Abstract
Background
Japanese cedar (Cryptomeria japonica) pollinosis is the most prevalent allergy in Japan. Recently, the Japanese cedar pollen allergen Cry j 3 was cloned as a homologue of Jun a 3, which is a major allergen from mountain cedar (Juniperus ashei) pollen. However, native Cry j 3 has not been isolated and there are no reports on its allergenic activity. The aims of this study were to isolate native Cry j 3 and assess its immunoglobulin E (IgE)-binding capacity in patients with Japanese cedar pollinosis.
Methods
Native Cry j 3 was purified from Japanese cedar pollen by multidimensional chromatography. We assessed the IgE-binding capacity using sera from patients allergic to Japanese cedar pollen by immunoblot analysis and ELISA. Moreover, we assayed the capacity of Cry j 3 to induce histamine release from the patient’s leukocytes. We cloned cDNA corresponding to purified Cry j 3 from a cDNA library of Japanese cedar pollen.
Results
We isolated native Cry j 3 as a 27-kDa protein. The IgE-binding frequency of Cry j 3 from the sera of patients allergic to Japanese cedar pollen was estimated as 27% (27/100) by ELISA. Cry j 3 induced the release of histamine from leukocytes. We cloned the cDNA and named it Cry j 3.8. Cry j 3.8 cDNA encoded 225 amino acids and had significant homology with thaumatin-like proteins.
Conclusions
Cry j 3 is a causative allergen in Japanese cedar pollinosis and may play crucial roles in the cross-reactivity with oral allergy syndrome.
Keywords: Japanese cedar, oral allergy syndrome, pathogenesis-related protein, pollinosis, thaumatin-like protein
Japanese cedar (Cryptomeria japonica) pollen is one of the most prevalent allergens in Japan. A nationwide survey using a cross-sectional random sampling method estimated that at least 13% of the Japanese population is suffering from Japanese cedar pollinosis (1). Because of the steady increase in this allergy, especially among school children, Japanese cedar pollinosis has become a severe social problem.
Two major allergens from Japanese cedar pollen, Cry j 1 and Cry j 2, have been shown to cause Japanese cedar pollinosis (2, 3). Cry j 1 was identified as a 41–46 kDa allergen with pectate lyase enzyme activity (2, 4, 5), while Cry j 2 was identified as a 45 kDa allergen with polymethylgalacturonase enzyme activity (3, 6–8). Both Cry j 1 and Cry j 2 have several isoforms that differ in their primary structures, post-translational modifications, or reactivity with antibodies (9–11).
Recently, we cloned seven isoforms of cDNA encoding Cry j 3 as homologues of Jun a 3 (12, 13). Jun a 3 is a major allergen from mountain cedar (Juniperus ashei) pollen and has been reported as a pathogenesis-related-5 group family (PR-5) protein allergen with an immunoglobulin E (IgE)-binding reactivity of 42.9% (6/14) in patients allergic to mountain cedar pollen. Moreover, Jun a 3 has been reported to cross-react in pollinosis patients allergic to Japanese cedar pollen at a rate of 33.3% (12/36) (14). This suggests that Cry j 3 has IgE-binding capacity in Japanese cedar pollinosis patients and cross-reacts with Jun a 3. However, the native protein of Cry j 3 has not been isolated, and thus its IgE-binding capacity has not been elucidated despite its importance as a PR-5 family allergen. Therefore, we isolated native Cry j 3 from Japanese cedar pollen and assessed its IgE-binding capacity.
In the present study, we determined the IgE-binding capacity of native Cry j 3 in patients allergic to Japanese cedar pollen using immunoblot, ELISA and histamine-release assays. We also cloned the cDNA corresponding to the isolated novel isoform of Cry j 3, and named it Cry j 3.8.
