Abstract
The feasibility of employing nonintegrating lentiviral vectors has been demonstrated by recent studies showing the ability of nonintegrating lentiviral vectors to maintain transgene expression in vitro and in vivo. Furthermore, HIV-1 vectors packaged with a mutated integrase were able to correct retinal disease in a mouse model. Interestingly, these results differ from earlier studies in which first-generation nonintegrating lentiviral vectors yielded insignificant levels of transduction. However, to date a rigorous characterization of transgene expression from the currently used self-inactivating (SIN) nonintegrating lentiviral vectors has not been published. Here we characterize transgene expression from SIN nonintegrating lentiviral vectors. Overall, we found that nonintegrating vectors express transgenes at a significantly lower level than their integrating counterparts. Expression from nonintegrating vectors was improved upon introducing a longer deletion in the vector’s U3 region. A unique shuttle-vector assay indicated that the relative abundance of the different episomal forms was not altered by the longer U3 deletion. Interestingly, the longer U3 deletion did not enhance expression in the corpus callosum of the rat brain, suggesting that the extent of silencing of episomal transcription is influenced by tissue-specific factors. Finally, and for the first time, episomal expression in the mouse liver was potent and sustained.
INTRODUCTION
A number of recent clinical trials have demonstrated the efficacy of retroviral vectors as a means of delivering therapeutic transgenes to patients [1–3]. However, retroviral vectors carry an inherent risk of insertional mutagenesis, leading to oncogenic transformation of target cells. This risk has been documented in animal models [4] and has been borne out in a recent clinical trial, in which four out of ten patients receiving retrovirally corrected cells as treatment for X-linked severe combined immunodeficiency (X-SCID) developed leukemia [5, 6].
Employing nonintegrating, HIV-1-based vectors provides a logical strategy to alleviate the risk of insertional mutagenesis associated with traditional retroviral vectors. This approach is premised on the fact that retroviral episomal forms were observed to constitute the vast majority of viral DNA following transduction, and were found stable in nondividing cells [7–12]. The episomal viral genomes appear in four forms: linear episomes, which may also serve as precursors to integration, and 2-LTR, 1-LTR, and aberrant circular episomes, which have been suggested to form through host-mediated DNA repair, incomplete reverse transcription, and autointegration, respectively [13–16]. Current findings in the field of retroviral vectors demonstrate that nonintegrating lentiviral vectors are capable of mediating significant gene expression and imparting clinically relevant correction of genetic disease models in vivo [17, 18]. These results are seemingly not in line with previous findings in retrovirus biology, which indicated that integration is essential for HIV replication [19, 20] and that integrase-deficient HIV is capable of relatively low levels of viral gene expression [21–23]. Furthermore, nonintegrating first-generation lentiviral vectors also displayed low to negligible transgene expression [24–27], Importantly, subsequently developed SIN vectors demonstrated measurable episomal transgene expression in vitro and in vivo [18, 28–30]. The improved efficiency of episomal expression exhibited by U3-truncated SIN vectors suggests that cis-acting sequences in the U3 may negatively regulate expression. Indeed, a sequence element in the 5’ U3 region has been shown to reduce both LTR-driven and internally promoted expression [31–33]. However, the level and efficiency of episomal lentiviral expression has not been rigorously characterized.
Here, we present an analysis of episomal expression from nonintegrating vectors. We report that, while the expression levels of episomal lentiviral genomes remained well below that of integrated lentiviral provirus, a long U3 deletion significantly improved expression from nonintegrating lentiviral vectors, both in vitro and in vivo. The deletion, however, did not change the relative abundances of the four episomal forms in transduced cells, suggesting that the increase in expression cannot be attributed to a distinct episomal vector form. Furthermore, the expression-enhancing effect of the U3 deletion showed tissue specificity in the rat brain, which is in line with earlier studies demonstrating cell type-dependent gene expression levels from nonintegrating HIV-1 [22]. Finally, we report for the first time that episomal HIV-1 vectors achieved strong and sustained transgene expression in the mouse liver. These findings demonstrate that nonintegrating lentiviral vectors may provide an effective means of delivery of therapeutic transgenes to nondividing and slowly dividing cells in vitro and in vivo.
RESULTS
Increased in vitro episomal expression from a vector with a large U3 deletion
Several groups recently demonstrated notable in vitro and in vivo expression from nonintegrating, self-inactivating (SIN) lentiviral vectors [17, 18, 28, 30, 34]. These results are not in line with previous studies on integrase-deficient lentiviral vectors, which exhibited low to negligible viral gene expression from genomes with full-length U3 regions [24, 26, 27]. Furthermore, as shown below, an HIV vector containing a short SIN deletion produced comparatively low episomal expression. To investigate the possibility that a larger U3 deletion on a SIN vector could enhance its expression, two SIN vectors were made, one bearing a short U3 deletion (vTK113, deleted from –7 to –141, as in the vector described by Miyoshi et al. [35]), and one containing a larger U3 deletion (vTK945, deleted from –48 to –396, thus retaining the TATA box)(Fig. 1a)(deletions outlined in Fig. S1, see Fig. S2 for a full list of vectors used for this study); both vectors were packaged with functional or deficient integrase and used to transduce 293T cells (Fig. 1b). To minimize the risk of expressing saturating quantities of GFP, thereby confounding subsequent GFP expression analysis, cells were transduced with decreasing amounts of vector, and target-cell populations exhibiting <60% transduction were analyzed by FACS analysis at five days posttransduction for GFP expression. As shown in Fig. 1b, a larger U3 deletion increased episomal expression of GFP nearly threefold, from an MFI of 28.34 for the nonintegrating short-deletion vector vTK113/IN- to an MFI of 82.10 for the nonintegrating long-deletion vector vTK945/IN-. However, regardless of the length of the U3 deletion, integrating vectors expressed more GFP than either nonintegrating vector, and the difference between the levels of transgene expression exhibited by vTK113/IN+ and vTK945/IN+ was not significant. Not surprisingly, after four passages GFP was not detected by FACS analysis in cells transduced with nonintegrating vectors (Fig. 1b), and less than 0.1% of cells displayed GFP measurable by fluorescence microscopy (data not shown).
Figure 1.
