Abstract
Recent evidence indicates that mitochondrial homeostasis is critical for myelination and maintenance of peripheral nerve function. Mice lacking the metabolic transcriptional coactivator peroxisome proliferator activated receptor γ coactivator 1α (PGC-1α) show reductions in expression of myelin-related proteins and exhibit myelin-associated lesions, so we identified PGC-1α target genes in Schwann cells (SCs) in vitro to determine potential roles for PGC-1α in glia and tested whether PGC-1α was sufficient for SC differentiation and myelination. Forskolin-induced differentiation was associated with an upregulation of PGC-1α mRNA and protein, and while overexpression of PGC-1α upregulated genes such as manganese superoxide dismutase and estrogen-related receptor α, it was not sufficient for induction of differentiation. Both PGC-1α overexpression and forskolin exposure caused an increase in the mitochondrial fusion-related protein Mitofusin 1. These studies suggest that PGC-1α might be a potential target to promote mitochondrial stability during differentiation and myelination.
INTRODUCTION
Recent evidence suggests that the maintenance of mitochondrial stability is critical for normal peripheral nerve function and neuronal-glial interactions. Charcot-Marie Tooth (CMT) disease, a devastating illness characterized by progressive peripheral nerve fiber deterioration, paralysis, and muscular atrophy, is frequently associated with mutations in genes controlling mitochondrial homeostasis (especially CMT2a, reviewed in [29]). Mitochondrial homeostasis requires a delicate balance between fission and fusion, the disturbance of which can lead to decreased cellular respiration, decreased cell growth, and cell death [2, 7, 8, 28]. Mitochondrial fusion is orchestrated by several groups of proteins, including the transcription factor estrogen-related receptor α (ERRα; [21]) and the mitofusins (Mfn1, Mfn2; [5, 18]), which regulate mitochondrial network formation, glucose and oxygen consumption, and mitochondrial membrane potential [1]. The process of mitochondrial proliferation (biogenesis), on the other hand, is associated with increases in the transcription factors nuclear respiratory 1 (NRF-1) and nuclear respiratory factor 2 (GA-binding protein α and β) and the expression of proteins involved in mitochondrial DNA transcription and oxidative phosphorylation [20, 26]. The ERRα and NRF-1/NRF-2-dependent pathways function in parallel to influence mitochondrial gene expression and function [25].
A series of studies have demonstrated that the transcriptional coactivator peroxisome proliferator activated receptor γ coactivator 1α (PGC-1α) can regulate mitochondrial function, ERRα and NRF-1 activity, and mitofusin expression [4, 14, 21]. Interestingly, myelin abnormalities have been observed in brains from mice lacking PGC-1α [17], but to date, there is no information about the roles of PGC-1α in glia. In the liver, PGC-1α expression is induced by glucocorticoids and agents that activate the cAMP/protein kinase A pathway, leading to the stimulation of gluconeogenesis via activation of the transcription factor CREB (cyclic AMP responsive element binding protein [10, 27]). In the peripheral nerve, the protein kinase A/CREB pathway is crucial for the differentiation of Schwann cells (SCs) [12], so it is possible that PGC-1α is involved in SC differentiation.
In this study, we hypothesized that differentiation of SCs by activation of the protein kinase A pathway involves the upregulation of PGC-1α and PGC-1α-target genes and sought to determine whether differentiation and/or mitochondrial pathways are regulated by PGC-1α in SCs.
EXPERIMENTAL METHODS
Primary Schwann Cell Culture
SCs were isolated from postnatal day 2 rat sciatic nerve and cultured as described [3] with modifications. Briefly, sciatic nerves were digested with Trypsin and collagenase and plated in DMEM with 10% fetal bovine serum (Gibco/Invitrogen, Carlsbad, CA). Cells were fed for 3 days with 10 μM cytosine arabinofurosine and fibroblasts were eliminated using mouse IgM anti-Thy 1.1 hybridoma supernatant (clone T11/D7/e2 from Bob Hyman, the Salk Institute) and rabbit complement (Research Diagnostics, Inc., Concord, MA). Cells were maintained and passaged in low glucose DMEM, 10% fetal bovine serum, 1% penicillin-streptomycin, forskolin (2μM), and pituitary extract (20 μg/ml). Only passages 4–7 were used for experiments. For all experiments involving forskolin-induced differentiation, cells were harvested, plated at ~80% confluency, and forskolin-starved for 48–72 hours before addition of media containing forskolin. Dideoxyforskolin (10 μM) was used as a negative control in differentiation experiments. All animal procedures were approved by the University of Michigan and Ann Arbor VA Medical Center committees for use of laboratory animals. Reagents are from Sigma-Aldrich (St. Louis, MO) unless otherwise noted.
