p = 0.02). The colony-forming ability of DBDmut-infected JSC-1 cells was reduced to 10% and that of deltaUR1-infected JSC-1 cells was reduced to 30% that of the level of control-infected cells (p < 0.01).
The EBNA1 derivatives, DBDmut and deltaUR1, do not inhibit EBV−, KSHV-hematopoietic cell lines from forming colonies
We tested whether these derivatives of EBNA1 could affect EBV-negative, KSHV-negative hematopoietic cells. Two B-cell lines derived from Burkitt's lymphomas (BJAB and DG75 (9)), two T-cell lymphoblastic leukemia lines (MOLT-4 and Jurkat), and one erythroleukemia cell line (K562) were infected with retroviruses expressing either DBDmut, deltaUR1, or a control retrovirus. Neither derivative of EBNA1 had detectable effects on the colony-forming ability of the five hematopoietic cell lines (Figure 1c and reference 9).
Those EBV+, KSHV+ JSC-1 cells that survive to form colonies contain non-functional mutants of deltaUR1
Surviving clones from control and deltaUR1-infected JSC-1 cells were expanded from colony-formation assays to investigate their escape from the inhibition of EBNA1. Nine control-infected JSC-1 clones and fourteen deltaUR1-infected clones from two independent infections were selected and expanded from the colony-formation assays (Figure 2a). The signal for intact deltaUR1 (lane B and C) was undetectable in most of the fourteen clones which instead expressed a truncated form of the protein (Figure 2b). The DNA sequences of three of the truncated forms contained a single adenine insertion located at amino acid position 460 of full-length EBNA1. This mutation creates a frame-shift resulting in a premature stop codon approximately 16 amino acids downstream of the insertion site to yield a protein incapable of binding DNA. Thus those EBV+, KSHV+ PEL cells that continue to proliferate in the presence of a dominant-negative derivative of EBNA1 do so only when EBNA1 failed to be inhibited.
Figure 2. Only JSC-1 cells expressing mutant forms of deltaUR1 and retaining EBV survive to form colonies.

(a) Shown is an illustration depicting the two different cell populations analyzed by Western blotting. The Parental population is the initial set of cells transduced with the highest levels of eGFP-expression while the Survivors are the transduced cells that successfully formed the clones recovered from colony-formation assays. (b) Six lysates chosen from a total of fourteen deltaUR1-JSC-1 clones that were the Survivors were analyzed by Western blotting and compared with the corresponding Parental populations, B and C. 3 × 104 cells were loaded in each lane. 721 cells were loaded as an EBNA1-positive control and tubulin served as a loading control. (c) Clones of Survivors have distributions of the number of EBV genomes per cell similar to that of uninfected JSC-1 cells. The number of viral DNAs per cell was measured by FISH. These distributions for three representative clones of Survivors are shown (B.1, B.3, and C.1) along with that of uninfected JSC-1 cells.
All deltaUR1-infected JSC-1 cells that survive to proliferate maintain EBV
We expanded clones of JSC-1 cells infected with deltaUR1 or with the control virus that survived the colony-formation assays and tested them for the presence of EBV. Though there was an increase in the number of parental cells lacking EBV when deltaUR1 was present, all fourteen deltaUR1-JSC-1 clones that survived limiting dilution maintained EBV as measured by FISH with their positive proportions being similar to the percent of EBV-positive cells found in JSC-1 cells infected with control virus (Supplementary Table 1). The distribution in the number of EBV genomes per cell paralleled that of JSC-1 clones infected with the control virus as well as that of untransduced JSC-1 cells (Figure 2c). This distribution results from defects in synthesis and partitioning of EBV plasmids (7). It and the fact that the probe used to detect EBV in these clones covers approximately one-third of the viral genome indicate that the EBV DNA in these clones is extra-chromosomal as it is in the parental JSC-1 cell. Our finding that all surviving clones of JSC-1 cells maintain EBV indicates that EBV contributes a function to these dually infected PEL cells required for their growth under the initial conditions of limiting dilution.
Analyzing the inhibition of colony-formation of EBV+, KSHV+ PEL cells
The reduced colony-forming ability resulting from inhibiting EBNA1 in EBV-infected PEL cells parallels that observed in EBV-infected BL-cell lines (9). Inhibiting EBNA1 to force the loss of EBV in BL cells induces apoptosis (ibid and Dave Vereide, unpublished). We tested whether inhibiting EBNA1 in EBV+, KSHV+ PEL cells would induce apoptosis using two independent assays and found that it did not (Supplementary figure, S1). In addition, we tested whether the inhibition of colony formation might result from defects in autocrine signaling that could be rescued by co-cultivation with parental PELs cells. These co-cultivation experiments did not rescue growth of dually infected PELs in which EBNA1 was inhibited (Supplementary figure, S2).