Comparative analysis of in vitro expression from lentiviral vectors with short or long deletions in the U3 sequence. (a) Schematic of vectors with partial (vTK113) or larger (vTK945) U3 deletions; deleted sequences are indicated by crosshatching. (b) FACS analysis of GFP expression generated by vTK113 (left) or vTK945 (right), with (upper) or without (middle and lower) functional integrase, in 293T cells. Mean fluorescence intensity (MFI) was measured 5 days posttransduction (p0) and after four passages (p4) (lower). (c) Northern-blot analysis of transcription mediated by short-deletion (lanes 1 and 3) or long-deletion (lanes 2 and 4) vectors, with (lanes 1 and 2) or without (lanes 3 and 4) functional integrase, in 293T cells. RNA was harvested 5 days posttransduction. Equal loading of RNA was verified through ethidium-bromide staining of ribosomal RNA (lower panel).
To support further the notion that U3 sequences inhibit transcription, a Northern-blot assay was employed to measure the effect of U3 deletion length on transcription in 293T cells. Cells were transduced as for FACS and RNA was harvested, hybridized to a radiolabeled, woodchuck hepatitis virus posttranscriptional regulatory element (WPRE)-specific probe, which recognizes transcripts both originating from the internal CMV promoter and LTR-derived transcripts, and quantified via phosphorimager. In keeping with FACS analysis, the Northern assay indicated that the larger U3 deletion enhanced episomal transcription nearly fourfold, that both integrating vectors produced significantly more RNA than either nonintegrating vector, and that short-deletion and long deletion integrating vectors generated comparable levels of RNA (Fig. 1c, Table S1). Also of note are the two transcripts produced by nonintegrating vTK113 and vTK945, in which the lower, stronger band corresponds to a normal, CMV-derived transcript, while the higher, weaker band, significantly higher for vTK113 than vTK945, is suggestive of transcriptional readthrough on 2-LTR circular episomes, such that termination occurs at the distant LTR’s polyA site; in such a case, vTK945’s larger U3 deletion would produce a smaller readthrough product than vTK113’s (Fig. 1c). Furthermore, full-length transcripts were not detected, indicating that vTK113 and vTK945 are SIN vectors, despite the presence of a TATA box in vTK945’s U3. These findings are in line with a prior study by Zufferey et al. that also failed to detect full-length transcription from SIN vectors with TATA-containing U3 sequences [36]. However, mindful of the possibility that the TATA box in vTK945 may contribute to its increased episomal expression, we constructed a new vector (vTK1125, Figs. S1 and S2) containing a state-of-the-art U3 deletion from (–18 to –418, as described in Zufferey et al.) that eliminates the TATA box and 5’ U3 sequences. Interestingly, extending the deletion in the U3 to include the TATA box did not improve episomal transgene expression beyond the levels obtained by vTK945 (Fig. S3). Finally, the notion that the larger U3 deletion mediates increased episomal expression was further confirmed by a qPCR-normalized luciferase assay comparing short-deletion (vTK464) and long-deletion (vTK993) vectors (Fig. S4).
Relative abundances of episomal forms are unaffected by U3 deletion length
These results were in line with earlier studies suggesting that the U3 sequence contains an expression-repressing element in its 5’ region [31–33, 37]. However, we could not rule out the possibility that other factors account for the increase in expression associated with the long U3 deletion in vTK945. To date, there is no indication that the various episomal forms (2-LTR circular, 1-LTR circular, mutant, and linear) express transgenes with differing efficiencies. Thus, the question remains of whether the increased episomal expression generated by nonintegrating long U3-deletion vectors is associated with a concomitant change in the relative abundance of episomal forms. Similar to earlier studies characterizing the relative abundance of vector episomes and integrated provirus [15, 29, 38], Southern analysis was employed on DNA extracted from transduced Jurkat and 293T cells at three days posttransduction and following three passages (Fig. 2a). As shown in Fig. 2b and quantified in Table 1, the relative abundance of episomal forms in integrating and nonintegrating vectors was not changed significantly by the longer U3 deletion. However, and in keeping with a prior study [39], differences in relative episomal abundance were caused by integrase status, with nonintegrating vectors producing significantly more 2-LTR circles than integrating vectors, and by cell type, with 293T cells exhibiting a notably greater proportion of linear episomes than Jurkat cells. Interestingly, and differing from a recent report [29], the relative abundance of linear episomes was not dramatically upregulated by integrase deficiency (Fig. 2b). Under the condition of harvesting DNA three days posttransduction, linear episomes were comparable to, and in some cases exceeded by, 1-LTR circles in terms of relative abundance (Table 1). Furthermore, integrated provirus accounted for between 21 and 33% of all vector genomes, regardless of U3 deletion size (Fig. 2b). While Southern-blot analysis did not show significant changes in episome formation mediated by U3 length, cell line-specific changes were noted, with all vectors producing a greater share of linear episomes and a smaller proportion of 1-LTR circles in 293T cells than in Jurkat cells (Fig. 2b). Prior studies on simple retroviruses demonstrated the existence of autointegrated lentiviral episomes [16]; however, the relative contribution of this unique episomal form to the total population of episomes in the context of lentiviral/retroviral vectors has not been elucidated, largely because autointegrated episomes, being variably sized, cannot be adequately detected by Southern blot. To overcome this technical difficulty, we developed an HIV-1-derived shuttle vector containing a bacterial origin of replication and a drug-resistance gene within the vector’s transcribed region. This vector configuration enables circular episomes produced in vector-transduced cells to be extracted by the Hirt protocol [40], and monoclonally isolated from bacteria (Fig. 2c). Simple restriction analysis allows individual episomal clones to be characterized either as 1-LTR, 2-LTR, or autointegrated circles. As shown in Fig. 2d (and in keeping with previous results [7, 39]), results of the shuttle-vector assay indicate that, in integrating-vector-transduced cells, 1-LTR circular episomes, 2-LTR circular episomes, and autointegrated circular episomes account for roughly 75%, 15%, and 10% of circular episomes, respectively, while in nonintegrating vector-transduced-cells, their relative amounts were 60%, 35%, and 5%, respectively. Shuttle vectors bearing a full-length LTR (vTK459, described in detail in Ma et al., [41]), a short U3 deletion (vTK1055), or a long U3 deletion (vTK1054)(Fig. S2) were assayed, leading to the finding that U3 length did not affect the relative abundances of circular episomes, with the notable exception that vTK1054 demonstrated significantly more autointegrated episomes than vTK459 (p-value of 0.0005 by chi2 test)(Fig. 2d, Table S2).