Adenoviral transfection
PGC-1α adenovirus was provided by Bruce M. Spiegelman (Dana Farber Cancer Research Center, Harvard University, [13, 15]), and was purified and amplified at the University of Michigan Cancer Center Vector Core (director, Thomas Lanigan). Cells were exposed for 48 hours to adenovirus containing the gene for GFP (5.1×1010 pfu/ml) or GFP and PGC-1α (1.4×1011 pfu/ml; the gene for GFP was in tandem with the PGC-1α gene [13]). The optimal multiplicity of infection (MOI) was determined to be 200:1 based on expression analysis and evidence of cell death at higher concentrations.
Quantitative RT-PCR
RNA was isolated from cells using the Trizol method, according to manufacturers instructions (Invitrogen Corporation, Carlsbad, CA), and RT-PCR was performed as previously described using Taqman gene expression assays [9]. Primer/probe sets (Applied Biosystems) included β-actin (Rn00667869), myelin protein zero (MPZ; Rn00566746), peripheral myelin protein 22 (PMP22; Rn00566835), Lamininβ2 (Rn00564264), PGC-1α (rat: Rn00580241; mouse: Mn00447183), cyclin D1 (Rn00432359), estrogen-related receptor (ERRα; Rn01479215), mitochondrial transcription factor A (TFAM; Rn00580051), mitofusin 1 (Mfn1; Rn00594496), mitofusin 2 (Mfn2; Rn00500120), superoxide dismutase 2 (MnSOD; Rn00566942), glutathione peroxidase (GPx; Rn00577994), and NRF-1 (Forward: TCTATCCGAAAGAGACAGCAGACA; Reverse: TCCCACTCGTGTCGTATATTCATCT; Probe: 6FAM-ttgcttcggaaactca-TAMRA). As a negative control for the RT reaction, reverse transcriptase was omitted in the reaction mix. For negative controls for the PCR reaction, either the primer sets or the cDNA were omitted from the reactions. Relative concentrations of cDNA were calculated with comparison to a standard curve made with dilutions (1:5, 1:10, 1:20) of cDNA from the sample with the highest dose in that particular experiment (calibrator method). Values were normalized to actin-β and expressed as arbitrary units or fold control, +/− standard error.
Western Blot Analysis
Cells were treated for three days with forskolin (0–10 μM), rinsed in cold PBS, and collected in RIPA buffer (150 mM NaCl, 1% Triton X-100, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate in 50 mM Tris, pH 8.0) and protease inhibitors (Complete Mini-tablets; Roche Diagnostics Corporation, Indianapolis, IN). Cell membranes were disrupted with a sonicator, and samples were spun at 14,000xg to remove cell debris. Supernatants were immediately mixed in SDS sample buffer and boiled. Protein content was determined using bovine serum albumin as a standard (Pierce Biotechnology Incorporated, Rockford, IL). Proteins were separated with SDS-PAGE electrophoresis (10 μg/lane; Invitrogen) and transferred to nitrocellulose membranes (Bio-Rad Laboratories, Hercules, CA), and Western blotting was performed as described by Cell Signaling Technology, Inc. (Danvers, MA), with slight modifications. Briefly, blocking was performed for 1 hour with 5% dry milk and 0.1% Tween in Tris-buffered saline (TBS), the membrane was washed 15 minutes with TBS, incubated with the primary antibody in 5% IgG-free BSA (Jackson Immunoresearch; West Grove, PA) overnight at 4° C, and incubated with the peroxidase-conjugated secondary antibody (Jackson Immunoresearch) in 5% milk for one hour. Signal was detected with enhanced chemiluminescence (Amersham Biosciences, Piscataway, NJ) and exposure to Biomax Kodak film (Eastman Kodak Company, Rochester, NY). Primary antibodies included mouse anti-myelin protein zero (Juan Archelos, Medical University, Graz, Austria), mouse anti-cyclin D1 (Millipore, Billerica, MA; cat#: 05-815), rabbit anti-PGC-1α (Daniel P. Kelly, Center for Cardiovascular Research, Washington University, St. Louis; [9, 13]), and mouse anti-β actin (Millipore; cat#: MAB1501R).