Levels of EBNA1 are atypically low in three EBV-positive PEL cell lines
EBV-positive normal B-cells and Burkitt's lymphoma cells usually express similar, average levels of EBNA1 per cell although they vary widely in their average number of viral genomes per cell (10). Dually-infected PEL cells have average numbers of EBV genomes per cell well within the range of other EBV-positive cells but express one-fifth to one-tenth the levels of EBNA1 as found in other EBV-positive B-cells studied (Figure 3). These findings raise the possibility that efficient expression of EBNA1 is deleterious to KSHV-infected cells and that its expression is likely regulated by the KSHV-encoded protein LANA1, for example, as has been reported (11).
Figure 3. Levels of EBNA1 are atypically low in three EBV-positive PEL cell lines.

Lysates from three EBV-infected PEL cell lines JSC-1, BC-2, and BC-1 were examined by Western blotting to determine the levels of EBNA1. The intensities of the signals for EBNA1 were determined using ImageQuant software version 5.2. The numbers at the bottom of each lane in the Western blots represent the amount of EBNA1 relative to the levels found in EBV-positive 721 cells. Antibody specificity was determined using the EBV-negative cell lines BJAB and BC-3 as well as the EBV-positive cell line 721. The size of EBNA1 detected in each of these cell lines fell within the estimated 72 kD range as indicated by the size marker located in the first lane of each panel.
DBDmut and deltaUR1 also inhibit three EBV−, KSHV+ PEL cell lines from forming colonies
We tested three EBV−, KSHV+ PEL cell lines BCBL-1, BCP-1, and BC-3 to determine if DBDmut or deltaUR1 inhibited their ability to form colonies. Surprisingly, these derivatives did inhibit the colony-formation of all three cell lines (p ≤ 0.05, Wilcoxon rank sum test; Figure 4a). The inhibition of colony-formation of one of these lines, BC-3, did not correlate with an increase in apoptosis or a defect in autocrine signaling (Supplementary figures, S1 and S2). There was no obvious, detectable decrease in the number of KSHV genomes per cell in DBDmut-infected BC-3 cells 10 days after infection as measured by FISH (Figure 4b).
Figure 4. EBV(−), KSHV(+) PEL cell lines are inhibited in their ability to form colonies by derivatives of EBNA1 and only BC-3 cells with mutant forms of these derivatives survive to form colonies.

(a) BCBL-1, BCP-1, and BC-3 cells were infected with retroviruses expressing DBDmut, deltaUR1, or the control retrovirus. Infected cells were sorted by FACs for the highest levels of eGFP, 5 − 10% of the total population, 48 hours post-infection into 96-well plates. Their colony-forming efficiency was determined using a Poisson Distribution. (b) KSHV DNA was detected in EBV−, KSHV+ BC-3 cells by FISH and no difference in the number of KSHV genomes in the presence or absence of deltaUR1 expression 10 days post-infection was observed. (c) Six lysates of Survivors from deltaUR1-BC-3 clones were analyzed by Western blotting and compared with a corresponding Parental population. Lysates from 3 × 104 cells were loaded for each lane representing Survivors and 1 × 104 cells for the Parent population. 1 × 104 (1x) and 3 × 104 (3x) 721 cells were loaded as an EBNA1-positive control and tubulin was included as a loading control.
DBDmut and deltaUR1 are themselves mutated in BC-3 cells that form colonies in their presence
Six surviving BC-3 clones expressing deltaUR1 were expanded from colony-formation assays and the deltaUR1 expressed in them was characterized. Five of these clones expressed little to undetectable levels of deltaUR1 which likely explained their ability to form colonies. All of these clones expressed mutant forms of deltaUR1 that resembled the truncated deltaUR1 mutants detected in the clones of JSC-1 cells that survived the colony-formation assays (Figure 4c). We also examined the DNA sequences of the derivatives of EBNA1 found in the two BC-3 clones expressing DBDmut that survived in colony-forming experiments. Sequencing revealed the same single adenine insertion in the EBNA1 sequence resulting in a frame-shift identified in the surviving deltaUR1-infected JSC-1 clones. These findings demonstrate that the tested EBV−, KSHV+ cells that survive limiting dilution express the derivatives of EBNA1 inefficiently and/or express ones defective in binding DNA.