Figure 2.
Analysis of episome formation in cells transduced with lentiviral vectors. (a) To simultaneously analyze integrated and nonintegrated vector genomes, total DNA from transduced cells was cut at sites (EcoNI and PflMI) flanking the LTR(s), and a radiolabeled probe complementary to a region spanning the EcoNI site was employed on vTK113 (left) and vTK945 (right) vector genomes. (b) Episome formation and integration efficiency was examined by Southern blot in Jurkat (upper) and 293T (lower) cells. Jurkat cells were transduced with vTK113, integrating (lanes 1 and 3) or nonintegrating (lanes 2 and 4), or with vTK945, integrating (lanes 5 and 7) or nonintegrating (lanes 6 and 8) and total DNA was extracted from transduced cells 3 days (no passages)(lanes 1–2 and 5–6) or ~14 days (3 passages)(lanes 3–4 and 7–8) posttransduction, digested as shown in (a), and analyzed by Southern blot, and the same procedure was followed using 293T cells (lanes 9–16). Size markers are shown in red. (c) Outline of the shuttle-vector assay for characterizing the relative abundance of circular episomes. (d) Episomes were harvested from 293T cells 16 hours posttransduction; each transduction was performed in triplicate.
Table 1.
Quantification of relative amounts of integrated lentiviral genomes and linear, 1-LTR, and 2-LTR episomal genomes produced by integrating and nonintegrating lentivectors in 293T and Jurkat cells.
| Vector and Cell Type | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DNA form |
Jurkat 113 IN+a |
Jurkat 113 IN-b |
Jurkat 113 IN+ integ.c |
Jurkat 945 IN+ |
Jurkat 945 IN- |
Jurkat 945 IN+ integ. |
293T 113 IN+ |
293T 113 IN- |
293T 113 IN+ integ. |
293T 945 IN+ |
293T 945 IN- |
945 IN+ integ. |
| 2-LTR | 8.97% | 16.29% | 8.48% | 17.69% | 5.42% | 19.58% | 4.78% | 15.76% | ||||
| 1-LTR | 46.21% | 43.50% | 41.16% | 41.26% | 40.16% | 26.77% | 26.64% | 30.42% | ||||
| linear | 44.82% | 40.21% | 50.36% | 41.05% | 54.41% | 53.65% | 68.58% | 53.82% | ||||
| integrated | 35.04% | 30.51% | 23.03% | 29.76% | ||||||||
IN+ = vector packaged with wild-type integrase
IN- = vector packaged with D64E-mutant integrase
integ. = proportion of lentivector genomes remaining after three passages of transduced cells
Increased episomal expression in the brain and liver from a vector with a large U3 deletion
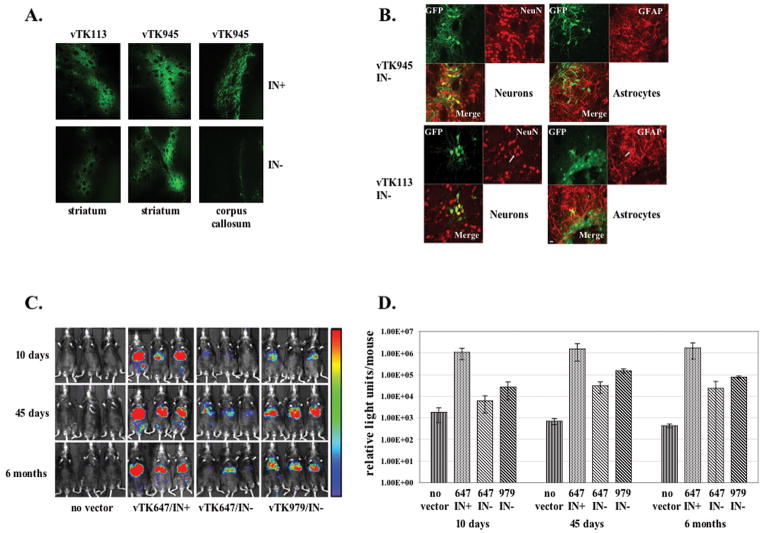
In accordance with the notion that lentiviral episome formation may vary across cell types, previous findings indicate that the level of episomal lentiviral gene expression may be cell type-specific [22]. These findings spurred an investigation into the ability of nonintegrating lentiviral vectors to transduce effectively a variety of cell types in vivo. Accordingly, rats were injected intracranially with equal transducing units of integrating and nonintegrating small-deletion (vTK113) and long-deletion (vTK945) vectors. Two weeks and three months after striatal infusion, brain tissues were harvested and imaged for GFP, as well as the neuron-specific marker NeuN and the astrocyte-specific marker GFAP. As shown in Fig. 3a, the U3 deletion notably increased episomal GFP expression in striatal tissue three months postinfusion (Fig. S5 shows similar results two weeks postinfusion). However, while nonintegrating vTK113 expressed GFP in both neurons and astrocytes, nonintegrating vTK945 appeared to transduce astrocytes to a significantly lesser degree (Fig. 3b). Furthermore, while vTK945 expressed GFP strongly in the striatum, episomal expression of vTK945 in the corpus callosum was relatively weak (Fig. 3a). Intrigued by the long U3 deletion’s effect on episomal expression in the rat brain, we sought to replicate the vector’s efficacy in the slowly dividing tissue of the liver, which prior findings had found resistant to sustained episomal lentiviral transduction [27]. To that end, mice were injected intraperitoneally with 100 μg p24 of small-deletion (vTK647) or large-deletion (vTK979) vector expressing luciferase from the liver-specific promoter hAAT. After imaging for luciferase activity at 10 days, 45 days, and 6 months postinjection, nonintegrating vTK979 was found to generate roughly fourfold more luciferase activity than nonintegrating vTK647, but still yielded approximately 40-fold less luciferase activity than integrating vTK647 (Fig. 3c and d). Furthermore, in disagreement with a previous report, we found that transgene expression in the liver was largely stable over a six-month time course [42](Fig. 3c and d). Taken together, the results in Fig. 3 indicate that the U3 deletion improves episomal expression in several tissues in vivo, and that the effect is sustained over time.