Immunocytochemistry
SCs (1 × 105) were plated on poly-L-lysine-coated coverslips in 24-well plates in forskolin-free media, and then treated 48 hours later with forskolin for 24 hours. Cells were fixed in warmed paraformaldehyde (4% in PBS, pH 7.4) and washed with PBS before storing at 4° C overnight. For visualization of cytoskeletal structure, cells were stained with alexa fluor 647-phalloidin for 2 hours at room temperature (0.1 μM; Molecular Probes, Eugene, OR) and mounted with Prolong anti-fade aqueous mounting media (Molecular Probes). For immunostaining, cells were blocked in PBS with 10% donkey serum (Jackson Immunoresearch, West Grove, PA) for one hour and then treated with 10 mM citrate buffer for 30 minutes at 37° C before incubation overnight with rabbit anti-mouse PGC-1α (1:10,000) and one hour at room temperature with Alexa fluor 568 donkey anti-rabbit IgG (1:2000; Molecular Probes). To rule out non-specific binding of the secondary antibody and autofluorescence, primary antibodies were replaced with species-matched non-specific IgG (Vector Labs, Burlingame, MA). In some cases, cells were counterstained with DAPI (300 nM; Molecular Probes) for 10 minutes after incubation with the secondary antibody.
Statistical Analysis
Western blotting and quantitative RT-PCR data were expressed as mean +/− standard error. Significant differences among groups were determined using T-tests. A Bonferroni correction for multiple comparisons was used for data regarding changes in gene and protein expression with increasing doses of forskolin (Figure 1). Statistical significance was set at p = 0.05.
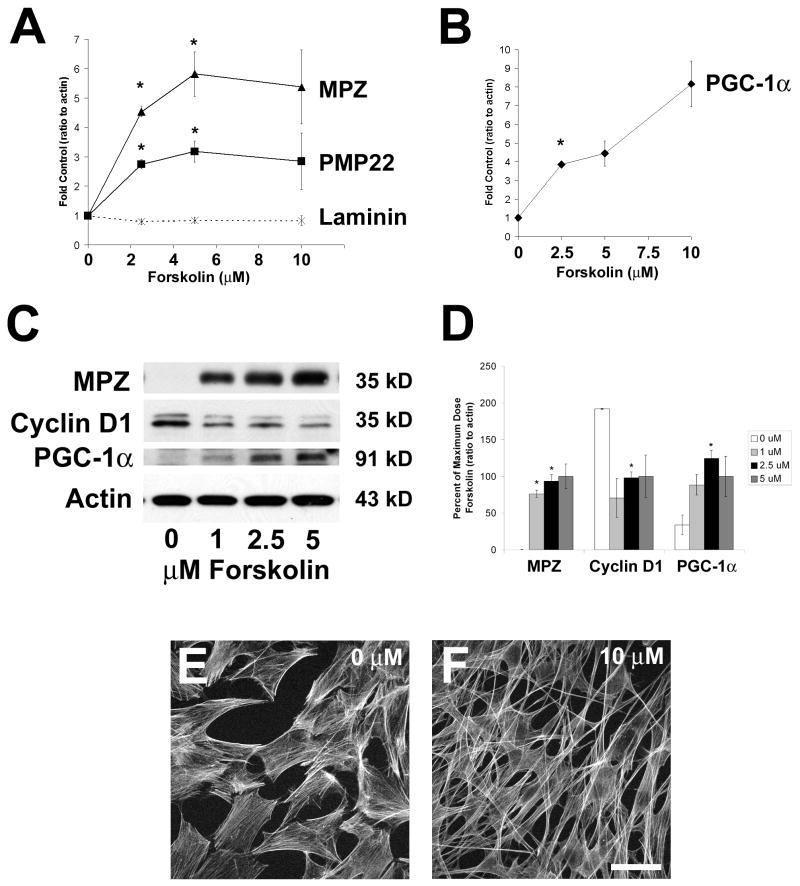
Figure 1. Forskolin-induced differentiation of primary Schwann cells (SCs) involves upregulation of PGC-1α.