Discussion
These experiments reveal a necessary role for EBV in supporting proliferation of dually infected PEL cells. They demonstrate that inhibiting EBNA1 to force the loss of EBV from these cells inhibits their proliferation under limiting dilution (Figure 1). When the EBNA1-derivatives, DBDmut or deltaUR1, were introduced into EBV+, KSHV+ PEL cells, most of the cells failed to grow under limiting dilution as measured by colony-forming assays. The infrequent surviving clones expressed non-functional levels of deltaUR1 or non-functional mutant forms of either DBDmut or deltaUR1 (Figure 2). These non-functional derivatives could not bind DNA. All 14 clones expanded from the JSC-1 survivors of colony-formation assays maintained EBV demonstrating a requirement for EBV for dually infected PELs to proliferate under limiting dilution (Figure 2c and Supplementary Table 1).
We also tested the effects of efficient expression of the two derivatives of EBNA1 in five EBV−, KSHV-hematopoietic cell lines and three EBV−, KSHV+ PEL cell lines on their growth under limiting dilution. The derivatives of EBNA1 had no effect on the growth of the virus-negative cells (Figure 1c and reference 9) but did inhibit colony formation of the KSHV+ PELs (Figure 4). Those KSHV+ PEL cells that survived these assays either expressed negligible levels of the EBNA1-derivatives or mutants of them defective in binding DNA. We have found that EBNA1 binds KSHV DNA specifically at multiple sites (Dresang and Sugden, unpublished observations) and hypothesize that the efficient expression of DNA-binding derivatives of EBNA1 on binding KSHV DNA inhibit one or more functions important for the proliferation of KSHV+ PELs. It is possible that the binding of these derivatives of EBNA1 to cellular DNA could affect the growth of KSHV+ PEL cells, but we find this a less likely hypothesis because these derivatives have no effects on five EBV−, KSHV− hematopoietic cell lines. The hypothesis that EBNA1's efficient binding to the KSHV genome inhibits proliferation of PEL cells is also consistent with the atypically low levels of EBNA1 found in dually infected PEL cells (Figure 3; (11)). These dually infected cells likely require low levels of EBNA1 to avoid its inhibiting functions encoded by KSHV but sufficient EBNA1 to maintain their EBV genomes on which they also depend for their growth. We hypothesize that efficient expression of the mutant derivatives of EBNA1 and their ability to bind DNA site-specifically are responsible for the loss of colony-forming ability of EBV+ and EBV− PEL cells. By extension, it is likely that the efficient expression of wild-type EBNA1 which binds DNA as do the derivatives of it that we used would similarly result in a reduced ability of PEL cells to form colonies. This likelihood is difficult to test, however, because efficient expression of intact EBNA1 is often toxic to cells while the derivatives of it we used are not (9).
At first glance our observation that the expression of two derivatives of EBNA1 inhibits EBV−, KSHV+ PELs from forming colonies might be inconsistent with our conclusion that EBV is necessary for EBV+, KSHV+ PELs to form colonies. For example, these derivatives may act in the latter cells by inhibiting KSHV gene expression as we speculate it does in EBV−, KSHV+ PELs. In fact, we think this notion is likely correct because the deltaUR1 derivative of EBNA1 blocks transcription but not replication of EBV and may force the loss of EBV indirectly. However, our finding that all clones of EBV+, KSHV+ PELs initially selected to express these derivatives can only grow under limiting dilution if the derivatives are non-functional and EBV is retained in the cells indicates that EBV is necessary for the proliferation of this class of PELs.
Multiple studies indicate that KSHV is also necessary to sustain dually infected PELs. For example, the inhibition of LANA1 with shRNAs in JSC-1 cells inhibits their proliferation while the inhibition of v-cyclin and v-flip induces apoptosis in them (12). The inhibition of the vIRF-3 gene of KSHV by shRNA also induces apoptosis in JSC-1 cells (13). The introduction of intact EBV into an EBV−, KSHV+ PEL, BC-3, renders this experimentally, dually infected PEL more tumorigenic in SCID mice than is the parental, singly infected PEL (14). All of these observations when coupled with our finding that the forced loss of EBV from naturally, dually infected PELs inhibits their proliferation following limiting dilution indicates that both KSHV and EBV contribute to the maintenance of these dually infected lymphomas.
Acknowledgements
We thank David Vereide for his valuable insights and critical reading of this manuscript. Our research was supported by grants from the NIH: CA22443 and T32 CA09135. Bill Sugden is an American Cancer Society Research Professor.