Materials and methods
Purification of Cry j 3
Crude extracts were obtained from Japanese cedar pollen as described previously (2). Briefly, Japanese cedar pollen was suspended in 125 mM sodium bicarbonate buffer (pH 8.0) containing 3 mM ethylenediamine tetraacetic acid for 4 h at 4°C, and the suspension was centrifuged at 9300 g for 35 min at 4°C. Ammonium sulfate was added to the supernatant until 80% saturation. The resultant precipitate was dialyzed against 5 mM phosphate buffer (pH 6.8) and then centrifuged at 10 000 g for 15 min at 4°C. The supernatant was filtered through a 1.0 μm pore membrane filter (Toyo Roshi Ltd, Tokyo, Japan) to obtain crude Japanese cedar pollen extract. The crude extract was applied directly to a DEAE-Cellulofine column (Seikagaku Corporation, Tokyo, Japan). The unadsorbed fraction was applied to a Micro-Prep® Ceramic Hydroxyapatite type I column (BioRad Laboratories Inc., Hercules, CA, USA), and then ammonium sulfate was added to the unadsorbed fraction until 80% saturation. The resultant precipitate was dialyzed against 20 mM acetate buffer (pH 5.0). The dialyzed solution was applied to a Hiload 26/10 SP Sepharose HP column (GE Healthcare Bio-Sciences Corporation, Piscataway, NJ, USA). The obtained unadsorbed fraction was dialyzed against 0.065% trifluoroacetic acid in 2% acetonitrile, and then applied to a Resource™ RPC column (GE Healthcare Bio-Sciences Corporation). The adsorbed fraction was obtained by gradient elution with 0.05% trifluoroacetic acid in 80% acetonitrile. The fractions containing native Cry j 3 were pooled and phosphate-buffered saline (PBS) was used as an exchange buffer in a PD-10 desalting column (GE Healthcare Bio-Sciences Corporation). The resultant single protein was named Cry j 3. N-terminal amino acid sequencing of purified Cry j 3 was performed by Toray Research Center Inc., Tokyo, Japan.
SDS-PAGE and immunoblotting
Protein samples (crude pollen extract and purified Cry j 3) were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 12.5% slab gel under reducing conditions with 50 mM dithiothreitol using the discontinuous buffer system of Laemmli (15). Proteins were then detected with Phast- Gel™ Blue R (GE Healthcare Bio-Sciences Corporation) or blotted onto a Hybond-P membrane (GE Healthcare Bio-Sciences Corporation) at 1 mA/cm2 for 1.5 h. The blot was probed with primary antibody (anti-Jun a 3 rabbit serum IgG, or sera from Japanese cedar pollinosis patients or healthy individuals). To detect rabbit IgG, the blot was incubated with horseradish peroxidase-conjugated anti-rabbit IgG (Zymed Laboratories Inc., San Francisco, CA, USA). In the case of human IgE, the blot was overlaid with biotinylated anti-human IgE (Vector Laboratories Inc., Burlingame, CA, USA), followed by reaction with horseradish peroxidase-conjugated streptavidin (Zymed Laboratories). After immunolabeling, the positive bands were visualized on the membrane using 3,3′,5,5′-tetramethylbenzidine or on a chemiluminescence imager (Atto Corp., Tokyo, Japan) or an X-ray film (Fuji Photo Film Co. Ltd, Tokyo, Japan) using an ECL-Plus Western blotting detection kit (GE Healthcare Bio-Sciences Corporation).
Analysis of glycosylation on Cry j 3
Five micrograms of protein samples (Cry j 1, Cry j 3, horseradish peroxidase as a positive control and soybean trypsin inhibitor as a negative control) were fractionated by SDS-PAGE, followed by blotting onto polyvinylidene difluoride (PVDF) membrane as described above. Glycoprotein was visualized by using a GelCode™ Glycoprotein staining kit (PIERCE Biotechnology, Inc., Rockford, IL, USA) according to the manufacturer’s instruction. Briefly, gel was incubated with oxidizing solution and then washed three times by gently agitating with 3% acetic acid. The gel was submerged in glycoprotein staining reagent, followed by reaction with reducing solution with gentle agitation. The gel was washed with 3% acetic acid, followed by ultrapure water.
ELISA for specific IgE to pollen allergens
Specific IgE to pollen allergens was measured by fluorometric ELISA. Briefly, purified antigen solutions (500 ng/ml of Cry j 1, Cry j 2 or Cry j 3) were applied to a 96-well microtiter plate (NUNC-Immuno® Plate Maxisorp F96; NalgeNunc International, Roskilde, Denmark) and incubated at 4°C overnight. After the plate was blocked with 1% (w/v) bovine serum albumin in PBS for 2 h at 37°C, 10-fold diluted patient’s sera were added and incubated for 4 h at room temperature. Diluted (1 : 10) β-galactosidase- conjugated anti-human IgE monoclonal antibody (Pharmacia Diagnostics AB, Uppsala, Sweden) was then added, followed by incubation at 4°C overnight. For enzymatic reaction, 0.2 mM 4-methylumbelliferyl β-D -galactopyranoside (Sigma Aldrich Corp., St Louis, MO, USA) was added, followed by incubation at 37°C for 2 h. The fluorescence intensity was measured using a fluorometric microplate reader (Fluoroscan; Flow Laboratories, McLean, VA, USA).