A–B. SCs were treated with different doses of forskolin (0–10 μM) for 3 days and cells were collected for quantitative RT-PCR. All genes were normalized to actin content, and expressed as fold control (0 μM forskolin). There was a dose-dependent increase in expression of differentiation-related genes myelin protein zero (MPZ) and PMP22 and no increase in β2-laminin expression (A). These changes were accompanied by an increase in PGC-1α RNA expression (B). C. As detected by Western blotting, treatment of Schwann cells (SCs) with 0–5 μM forskolin for 3 days increased the expression of myelin protein zero (MPZ) and PGC-1α and reduced cyclin D1 protein expression. Immunoblotting for β-actin confirmed equal loading of all lanes. The blot shown is representative of 3 separate sample sets. D. The optical density of each band (2–3 samples per dose) was measured and normalized to actin content. Values are expressed as percent of expression at the maximum dose (5 μM) +/− standard error. With respect to untreated cells (0 μM), SCs treated with forskolin show increased expression for MPZ and PGC-1α and decreased expression for cyclin D1. E–F. Actin staining with phalloidin revealed that untreated immature SCs were trapezoidal (fibroblast-like) in shape (E) while SCs treated with 10 μM forskolin for 24 hours were elongated with spindle-like processes (F). Scale bar: 15 μm. For all data: n=3/group, T-test with Bonferroni correction, *p<0.017).
RESULTS
To study gene regulation in an in vitro model of SC differentiation, we isolated and purified primary SCs from postnatal day 2 rat sciatic nerve (see Methods). Initial experiments determined that forskolin induced proliferation when cells were less confluent, so for all differentiation experiments, cells were plated at high density (80–90% confluency). Forskolin-induced differentiation was confirmed by several methods. Based on preliminary experiments indicating that 2–3 days of forskolin treatment reduced proliferation of SCs as measured by the MTT assay (not shown), for mRNA expression analysis, we exposed cells to different doses of forskolin for 3 days (Figure 1). There was a dose-dependent increase in the expression of the myelin-related gene products myelin protein zero (MPZ) and peripheral myelin protein 22 (PMP22)(Figure 1A), with no effect on β2-laminin expression (Figure 1A).
Based on reports in the literature indicating that PGC-1α expression is stimulated by adenyl cyclase activators [10, 27] and that PGC-1α is involved in the differentiation of adipocytes [23], we hypothesized that SC differentiation also involves PGC-1α upregulation. PGC-1α mRNA expression was robustly stimulated by forskolin in a dose-dependent manner, with almost an 8-fold increase in expression noted with 10 μM forskolin treatment (Figure 1B). Forskolin was also able to dose-dependently upregulate MPZ and PGC-1α protein expression and downregulate cyclin D1 protein expression (Figure 1C, D), and forskolin-stimulated changes in protein expression were accompanied by a change in SC morphology as detected by CY-5-labeled phalloidin. Forskolin-starved SCs and their associated processes were trapezoidal in shape (Figure 1E) whereas forskolin-treated SCs showed elongated cell bodies and long, filamentous processes (Figure 1F). Dideoxyforskolin, a biologically inactive analog of forskolin, did not increase the expression of MPZ or PGC-1α (not shown).
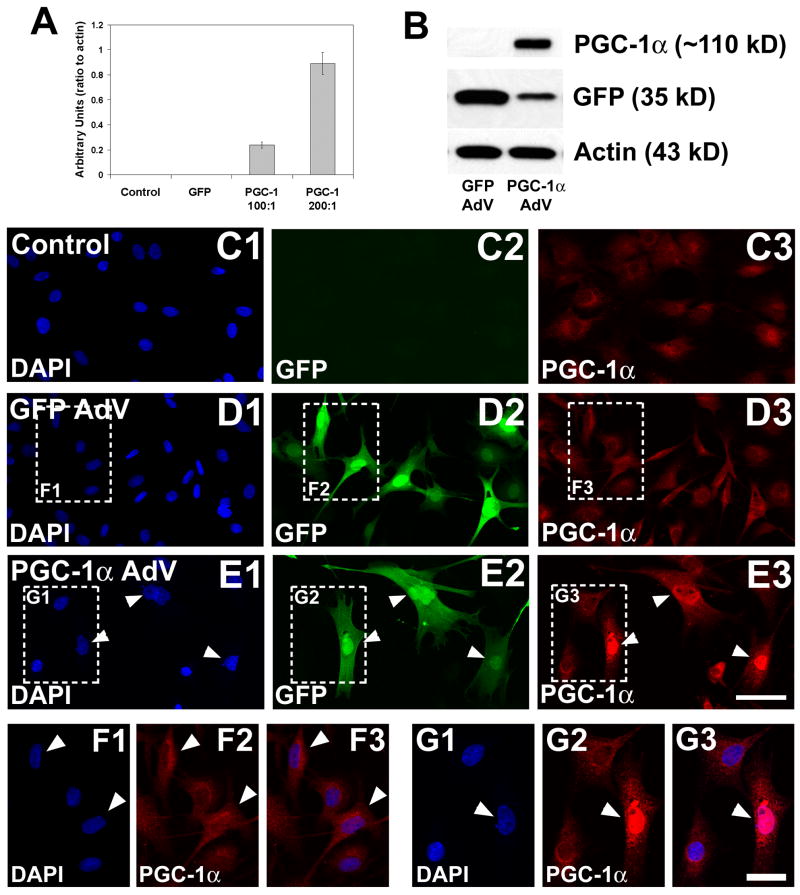