Figure 3.
Analysis of in vivo episomal expression from lentiviral vectors with short or large deletions in the U3 sequence. (a) Tissue compartment-specific GFP expression mediated by short-deletion or long-deletion vectors, with (top) or without (bottom) functional integrase, in the striatum and corpus callosum of the rat brain, measured three months after vector infusion. (b) Cell type-specific GFP expression mediated by short-deletion (lower) and long-deletion (upper) nonintegrating vectors in neural (fluorescing with NeuN, left side) and glial (fluorescing with GFAP, right side) cells of the rat brain, measured three months after vector infusion. A 20-micron size bar appears in the 113/IN- GFAP/GFP merge panel. (c) Long-term luciferase expression produced by short-deletion or long-deletion vectors, with or without functional integrase, in the mouse liver. Animals were imaged 10 days, 45 days, and 6 months after intraperitoneal injection of vector. (d) Quantification of luciferase expression described in (c). Each bar represents the average of luciferase measurements from three mice.
DISCUSSION
The overall goal of this study was to characterize episomally derived transgene expression from SIN lentiviral vectors, in vitro and in vivo. In general, while all vectors demonstrated measurable levels of transgene expression regardless of the SIN vector used, nonintegrating vectors exhibited significantly less transgene expression than their integrating counterparts. This finding represents a notable discrepancy with recent reports concerning nonintegrating vectors, which do not indicate any major difficulty in obtaining robust expression from nonintegrating vectors [17, 30]. Differences of methodology in quantifying expression may explain the discrepancy. Specifically, in the present study efforts were made to evaluate the efficiency of expression by Northern analysis and direct measurement of transgene abundance, either by mean fluorescence index (MFI) or by relative light units (RLU); furthermore, to avoid saturation, transduction was carried out at low m.o.i. and expression was normalized to vector copy number. Conversely, previous reports measured the percentage of cells expressing a given transgene; this method does not measure the degree of expression in a transgene-positive cell. Another methodological concern is that expression from integrating vectors, if measured shortly after transduction or from nondividing cells, is generated primarily from episomes, which account for 70–95% of vector genomes [7, 9, 12]. Therefore, early expression seen from cells transduced with integrating vectors may still be largely episome-derived and not as efficient, on average, as expression from exclusively integrated genomes, possibly leading to the undervaluation of expression generated from integrated provirus. However, the mechanism by which expression from unintegrated lentiviral genomes is repressed is not clear. The fact that the earliest lentiviral vectors exhibited the lowest level of episomal expression raised the possibility that a cis element in the U3 region may inhibit transcription from lentiviral vectors, and, in fact, early studies on HIV-1 suggest the existence of such an element [32, 33, 37]. While the effect of the long U3 deletion on episomal expression implies that cis–acting elements in the lentiviral vector genome influence extrachromosomal transcriptional activity, the possibility exists that cell-specific trans-acting factors are also involved in the mechanism of downregulated transgene expression from lentiviral episomes. In keeping with the concept of cell-specific factors influencing episomal expression, previous findings indicate varying levels of gene expression across a number of cell lines infected with nonintegrating lentivirus [22]. Interestingly, transduction of murine dorsal root ganglia in culture by wild-type U3 or long U3-deletion lentiviral vectors resulted in strong expression exclusively in neurons, or both neurons and stromal cells, respectively, further suggesting that the silencing mechanism of retroviral vector expression bears cell-specific characteristics [31].
The potentially variable nature of episomal expression and U3-mediated transcriptional repression across cell types, coupled with the clinical relevance of in vivo transduction, underscored the importance of measuring gene expression in a variety of cell types in vivo. Here we show that the large U3 deletion had a pronounced effect on episomal expression in rat-brain striatal cells; however, this improvement in SIN vector design did not confer robust episomal expression in corpus callosum cells, suggesting that additional mechanistic elements contribute to the silencing of episomal expression. The possibility that increased efficiency of expression from long U3-deletion vectors is due to differences in episome formation is ruled out by the present study, which indicates that no significant change in the relative abundances of episomal forms is associated with the large U3 deletion. However, analysis of episome formation revealed discrepancies with published results. In contrast to a prior report, the relative abundance of linear episomes did not increase in the absence of functional integrase [29]. In keeping with the findings of Svarovskaia et al. [39], the relative abundance of 2-LTR circular episomes increased in the absence of integrase activity. Two putative mechanisms proposed earlier by Engelman et al. [43] to explain this phenomenon suggest either that blunt-ended, linear DNA, unprocessed by mutant integrase, is more amenable to circularization by end-joining, or that more linear DNA is available for circularization in the absence of integrase activity. Interestingly, the efficiency of integration in the present study, as measured by Southern blot, ranged from 23% to 35% of all integrase-functional vector genomes, which is higher than the 5–16% measured previously by PCR [7]. The present study employed a novel shuttle-vector assay to quantify the autointegrated circular episomal form produced by lentivectors, and observed that integrase-deficient vectors produced a measurable number of autointegrated circles, suggesting that the mechanism of their formation may be partly integrase-independent. Furthermore, integrating long U3-deletion vectors were found to produce notably more autointegrated circular episomes than did other vectors, implying that U3 sequences help prevent the formation of autointegrated circles.
Prior studies administered nonintegrating vectors in vivo by direct injection into nondividing target organs, including eye, brain or muscle [17, 18, 34], thereby delivering potentially saturating quantities of vector particles; however, the present study, for the first time, administered nonintegrating lentiviral vectors systemically to slowly dividing liver cells, which allowed testing of the vector’s efficacy in a condition of low concentration and, by extension, relatively low multiplicity of infection in the target organ. Intriguingly, a long U3-deletion vector expressed four times more liver-specific luciferase than a short U3-deletion vector, in keeping with in vitro results and indicating that cells in the liver may express transcriptionally repressive factors binding to the 5’ U3 region eliminated from the large U3-deletion vector. The significant, hepatocyte-specific luciferase expression noted by the present study contrasts with previous reports characterizing episomal lentiviral expression in the liver as insignificant [27]; discrepancies with the prior study may be due to differences in transgene employed and LTR length. Furthermore, in divergence from prior findings suggesting that liver-specific lentiviral vector transgene expression induces an effective immune response [42], the present study found no significant loss of expression in immunocompetent mice. In fact, transgene expression from every vector assayed was sustained for at least six months after administration, in agreement with prior studies stably expressing transgenes episomally in the liver from adeno-associated viral vectors and plasmid DNA [44, 45]. A slight reduction over time of episomal expression in mouse livers observed in this study may be due to the slow division of liver cells, and could eventually necessitate readministration of alternately pseudotyped vector.