Assay of histamine release from human leukocytes
Histamine release experiments from washed leukocytes were conducted using the same method as described previously (16). Washed leukocytes were obtained from the venous blood of donors and suspended in PIPES buffer (25 mM piperazine-N,N′-bis-2-ethane-sulfonic acid, 110 mM sodium chloride and 5 nM potassium chloride) containing 1 mM Ca2+, 1 mM Mg2+ and 0.03% (w/v) human serum albumin (pH 7.4). Allergen preparations diluted in the same PIPES buffer were mixed at a series of concentrations with washed leukocytes at 37°C for 45 min. The histamine released into the supernatant was measured by automated fluorometry (17).
Molecular cloning of Cry j 3.8 from a cDNA library
We constructed a full-length cDNA library from male flowers and pollen of Japanese cedar at several stages during the development (N. Futamura et al., unpublished data). In addition, the 5′ ends of cDNA sequences from approximately 20 000 clones obtained from the cDNA library were analyzed. We obtained a clone in which the 5′-end sequence coincided completely with the sequence deduced from the N-terminal amino acid sequence of purified native Cry j 3. Thus, we determined the complete cDNA sequence from the clone and named it Cry j 3.8.
Results
Purification of Cry j 3 from Japanese cedar pollen
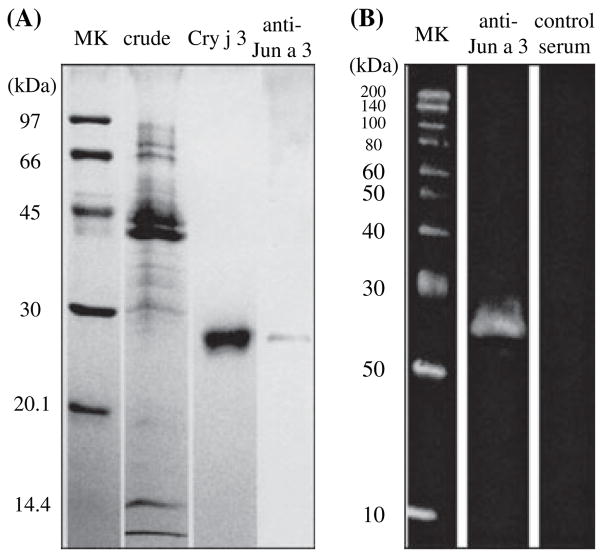
As in the many cases of purification of thaumatin-like proteins (TLPs), we isolated a single protein after gradient elution with acetonitrile on reverse phase chromatography following anion exchange, hydroxyapatite and cation exchange chromatography. The purified protein migrated at 27.3 kDa under reducing conditions on SDS-PAGE (Fig. 1A). The homologous purified protein reacted with anti-Jun a 3 polyclonal antibody on Western blotting (Fig. 1B). To confirm that this positive protein was Cry j 3, we analyzed the N-terminal amino acid sequence, which was found to coincide completely with that of Jun a 3. Therefore, we concluded that the purified protein was native Cry j 3 (Fig. 2).
Figure 1.
Purification of native Cry j 3 from crude Japanese cedar pollen extract. Crude extract from Japanese cedar pollen and purified Cry j 3 were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The molecular weight marker (lane 1), crude soluble extract (lane 2) and purified Cry j 3 (lane 3) were separated. Crude soluble extract was then transferred onto polyvinylidene difluoride (PVDF) membrane, followed by staining with anti-Jun a 3 rabbit serum (lane 4) (A). Cry j 3.8 was subjected to SDS-PAGE and transferred onto a PVDF membrane. The blots were stained with anti-Jun a 3 rabbit serum or control rabbit serum (B).
Figure 2.
Alignment of N-terminal amino acid sequences from Cry j 3 isoforms and Jun a 3.