To identify the specific gene targets for PGC-1α in SCs, we transfected immature SCs with an adenovirus encoding mouse PGC-1α. The PGC-1α adenovirus contained the gene for green fluorescent protein (GFP) in tandem with PGC-1α (expressed independently of PGC-1α), allowing for confirmation of efficient transfection by RT-PCR, Western blotting, and immunocytochemistry. Mouse PGC-1α expression increased with the amount of virus added to the media [multiplicity of infection (MOI) 100:1 and 200:1; Figure 2A]. Cells treated with the PGC-1α adenovirus showed a large increase in PGC-1α protein expression (~110 kD, tagged; Figure 2B). To confirm that PGC-1α was localized to the nucleus of transfected cells, we used a PGC-1α-specific antibody and immunofluorescence and visualized the staining with confocal microscopy (Figure 2C–G). While little PGC-1α immunoreactivity was detectable in untreated immature SCs (Figure 2C) and SCs transfected with GFP alone (Figure 2D, F), PGC-1α immunoreactivity was concentrated in the nucleus of SCs transfected with GFP-PGC-1α (arrowheads, Figure 2E, G). While both GFP and PGC-1α-transfected cells showed a subtle increase in the extension of processes, possibly due to transfection alone, overexpression of PGC-1α did not cause cells to become elongated or bi/tri-polar, as seen in Figure 1F.
Figure 2. Transient transfection of Schwann cells with PGC-1α.
To determine how the upregulation of PGC-1α influenced gene expression in SCs, we transfected immature SCs with an adenovirus encoding mouse PGC-1α and green fluorescent protein (GFP), in tandem. As a transfection control, cells were treated with an adenovirus encoding GFP alone. A. The overexpression of PGC-1α was confirmed using RT-PCR; the amount of PGC-1α RNA increased with the multiplicity of infection (MOI) from 100:1 to 200:1. B. Overexpression was also confirmed with an antibody to PGC-1α. Immature SCs transfected with GFP alone showed strong expression of GFP (~35 kD) but no detectable PGC-1α, and cells transfected with GFP-PGC-1α expressed both GFP and PGC-1α (~110 kD). C–E. To confirm that the overexpressed protein was localized to the nucleus, untransfected, GFP-transfected, and GFP-PGC-1α-transfected cells were fixed and processed for PGC-1α immunocytochemistry. Control (undifferentiated) and GFP-transfected immature Schwann cells exhibited little immunoreactivity for PGC-1α (C3, D3) while immature SCs transfected with the PGC-1α adenovirus expressed GFP throughout the entire cell body (E2, arrowheads) and PGC-1α in the nucleus (E3, arrowheads). F–G. Higher magnified regions of figures D and E (boxes) demonstrate that no PGC-1α immunoreactivity overlapped with DAPI fluorescence in the GFP-transfected cells (F1–F3), but PGC-1α immunoreactivity was concentrated in DAPI-stained nuclei in PGC-1α-transfected cells. Arrowheads indicate transfected cells, as shown in D–E. Scale bars: 25 μm (C–E), 12.5 μm (F–G).
Based on the literature suggesting that there are white matter abnormalities in PGC-1α knockout mice [17], we first wanted to test whether PGC-1α alone was sufficient for differentiation. Also, in light of the fact that PGC-1α can interact with a variety of transcription factors to regulate gene transcription, we chose other sets of genes that have been shown to be regulated by PGC-1α in other cell types and measured gene expression using Q-RT-PCR (Table 1). We included genes for proliferation and differentiation (cyclin D1, MPZ, PMP22, laminin), PGC-1α-interacting transcription factors (NRF-1 and ERRα), genes involved in mitochondrial biogenesis (TFAM) and fusion (Mfn1, Mfn2), and genes for antioxidant enzymes (SOD2 and glutathione peroxidase). Transfection with PGC-1α did not influence the expression of genes involved in proliferation/differentiation; however, PGC-1α upregulated the expression of ERRα, Mfn1, and SOD2 2–4 fold (Table 1). There was also a slight but significant increase in the expression of Mfn2.