In summary, the present study showed that a nonintegrating lentiviral vector with an extensively truncated U3 region can be an efficacious means of delivering transgenes to target cells in vivo. However, the fact that nonintegrating long-deletion lentiviral vectors still do not express transgenes as efficiently as their integrating counterparts indicates that an additional mechanism of episomal silencing may be involved, possibly one that is inherent to unintegrated retroviral genomes. The transcriptional downregulation of lentiviral episomes, combined with the fact that they are not influenced by the chromosomal transcriptional regulatory environment, indicate that the characteristics of integrated lentiviral genomes cannot be extrapolated to lentiviral episomes, and that further characterization of nonintegrating lentiviral vectors is required to optimize their use in basic research and gene therapy applications.
MATERIALS AND METHODS
Lentiviral vector constructs
The vector cassette pTK113 has been described previously [46] and contains a U3 deletion extending from –7 to –141. The vector cassette pTK945 bears a U3 deletion spanning –48 to –396, and is otherwise identical to pTK113. The vector cassette pTK1125 bears a U3 deletion extending from –18 to –418, and is otherwise identical to pTK113. Shuttle vectors were derived from the vector plasmid pTK459 [41], which includes a bacterial origin of replication and ampicillin-resistance gene in the transcribed region of the plasmid, as well as a full-length U3 (in pTK459), short-deletion U3 (pTK1055), or long-deletion U3 (pTK1054). The vector cassettes pTK647 and pTK979 were constructed by inserting the U3 sequence from pTK113 and pTK945, respectively, into pTK646, which bears the liver-specific promoter hAAT and the firefly luciferase gene, by standard cloning procedures. The Flp9 cell line was prepared as follows: pTK113 was cloned into Flp-In expression vector (Invitrogen, Carlsbad, CA) and the resulting plasmid was cotransfected with pOG44 (Flp recombinase) into the Flp-InTM host cell line, such that pTK113 integrated into the genomic FRT site (Invitrogen). The single-copy incorporation of the expression cassette per diploid genome was verified by Southern blot, following digestion of DNA isolated from Flp9 cells with AflII (New England Biolabs, Ipswich, MA, and all restriction enzymes to follow), which recognizes 2 sites in the LTR of the expression vector, or XbaI, which recognizes a single site in each FRT sequence.
Viral vector production
All lentiviral vectors were prepared as previously described [46], transiently transfecting 107 293T cells with 15μg vector cassette, 10μg packaging cassette, and 5μg envelope cassette. Integrating vectors were made using the packaging cassette ΔNRF [46], which expresses functional integrase, while nonintegrating vectors were made using the packaging cassette pTK939, which was made by inserting the D64E-mutant integrase from pD64E into ΔNRF by standard cloning procedures. All vectors were pseudotyped with the VSV-G envelope cassette. For vectors constitutively expressing GFP, titers were assessed by serial dilution in 293T cells followed by visual analysis of GFP expression by fluorescence microscope. For other vectors, concentrations were determined by p24gag ELISA. The absence of replication-competent retroviruses was determined by three independent methods: tat transfer assay, vector rescue assay, and p24gag ELISA, as described previously [47].
Cell culture
293T cells were grown in Dulbecco's modified Eagle medium (Mediatech, Herndon, VA) supplemented with 9% fetal bovine serum and 1% penicillin/streptomycin solution. Jurkat cells were cultured in Roswell Park Memorial Institute medium (Mediatech) supplemented with 9% fetal bovine serum and 1% penicillin/streptomycin solution.
FACScan analysis
293T cells were transduced at an m.o.i. of 1. At 3–5 days (p0) and four passages (p4) posttransduction, cells were harvested, fixed in 1X PBS containing 2% formaldehyde/0.2% glutaraldehyde, and analyzed by FACscan as peviously described [48].
Northern blot analysis
293T cells were transduced with the indicated vectors at an m.o.i. of 1. Total cell lysates were prepared 3–5 days posttransduction for RNA isolation using the PARISTM kit (Ambion, Austin, TX) or Rneasy® Plus Mini kit (Qiagen, Hilden, Germany). 3mg of total RNA was denatured at 70°C and resolved on a 1.2% denaturing formaldehyde/agarose gel. RNA was transferred to a Zeta-Probe GT® membrane (BioRad, Hercules, CA). Hybridization was executed at 68°C for ~18 hours with a 32P-labeled probe comprising a 595-base region spanning the woodchuck hepatitis virus posttranscriptional regulatory element (WPRE) that was cut from pTK113 with ClaI. The Northern blot image was imaged with BioMax MR film (Kodak, Rochester, NY). Relative quantification of RNA species was obtained via Storm phosphorimager (GE Healthcare, Chalfont St. Giles, UK) using ImageQuant 5.2 software (GE Healthcare).
Southern blot analysis
Cells were harvested three days and three passages after transduction with vTK113 and vTK945 and their total DNA was extracted using the Qiagen Blood and Cell Culture DNA Midi Kit (Qiagen). 10μg total DNA was digested with DpnI and either EcoNI and PflMI (for 293T cells) or NotI and PflMI (Jurkat cells), electrophoresed in a 1% gel, transferred to a nylon membrane (GE Healthcare), UV-crosslinked, and probed with the 1.4kb KasI/BamHI fragment of vTK945. To control for loading, membranes were also probed with a 731bp region of the endogenous gene BDNF, produced with the PCR primers 5'CGTTTGACCAATCGAAGC3' (forward) and 5'TCCCCTCAGTCAGGACCCTCG3' (reverse). Quantification of DNA density was achieved on a Storm phosphorimager using ImageQuant 5.2 software.