Determination of Cry j 3-specific IgE levels in the sera of patients allergic to Japanese cedar pollen
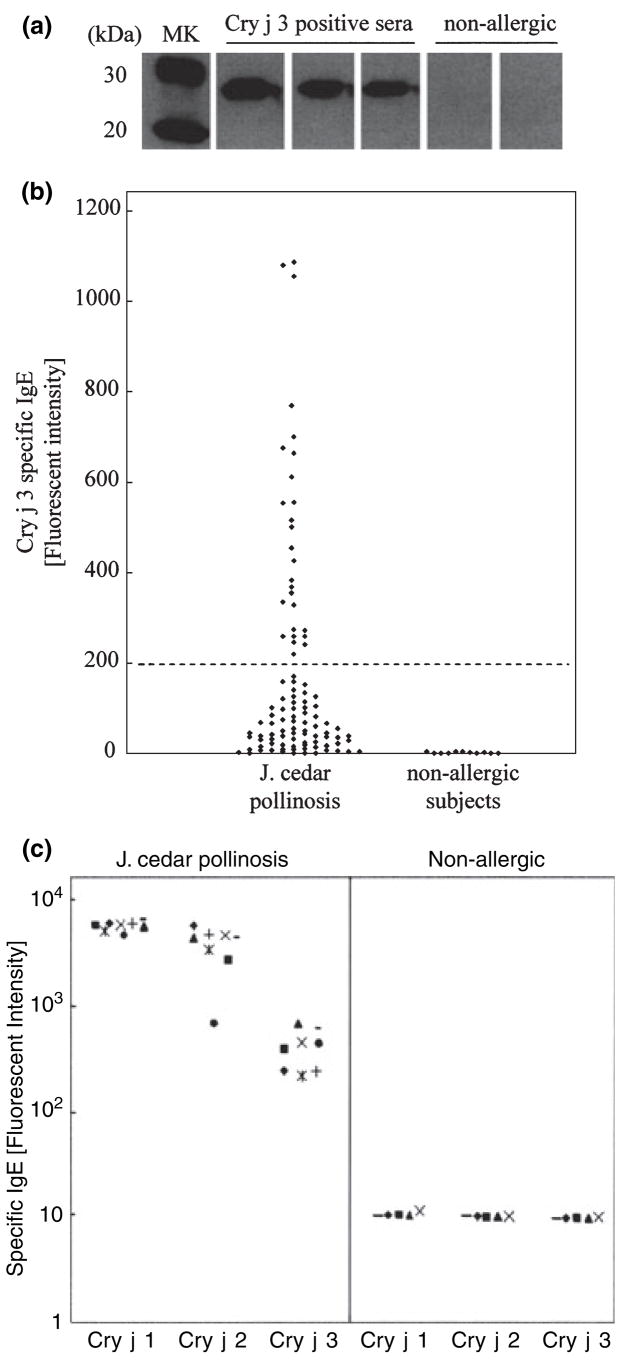
Purified Cry j 3 on a blot was reactive with the sera from patients allergic to Japanese cedar pollen but not with the sera from nonallergic individuals (Fig. 3A). ELISA was performed using the sera of 100 Japanese cedar pollinosis patients to analyze the IgE-binding frequency and specific IgE levels against Cry j 3. Twenty-seven percent (27/100) of these patients exhibited significantly greater IgE-binding ability against Cry j 3 compared with healthy volunteers (Fig. 3B). The IgE-binding intensity of Cry j 3 in the patient’s sera was almost one 14th and one 10th of the IgE-binding intensities of Cry j 1 and Cry j 2, respectively (the mean fluorescence intensities with patient’s IgE against Cry j 1, Cry j 2 and Cry j 3 were 5800, 3900 and 410, respectively; Fig. 3C).
Figure 3.
Immunoglobulin E (IgE)-binding ability of Cry j 3 in patients allergic to Japanese cedar pollen. Purified Cry j 3 was subjected to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred onto a polyvinylidene difluoride (PVDF) membrane. The blots were stained with serum IgE from Japanese cedar pollen-sensitized patients or healthy subjects as controls (A). ELISA was performed using IgE in the sera from 100 patients with Japanese cedar pollinosis and 11 healthy subjects. The cut line shows the cut-off value for a positive result (B). IgE-binding capacity was compared among Cry j 1, Cry j 2 and Cry j 3 (C). The specific IgE level against each allergen was assayed by ELISA using sera from eight Cry j 3.8-positive allergic patients with Japanese cedar pollinosis and five nonallergic subjects as controls. Identical symbols show IgE titers from the same patient or healthy subject.