Table 1.
Comparison of PGC-1α- and forskolin-induced changes in mRNA expression.
| Fold Control | |||
|---|---|---|---|
| PGC-1α AdV | Forskolin (5 μM) | ||
| Proliferation or differentiation | Cyclin D1 | 1.1 +/− 0.2 | 0.6 +/− 0.1 * |
| MPZ | 0.8 +/− 0.1 | 5.8 +/− 0.8 * | |
| PMP22 | 0.9 +/− 0.1 | 3.2 +/− 0.4 ** | |
| Laminin | 1.2 +/− 0.1 | 0.8 +/− 0.1 | |
| PGC-1α-interacting transcription factors | NRF-1 | 1.1 +/− 0.1 | 1.0 +/− 0.1 |
| ERRα | 4.0 +/− 0.6 * | 0.5 +/− 0.1 | |
| Mitochondrial biogenesis and fusion | TFAM | 1.4 +/− 0.2 | 1.2 +/− 0.1 |
| Mfn1 | 2.3 +/− 0.3 * | 2.2 +/− 0.2 ** | |
| Mfn2 | 1.8 +/− 0.1 * | 0.8 +/− 0.1 | |
| Antioxidant enzymes | SOD2 | 4.9 +/− 0.8 ** | 0.8 +/− 0.1 |
| GPx | 1.2 +/− 0.1 | 0.8 +/− 0.1 | |
Immature Schwann cells were treated with either GFP or GFP-PGC-1α-expressing adenovirus for 48 hours (MOI: 200:1) or with 5 μM forskolin for 3 days, and RNA was isolated and reverse transcribed for quantitative PCR using Taqman primer/probe sets. Results are grouped by gene function, and data are expressed as fold of GFP or 0 μM forskolin control, with normalization to β-actin. (n=3/group,
p < 0.05,
p < 0.01, T-test).
To determine whether similar genes were regulated by forskolin-stimulated expression of endogenous PGC-1α, we evaluated the expression of PGC-1α-responsive genes after 3 days of forskolin exposure (Table 1). Interestingly, the only gene increased with forskolin treatment was Mfn1 (2.2 +/− 0.2 fold control; n = 3/group, T-test, p = 0.002).
DISCUSSION
In this paper, we present the first evidence that the differentiation of SCs in vitro is associated with an increase in the expression of PGC-1α and Mfn1. Outside the nervous system, PGC-1α is involved in a variety of tissue and cell-specific responses to demands for energy during development and in adulthood [14]. For example, PGC-1α plays a role in mitochondrial biogenesis during adipocyte differentiation and adaptive thermogenesis [11, 24], and PGC-1α can also specify fiber type in muscle [16]. With respect to glia, myelin abnormalities have been observed in brains from mice lacking PGC-1α; in total brain homogenates, there is reduced expression of myelin-associated oligodendrocytic basic protein (MOBP) and the appearance of myelin-associated lesions in the striatum [17]. Our data indicate that PGC-1α alone is not sufficient for SC differentiation or induction of myelination; while the processes governing differentiation and myelination in the central nervous system differ from those in the peripheral nervous system, it is possible that the white matter abnormalities in the PGC-1α knockout animals result from a deficiency in the PGC-1α-target gene Mfn1. Preliminary analyses performed in our laboratory indicate that nerve conduction velocities are reduced in mice lacking PGC-1α (data not shown), but further testing is required to determine whether this is due to a disruption in Mfn1 expression and mitochondrial integrity.
In light of these data, it is interesting to consider the potential role for Mfn1 in the process of differentiation. Changes in mitochondrial morphology have been shown to accompany differentiation in other cell types, such as muscle and liver cells. During the process of differentiation in these cells, mitochondria change from small and spherical to long and filamentous, possibly allowing for the transport of biochemical substrates throughout the cell body (reviewed in [6]). In fact, overexpression of mitochondrial fusion proteins can promote the formation of long, tubular networks of mitochondria [19], so it is possible that PGC-1α and Mfn1 are involved in maintaining mitochondrial integrity during differentiation.
In addition to a change in Mfn1 expression, PGC-1α overexpression caused an increase in expression of ERRα, Mfn2, and SOD2, as has been observed for other cell types [4, 22]. While these genes were not regulated by forskolin, these results suggest that inducing endogenous PGC-1α by other mechanisms in glial cells may regulate mitochondrial function and/or antioxidant defense. Thus, PGC-1α may be a good target to promote mitochondrial fusion and cell survival in situations in which mitochondrial integrity is compromised (i.e. CMT2a). Further experiments are needed to investigate this possibility.