Shuttle vector assay
293T cells were transduced with integrating or nonintegrating vTK459, vTK1055, and vTK1054 (Fig. S2). Transduced cells were harvested 16 hours posttransduction using 0.85mL Hirt lysis buffer and 0.25mL 5M NaCl per 10cm plate of transduced cells. Episomal DNA was subsequently subjected to phenol/chloroform extraction and DpnI digestion and electroporated into bacteria. Bacterial colonies were cultured monoclonally, pelleted, and their episomes extracted by boiling in a hotprep buffer made with 2g sucrose, 0.4mL 0.5M Tris/EDTA solution, 40mL 10% Triton X-100 (Sigma, St. Louis, MO), 5mL 1M Tris/HCl pH 8.0 and water to 100mL. 125μL hotprep buffer and 2.5μg RNase A (Sigma) were added to each bacterial pellet before boiling. After boiling, samples were centrifuged at 14K for 10 minutes to dispose of cell debris. Episomes were digested with NotI and SacII and electrophoresed in a 1% agarose gel to be characterized as 1-LTR, 2-LTR, or mutant circular episomes. For every transduction, between 73 and 112 episome-bearing bacterial colonies were characterized.
In vivo experiments in rat brain
All of the animals were pathogen-free male Sprague-Dawley rats obtained from Charles Rivers. All care and procedures were in accordance with the Guide for the Care and Use of Laboratory Animals (DHHS Publication No. [NIH]85-23), and all procedures received prior approval by the University of North Carolina Institutional Animal Care and Usage Committee. Virus vector infusions were performed as previously described [49]. Briefly, rats first were anesthetized with 50 mg/kg pentobarbital and then placed into a stereotaxic frame. Using a 32 gauge stainless steel injector and a Sage infusion pump, the rats received 1 ml (at 6 x 109 GFP transducing units/mL) of the integrating and nonintegrating vTK113 and vTK945 vectors over a 10 minute period into the striatum (1.0mm anterior to bregma, 3.0mm lateral, 5.5mm vertical) according to the atlas of Paxinos and Watson [50]. In all cases, the injector was left in place 3 minutes postinfusion to allow diffusion from the injector.
Immunohistochemistry
Two weeks or 3 months after the vector infusion, rats received an overdose of pentobarbital (100 mg/kg pentobarbital, i.p.) and subsequently were perfused transcardially with ice-cold 100 mM sodium phosphate buffered saline (PBS) (pH=7.4), followed by 4% paraformaldehyde in 100 mM phosphate buffer (pH=7.4). After overnight fixation in paraformaldehyde-phosphate buffer, vibratome sections (40 mm thick) were taken through the striatum and rinsed in PBS. For immunohistochemistry, tissue sections were incubated in 10% normal goat serum and 0.1% Triton X-100 in PBS for 45 minutes. Next, sections were incubated with a primary antibody to NeuN (1:500, Chemicon, Temecula, CA) or glial fibrillary acidic protein (GFAP)(1:4,000, DAKO A/S, Denmark) overnight in 3% normal goat serum, 0.2 % Triton X-100 and PBS. Tissue sections were then rinsed in PBS, incubated in blocking serum (10% normal goat serum, 0.1% Triton X-100, PBS) for 1hr. and then incubated with a secondary fluorescent antibody (Alexa-fluor 594 goat anti-mouse (NeuN), goat anti-rabbit (GFAP), (Molecular Probes, Eugene, OR) for 1 hour at 40C. Following 3 rinses in PBS, the sections were mounted on slides and coverslipped with fluorescent mounting media. eGFP fluorescence initially was visualized on an Olympus IX 71 fluorescence microscope (Olympus, Center Valley, PA), and digital pictures were taken. In the cases where co-localization was to be determined, both the GFP and the Alexa 594 fluorescence was visualized with a Zeiss 510 Meta laser scanning confocal microscope (Zeiss, Oberkochen, Germany). Co-localization was determined by multiple scans through the Z axis of the sample.
In vivo experiments in mouse liver
All procedures received prior approval by the University of North Carolina Institutional Animal Care and Usage Committee. C57BL/6NHsd mice (Harlan Sprague Dawley, Indianapolis, IA) were injected intraperitoneally at eight weeks of age with 100μg p24 of lentivector. At 10 days, 45 days, and 6 months postinjection mice were assayed for luciferase expression following luciferin (NanoLight, Pinetop, AZ) injection and using the Xenogen IVIS imaging system (Xenogen, Hopkinton, MA).
Supplementary Material
Acknowledgments
The following reagents were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: pD64E from Dr. Vinay K. Pathak and HIV-1 p24 monoclonal antibody (183-H12-5C) from Dr. Bruce Chesebro and Kathy Wehrly. We thank Stephen Marron and Feng Liu for providing assistance with statistical analysis. We are grateful to Aristides de Sousa Mendes. The study was supported by the UNC Center for AIDS Research (CFAR) and by NIH grants 2-R01-DK058702-06 to T.K., 5-R21-HL086406 to T.K., 5-PO1-HL066973-05 to T.K and B.K., and P30-DK065988 to A.C. The authors have no conflicts of interest to disclose.