Footnotes
Publisher's Disclaimer: This PDF receipt will only be used as the basis for generating PubMed Central (PMC) documents. PMC documents will be made available for review after conversion (approx. 2−3 weeks time). Any corrections that need to be made will be done at that time. No materials will be released to PMC without the approval of an author. Only the PMC documents will appear on PubMed Central -- this PDF Receipt will not appear on PubMed Central.
References
- 1.Jenner RG, Boshoff C. The molecular pathology of Kaposi's sarcoma-associated herpesvirus. Biochim Biophys Acta. 2002;1602:1–22. doi: 10.1016/s0304-419x(01)00040-3. [DOI] [PubMed] [Google Scholar]
- 2.Walts AE, Shintaku IP, Said JW. Diagnosis of malignant lymphoma in effusions from patients with AIDS by gene rearrangement. Am J Clin Pathol. 1990;94:170–5. doi: 10.1093/ajcp/94.2.170. [DOI] [PubMed] [Google Scholar]
- 3.Knowles DM, Inghirami G, Ubriaco A, Dalla-Favera R. Molecular genetic analysis of three AIDS-associated neoplasms of uncertain lineage demonstrates their B-cell derivation and the possible pathogenetic role of the Epstein-Barr virus. Blood. 1989;73:792–9. [PubMed] [Google Scholar]
- 4.Green I, Espiritu E, Ladanyi M, et al. Primary lymphomatous effusions in AIDS: a morphological, immunophenotypic, and molecular study. Mod Pathol. 1995;8:39–45. [PubMed] [Google Scholar]
- 5.Cesarman E, Chang Y, Moore PS, Said JW, Knowles DM. Kaposi's sarcoma-associated herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–91. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 6.Lee DY, Sugden B. The latent membrane protein 1 oncogene modifies B-cell physiology by regulating autophagy. Oncogene. Epub. 2007 Nov 26; doi: 10.1038/sj.onc.1210946. [DOI] [PubMed] [Google Scholar]
- 7.Nanbo A, Sugden A, Sugden B. The coupling of synthesis and partitioning of EBV's plasmid replicon is revealed in live cells. EMBO J. 2007;26:4252–62. doi: 10.1038/sj.emboj.7601853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Russo JJ, Bohenzky RA, Chien MC, et al. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc Natl Acad Sci USA. 1996;93:14862–7. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kennedy G, Komano J, Sugden B. Epstein-Barr virus provides a survival factor to Burkitt's lymphomas. Proc Natl Acad Sci USA. 2003;100:14269–74. doi: 10.1073/pnas.2336099100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sternas L, Middleton T, Sugden B. The average number of molecules of Epstein-Barr nuclear antigen 1 per cell does not correlate with the average number of Epstein-Barr virus (EBV) DNA molecules per cell among different clones of EBV-immortalized cells. J Virol. 1990;64:2407–10. doi: 10.1128/jvi.64.5.2407-2410.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krithivas A, Young DB, Liao G, Greene D, Hayward SD. Human herpesvirus 8 LANA interacts with proteins of the mSin3 corepressor complex and negatively regulates Epstein-Barr virus gene expression in dually infected PEL cells. J Virol. 2000;74:9637–45. doi: 10.1128/jvi.74.20.9637-9645.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Godfrey A, Anderson J, Papanastasiou A, Takeuchi Y, Boshoff C. Inhibiting primary effusion lymphoma by lentiviral vectors encoding short hairpin RNA. Blood. 2005;105:2510–8. doi: 10.1182/blood-2004-08-3052. [DOI] [PubMed] [Google Scholar]
- 13.Weis E, Yasuko M, Hahn A, et al. The viral interferon-regulatory factor-3 is required for the survival of KSHV-infected primary effusion lymphoma cells. Blood. 2008;111:320–7. doi: 10.1182/blood-2007-05-092288. [DOI] [PubMed] [Google Scholar]
- 14.Trivedi P, Takazawa K, Zompetta C. Infection of HHV-8+ primary effusion lymphoma cells with a recombinant Epstein-Barr Virus leads to restricted EBV latency, altered phenotype, and increased tumorigenicity without affecting TCL1 expression. Blood. 2004;103:313–6. doi: 10.1182/blood-2003-05-1710. [DOI] [PubMed] [Google Scholar]
- 15.Lindner SE, Sugden B. The plasmid replicon of Epstein-Barr Virus: Mechanistic insights into efficient, licensed, extrachromosomal replication in human cells. Plasmid. 2007;58:1–12. doi: 10.1016/j.plasmid.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]