Acknowledgments
We would like to thank Ed Kim for isolation and purification of primary Schwann cell cultures. This work was supported by NRSA awards T32 NS07222 and F32 NS049863 from NINDS (RMC), NIH NS42056, the Office of Research Development (Medical Research Service), the Department of Veterans Affairs, and the Juvenile Diabetes Research Foundation Center for the Study of Complications in Diabetes (JWR). Fluorescence microscopy was performed at the Morphology and Image Analysis Core of the Michigan Diabetes Research and Training Center (NIH5P60 DK20572).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bach D, Pich S, Soriano FX, Vega N, Baumgartner B, Oriola J, Daugaard JR, Lloberas J, Camps M, Zierath JR, Rabasa-Lhoret R, Wallberg-Henriksson H, Laville M, Palacin M, Vidal H, Rivera F, Brand M, Zorzano A. Mitofusin-2 determines mitochondrial network architecture and mitochondrial metabolism. A novel regulatory mechanism altered in obesity. J Biol Chem. 2003;278:17190–7. doi: 10.1074/jbc.M212754200. Epub 2003 Feb 21. [DOI] [PubMed] [Google Scholar]
- 2.Barsoum MJ, Yuan H, Gerencser AA, Liot G, Kushnareva Y, Graber S, Kovacs I, Lee WD, Waggoner J, Cui J, White AD, Bossy B, Martinou JC, Youle RJ, Lipton SA, Ellisman MH, Perkins GA, Bossy-Wetzel E. Nitric oxide-induced mitochondrial fission is regulated by dynamin-related GTPases in neurons. Embo J. 2006;25:3900–11. doi: 10.1038/sj.emboj.7601253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brockes JP, Fields KL, Raff MC. Studies on cultured rat Schwann cells. I. Establishment of purified populations from cultures of peripheral nerve. Brain Res. 1979;165:105–18. doi: 10.1016/0006-8993(79)90048-9. [DOI] [PubMed] [Google Scholar]
- 4.Cartoni R, Leger B, Hock MB, Praz M, Crettenand A, Pich S, Ziltener JL, Luthi F, Deriaz O, Zorzano A, Gobelet C, Kralli A, Russell AP. Mitofusins 1/2 and ERRalpha expression are increased in human skeletal muscle after physical exercise. J Physiol. 2005;567:349–58. doi: 10.1113/jphysiol.2005.092031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell. 2006;125:1241–52. doi: 10.1016/j.cell.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 6.Chen H, Chan DC. Mitochondrial dynamics in mammals. Curr Top Dev Biol. 2004;59:119–44. doi: 10.1016/S0070-2153(04)59005-1. [DOI] [PubMed] [Google Scholar]
- 7.Chen H, Chomyn A, Chan DC. Disruption of fusion results in mitochondrial heterogeneity and dysfunction. J Biol Chem. 2005;280:26185–92. doi: 10.1074/jbc.M503062200. [DOI] [PubMed] [Google Scholar]
- 8.Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160:189–200. doi: 10.1083/jcb.200211046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cowell RM, Blake KR, Russell JW. Localization of the transcriptional coactivator PGC-1alpha to GABAergic neurons during maturation of the rat brain. J Comp Neurol. 2007;502:1–18. doi: 10.1002/cne.21211. [DOI] [PubMed] [Google Scholar]
- 10.Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, Spiegelman B, Montminy M. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature. 2001;413:179–83. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- 11.Ijichi N, Ikeda K, Horie-Inoue K, Yagi K, Okazaki Y, Inoue S. Estrogen-related receptor alpha modulates the expression of adipogenesis-related genes during adipocyte differentiation. Biochem Biophys Res Commun. 2007;358:813–8. doi: 10.1016/j.bbrc.2007.04.209. [DOI] [PubMed] [Google Scholar]
- 12.Jessen KR, Mirsky R. Schwann cell precursors and their development. Glia. 1991;4:185–94. doi: 10.1002/glia.440040210. [DOI] [PubMed] [Google Scholar]
- 13.Lehman JJ, Barger PM, Kovacs A, Saffitz JE, Medeiros DM, Kelly DP. Peroxisome proliferator-activated receptor gamma coactivator-1 promotes cardiac mitochondrial biogenesis. J Clin Invest. 2000;106:847–56. doi: 10.1172/JCI10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–70. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 15.Lin J, Tarr PT, Yang R, Rhee J, Puigserver P, Newgard CB, Spiegelman BM. PGC-1beta in the regulation of hepatic glucose and energy metabolism. J Biol Chem. 2003;278:30843–8. doi: 10.1074/jbc.M303643200. [DOI] [PubMed] [Google Scholar]