References
- 1.Aiuti A, Slavin S, Aker M, Ficare F, Deola S, Mortellaro A, et al. Correction of ADA-SCID by Stem cell gene therapy combined with nonmyeloablative conditioning. Science. 2002;296:2410–2413. doi: 10.1126/science.1070104. [DOI] [PubMed] [Google Scholar]
- 2.Cavazzana-Calvo M, Hacein-Bey S, de Saint Basile G, Gross F, Yvon E, Nusbaum P, et al. Gene therapy of human severe combined immunodeficiency (SCID)–X disease. Science. 2000;288:669–672. doi: 10.1126/science.288.5466.669. [DOI] [PubMed] [Google Scholar]
- 3.Gaspar HB, Parsley KL, Howe S, King D, Gilmour KC, Sinclair J, et al. Gene therapy of X-linked severe combined immunodeficiency by use of a pseudotyped gammaretroviral vector. Lancet. 2004;364:2181–2187. doi: 10.1016/S0140-6736(04)17590-9. [DOI] [PubMed] [Google Scholar]
- 4.Seggewiss R, Pittaluga S, Adler RL, Guenaga FJ, Ferguson C, Pilz IH, et al. Acute myeloid leukemia is associated with retroviral gene transfer to hematopoietic progenitor cells in a rhesus macaque. Blood. 2006;107:3865–3867. doi: 10.1182/blood-2005-10-4108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baum C. What are the consequences of the fourth case? Mol Ther. 2007;15:1401–1402. doi: 10.1038/sj.mt.6300263. [DOI] [PubMed] [Google Scholar]
- 6.Hacein-Bey-Abina S, von Kalle C, Schmdt M, McCormack MP, Wulffraat N, Leboulch P, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 7.Butler SL, Hansen MST, Bushman FD. A quantitative assay for HIV DNA integration in vivo. Nat Med. 2001;7:631–634. doi: 10.1038/87979. [DOI] [PubMed] [Google Scholar]
- 8.Butler SL, Johnson EP, Bushman FD. Human immunodeficiency virus cDNA metabolism: notable stability of two-long terminal repeat circles. J Virol. 2002;76:3739–3747. doi: 10.1128/JVI.76.8.3739-3747.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chun T-W, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 10.Gillim-Ross L, Cara A, Klotman ME. HIV-1 extrachromosomal 2-LTR circular DNA is long-lived in human macrophages. Viral Immunol. 2005;18:190–196. doi: 10.1089/vim.2005.18.190. [DOI] [PubMed] [Google Scholar]
- 11.Pierson TC, Kieffer TL, Ruff CT, Buck C, Gange SJ, Siliciano RF. Intrinsic stability of episomal circles formed during human immunodeficiency virus type 1 replication. J Virol. 2002;76:4138–4144. doi: 10.1128/JVI.76.8.4138-4144.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shaw GM, Hahn BH, Arya SK, Groopman JE, Gallo RC, Wong-Staal F. Molecular characterization of human T-cell leukemia (lymphotropic) virus type III in the acquired immune deficiency syndrome. Science. 1984;226:1165–1171. doi: 10.1126/science.6095449. [DOI] [PubMed] [Google Scholar]
- 13.Dina D, Benz E. Structure of murine sarcoma virus DNA replicative intermediates synthesized in vitro. J Virol. 1980;33:377–389. doi: 10.1128/jvi.33.1.377-389.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farnet CM, Haseltine WA. Circularization of human immunodeficiency virus type 1 DNA in vitro. J Virol. 1991;65:6942–6952. doi: 10.1128/jvi.65.12.6942-6952.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li L, Olvera JM, Yoder KE, Mitchell RS, Butler SL, Lieber M, et al. Role of the non-homologous DNA end joining pathway in the early steps of retroviral infection. EMBO J. 2001;20:3272–3281. doi: 10.1093/emboj/20.12.3272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shoemaker C, Goff S, Gilboa E, Paskind M, Mitra S, Baltimore D. Structure of a cloned circular Moloney murine leukemia virus DNA molecule containing an inverted segment: Implications for retrovirus integration. Proc Natl Acad Sci USA. 1980;77:3932–3936. doi: 10.1073/pnas.77.7.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Philippe S, Sarkis C, Barkats M, Mammeri H, Ladroue C, Petit C, et al. Lentiviral vectors with a defective integrase allow efficient and sustained transgene expression in vitro and in vivo. Proc Natl Acad Sci USA. 2006;103:17684–17689. doi: 10.1073/pnas.0606197103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yanez-Munoz RJ, Kamaljit SB, MacNeil A, Howe1 SJ, Schmidt M, Smith AJ, et al. Effective gene therapy with nonintegrating lentiviral vectors. Nat Med. 2006;12:348–353. doi: 10.1038/nm1365. [DOI] [PubMed] [Google Scholar]
- 19.LaFemina RL, Schneider CL, Robbins HL, Callahan PL, LeGrow K, Roth E, et al. Requirement of active human immunodeficiency virus type 1 integrase enzyme for productive infection of human T-lymphoid cells. J Virol. 1992;66:7414–7419. doi: 10.1128/jvi.66.12.7414-7419.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakai H, Kawamura M, Sakuragi J-I, Sakuragi S, Shibata R, Ishimoto A, et al. Integration is essential for efficient gene expression of human immunodeficiency virus type 1. J Virol. 1993;67:1169–1174. doi: 10.1128/jvi.67.3.1169-1174.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masuda T, Kuroda M, Harada S. Specific and independent recognition of U3 and U5 att sites by human immunodeficiency virus type 1 integrase in vivo. J Virol. 1998;72:8396–8402. doi: 10.1128/jvi.72.10.8396-8402.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakajima N, Lu R, Engelman A. Human immunodeficiency virus type 1 replication in the absence of integrase-mediated DNA recombination: definition of permissive and nonpermissive T-cell lines. J Virol. 2001;75:7944–7955. doi: 10.1128/JVI.75.17.7944-7955.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiskerchen M, Muesing MA. Human immunodeficiency virus type 1 integrase: effects of mutations on viral ability to integrate, direct viral gene expression from unintegrated viral DNA templates, and sustain viral propagation in primary cells. J Virol. 1995;69:376–386. doi: 10.1128/jvi.69.1.376-386.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blomer U, Naldini L, Kafri T, Trono D, Verma I, Gage FH. Highly efficient and sustained gene transfer in adult neurons with a lentivirus vector. J Virol. 1997;71:6641–6649. doi: 10.1128/jvi.71.9.6641-6649.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitrophanous KA, Yoon S, Rohl JB, Patil D, Wilkes FJ, Kim VN, et al. Stable gene transfer to the nervous system using a non-primate lentiviral vector. Gene Ther. 1999;6:1808–1818. doi: 10.1038/sj.gt.3301023. [DOI] [PubMed] [Google Scholar]
- 26.Naldini L, Blomer U, Gage FH, Trono D, Verma IH. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc Natl Acad Sci USA. 1996;93:11382–11388. doi: 10.1073/pnas.93.21.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park F, Ohashi K, Kay MA. Therapeutic levels of human factor VIII and IX using HIV-1–based lentiviral vectors in mouse liver. Blood. 2000;96:1173–1176. [PubMed] [Google Scholar]
- 28.Negri DRM, Michelini Z, Baronelli S, Spada M, Vendetti S, Buffa V, et al. Successful immunization with a single injection of non-integrating lentiviral vector. Mol Ther. 2007;15:1716–1723. doi: 10.1038/sj.mt.6300241. [DOI] [PubMed] [Google Scholar]
- 29.Nightingale SJ, Hollis RP, Pepper KA, Petersen D, Yu X-J, Yang C, et al. Transient gene expression by nonintegrating vectors. Mol Ther. 2006;13:1121–1132. doi: 10.1016/j.ymthe.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 30.Saenz DT, Loewen N, Peretz M, Whitwam T, Barraza R, Howell KG, et al. Unintegrated lentivirus DNA persistence and accessibility to expression in nondividing cells: analysis with class I integrase mutants. J Virol. 2004;78:2906–2920. doi: 10.1128/JVI.78.6.2906-2920.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ginn SL, Fleming J, Rowe PB, Alexander IE. Promoter interference mediated by the U3 region in early-generation HIV-1–derived lentivirus vectors can influence detection of transgene expression in a cell-type and species-specific manner. Hum Gene Ther. 2003;14:1127–1137. doi: 10.1089/104303403322167975. [DOI] [PubMed] [Google Scholar]
- 32.Lu Y, Stenzel M, Sodroski JG, Haseltine WA. Effects of long terminal repeat mutations on human immunodeficiency virus type 1 replication. J Virol. 1989;63:4115–4119. doi: 10.1128/jvi.63.9.4115-4119.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rosen CA, Sodroski JG, Haseltine WA. The location of cis-acting regulatory sequences in the human T cell lymphotropic virus type III (HTLV-III/LAV) long terminal repeat. Cell. 1985;41:813–823. doi: 10.1016/s0092-8674(85)80062-3. [DOI] [PubMed] [Google Scholar]
- 34.Apolonia L, Waddington SN, Ferdandes C, Bouma G, Blundell MP, Thrasher AJ, et al. Stable gene transfer to muscle using non-integrating lentiviral vectors. Mol Ther. 2007;15:1947–1954. doi: 10.1038/sj.mt.6300281. [DOI] [PubMed] [Google Scholar]
- 35.Miyoshi H, Blomer U, Takahashi M, Gage FH, Verma I. Development of a self-inactivating lentivirus vector. J Virol. 1998;72:8150–8157. doi: 10.1128/jvi.72.10.8150-8157.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zufferey R, Dull T, Mandel RJ, Bukovsky A, Quiroz D, Naldini L, et al. Self- inactivating lentivirus vector for safe and efficient in vivo gene delivery. J Virol. 1998;72:9873–9880. doi: 10.1128/jvi.72.12.9873-9880.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hoover T, Mikovits J, Court D, Liu Y-l, Kung H-f, Raziuddin A nuclear matrix-specific factor that binds a specific segment of the negative regulatory element (NRE) of HIV-1 LTR and inhibits NF- B activity. Nucleic Acids Res. 1996;24:1895–1900. doi: 10.1093/nar/24.10.1895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kilzer JM, Stracker T, Beitzel B, Meek K, Weitzman M, Bushman FD. Roles of host cell factors in circularization of retroviral DNA. Virology. 2003;314:460–467. doi: 10.1016/s0042-6822(03)00455-0. [DOI] [PubMed] [Google Scholar]
- 39.Svarovskaia ES, Barr R, Zhang X, Pais Godwin CG, Marchand C, Pommier Y, et al. Azido-containing diketo acid derivatives inhibit human immunodeficiency virus type 1 integrase in vivo and influence the frequency of deletions at two-long- terminal-repeat-circle junctions. J Virol. 2004;78:3210–3222. doi: 10.1128/JVI.78.7.3210-3222.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 41.Ma H, Kafri T. A single-LTR HIV-1 vector optimized for functional genomics applications. Mol Ther. 2004;10:139–149. doi: 10.1016/j.ymthe.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 42.Brown BD, Sitia G, Annoni A, Hauben E, Sergi Sergi L, Zingale A, et al. In vivo administration of lentiviral vectors triggers a type I interferon response that restricts hepatocyte gene transfer and promotes vector clearance. Blood. 2007;109:2797–2805. doi: 10.1182/blood-2006-10-049312. [DOI] [PubMed] [Google Scholar]
- 43.Engelman A, Englund G, Orenstein JM, Martin MA, Craigie R. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J Virol. 1995;69:2729–2736. doi: 10.1128/jvi.69.5.2729-2736.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miao CH, Thompson AR, Loeb K, Ye X. Long-term and therapeutic-level hepatic gene expression of human factor IX after naked plasmid transfer in vivo. Mol Ther. 2001;3:947–957. doi: 10.1006/mthe.2001.0333. [DOI] [PubMed] [Google Scholar]
- 45.Nakai H, Herzog RW, Hagstrom JN, Walter J, Kung S-H, Yang EY, et al. Adeno-associated viral vector-mediated gene transfer of human blood coagulation factor IX into mouse liver. Blood. 1998;91:4600–4607. [PubMed] [Google Scholar]
- 46.Xu K, Ma H, McCown TJ, Verma I, Kafri T. Generation of a stable cell line producing high-titer self-inactivating lentiviral vectors. Mol Ther. 2001;3:97–104. doi: 10.1006/mthe.2000.0238. [DOI] [PubMed] [Google Scholar]
- 47.Kafri T, van Praag H, Gage FH, Verma IM. Lentiviral vectors: regulated gene expression. Mol Ther. 2000;1:516–521. doi: 10.1006/mthe.2000.0083. [DOI] [PubMed] [Google Scholar]
- 48.Cockrell AS, Ma H, Fu K, McCown TJ, Kafri T. A trans-lentiviral packaging cell line for high-titer conditional self-inactivating HIV-1 vectors. Mol Ther. 2006;14:276–284. doi: 10.1016/j.ymthe.2005.12.015. [DOI] [PubMed] [Google Scholar]
- 49.McCown TJ. Adeno-associated virus-mediated expression and constitutive secretion of galanin suppresses limbic seizure activity in vivo. Mol Ther. 2006;14:63–68. doi: 10.1016/j.ymthe.2006.04.004. [DOI] [PubMed] [Google Scholar]
- 50.Paxinos G, Watson C. The rat brain in stereotaxic coordinates. Academic Press; Orlando, FL: 1986. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.