- 16.Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, Michael LF, Puigserver P, Isotani E, Olson EN, Lowell BB, Bassel-Duby R, Spiegelman BM. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 17.Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, Mootha VK, Jager S, Vianna CR, Reznick RM, Cui L, Manieri M, Donovan MX, Wu Z, Cooper MP, Fan MC, Rohas LM, Zavacki AM, Cinti S, Shulman GI, Lowell BB, Krainc D, Spiegelman BM. Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1alpha null mice. Cell. 2004;119:121–35. doi: 10.1016/j.cell.2004.09.013. [DOI] [PubMed] [Google Scholar]
- 18.Olichon A, Guillou E, Delettre C, Landes T, Arnaune-Pelloquin L, Emorine LJ, Mils V, Daloyau M, Hamel C, Amati-Bonneau P, Bonneau D, Reynier P, Lenaers G, Belenguer P. Mitochondrial dynamics and disease, OPA1. Biochim Biophys Acta. 2006;1763:500–9. doi: 10.1016/j.bbamcr.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Santel A, Fuller MT. Control of mitochondrial morphology by a human mitofusin. J Cell Sci. 2001;114:867–74. doi: 10.1242/jcs.114.5.867. [DOI] [PubMed] [Google Scholar]
- 20.Scarpulla RC. Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochim Biophys Acta. 2002;1576:1–14. doi: 10.1016/s0167-4781(02)00343-3. [DOI] [PubMed] [Google Scholar]
- 21.Soriano FX, Liesa M, Bach D, Chan DC, Palacin M, Zorzano A. Evidence for a Mitochondrial Regulatory Pathway Defined by Peroxisome Proliferator-Activated Receptor-{gamma} Coactivator-1{alpha}, Estrogen-Related Receptor-{alpha}, and Mitofusin 2. Diabetes. 2006;55:1783–91. doi: 10.2337/db05-0509. [DOI] [PubMed] [Google Scholar]
- 22.St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jager S, Handschin C, Zheng K, Lin J, Yang W, Simon DK, Bachoo R, Spiegelman BM. Suppression of Reactive Oxygen Species and Neurodegeneration by the PGC-1 Transcriptional Coactivators. Cell. 2006;127:397–408. doi: 10.1016/j.cell.2006.09.024. [DOI] [PubMed] [Google Scholar]
- 23.Tiraby C, Tavernier G, Lefort C, Larrouy D, Bouillaud F, Ricquier D, Langin D. Acquirement of brown fat cell features by human white adipocytes. J Biol Chem. 2003;278:33370–6. doi: 10.1074/jbc.M305235200. [DOI] [PubMed] [Google Scholar]
- 24.Uldry M, Yang W, St-Pierre J, Lin J, Seale P, Spiegelman BM. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006;3:333–41. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 25.Villena JA, Hock MB, Chang WY, Barcas JE, Giguere V, Kralli A. Orphan nuclear receptor estrogen-related receptor alpha is essential for adaptive thermogenesis. Proc Natl Acad Sci U S A. 2007;104:1418–23. doi: 10.1073/pnas.0607696104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Villena JA, Vinas O, Mampel T, Iglesias R, Giralt M, Villarroya F. Regulation of mitochondrial biogenesis in brown adipose tissue: nuclear respiratory factor-2/GA-binding protein is responsible for the transcriptional regulation of the gene for the mitochondrial ATP synthase beta subunit. Biochem J. 1998;331:121–7. doi: 10.1042/bj3310121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK, Newgard CB, Spiegelman BM. Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature. 2001;413:131–8. doi: 10.1038/35093050. [DOI] [PubMed] [Google Scholar]
- 28.Yuan H, Gerencser AA, Liot G, Lipton SA, Ellisman M, Perkins GA, Bossy-Wetzel E. Mitochondrial fission is an upstream and required event for bax foci formation in response to nitric oxide in cortical neurons. Cell Death Differ. 2007;14:462–71. doi: 10.1038/sj.cdd.4402046. [DOI] [PubMed] [Google Scholar]
- 29.Zuchner S, Vance JM. Emerging pathways for hereditary axonopathies. J Mol Med. 2005;83:935–43. doi: 10.1007/s00109-005-0694-9. [DOI] [PubMed] [Google Scholar]