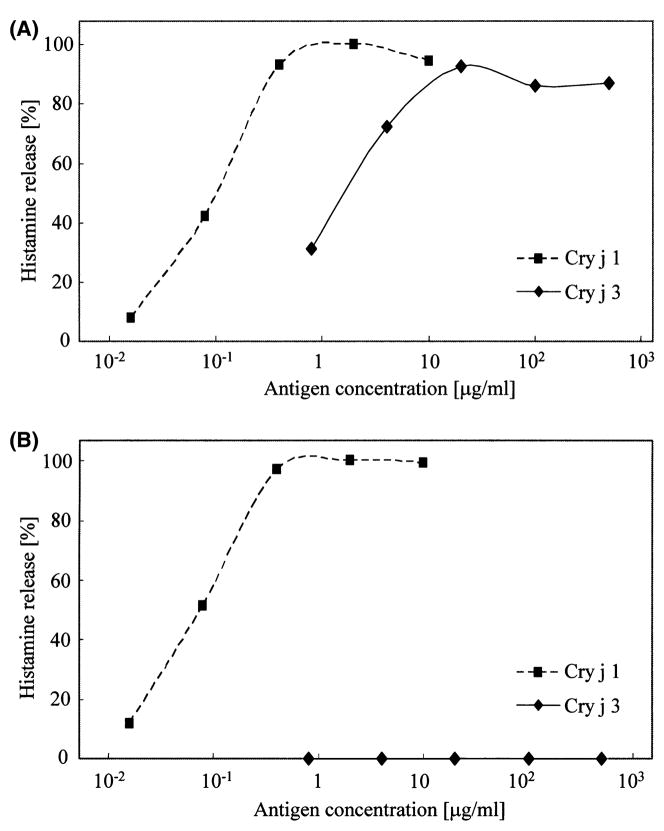
Purified Cry j 3 induced histamine release from leukocytes obtained from Cry j 3-positive patients in a dose-dependent manner, but not from Cry j 3-negative patients (Fig. 4). The sensitivity (the dose of allergen that induced 25% histamine release) was one 15th that of Cry j 1 (32 ng/ml for Cry j 1 and 480 ng/ml for Cry j 3; Fig. 4A). These data were consistent with the difference in IgE-binding intensity between Cry j 3 and Cry j 1 (1 : 14).
Figure 4.
The amount of histamine released into the culture supernatant from leukocytes obtained from Cry j 3-positive (A) and Cry j 3-negative (B) Japanese cedar pollinosis patients stimulated with Cry j 1 or Cry j 3 was assayed by automated fluorometry.
Molecular cloning of Cry j 3.8 from Japanese cedar pollen
The N-terminal sequence of the purified protein did not coincide with the deduced amino acid sequences from any of the previously cloned Cry j 3 isoforms, Cry j 3.1 to Cry j 3.6 (12, 13) and Cry j 3.7 (accession number AB212218) (Fig. 2). Therefore, we again cloned the cDNA from the cDNA library of Japanese cedar and named the novel isoform Cry j 3.8 (accession number AB254807).
Sequence analysis revealed that the Cry j 3.8 cDNA consisted of 943 nucleotides containing a 678 bp open reading frame (ORF). The ORF encoded a 225 amino acid polypeptide. In addition, we found two potential N-glycosylation sites, one consensus pattern of the TLP family, and 16 consensus cysteine residues in the ORF (18, 19) (Fig. 5). By glycoprotein staining of Cry j 3, we detected a faint positive band on the blot, indicating that native Cry j 3 was actually glycosylated (data not shown). The deduced amino acid sequence of whole Cry j 3.8 had 45–76% identity with other Cry j 3 isoforms and 86% identity with Jun a 3. Multiple sequence alignment showed a high degree of identity between Cry j 3.8 and other TLPs, especially with previously described TLP allergens from other plant species (Fig. 6). The amino acid identities between Cry j 3.8 and the other TLP allergens, Jun r 3.1 (J. rigida), Jun r 3.2, Jun a 3, Cup s 3.2 (Cupressus sempervirens) and Cup s 3.3 were 86.2%, 85.8%, 85.8%, 84.9% and 84.9%, respectively.
Figure 5.
Nucleotide sequence of Cry j 3.8 cDNA and its deduced amino acid sequence. A consensus pattern of TLRs (residues 82– 97), G-x-[GF]-x-C-x-T-[GA]-D-C-x(1,2)-G-x(2,3)-C, is shown in boldface, and conserved cysteine residues are indicated as C*. Two potential N-glycosylation sites (residues 133 and 188) are double-underlined, and the N-terminal sequence determined is single-underlined.
Figure 6.
Multiple sequence alignment of Cry j 3.8 with the TLPs, Jun r 3.1 (Juniperus rigida, accession number AY353703), Jun a 3 (J. ashei, AF121776), Cup s 3.2 (Cupressus sempervirens, AY353706), and T. occidentalis (Thuja occidentalis, AY795850). Identical amino acid residues are indicated in boldface.
Discussion
The IgE-binding frequency of Cry j 3 in pollinosis patients allergic to Japanese cedar pollen was estimated as 27%. The IgE-binding capacity was estimated as one 15th when compared with Cry j 1 by ELISA and histamine-release assay from leukocytes, and one 10th when compared with Cry j 2 by ELISA. In one patient, the specific IgE titer against Cry j 3 was comparable to that against Cry j 2 (filled circles in Fig. 3C). This suggests that Cry j 3 is an important allergen in some patients. It has been reported that the expression level and allergenic activity of TLPs vary under different environmental conditions (20). Therefore, the importance of Cry j 3 as a Japanese cedar allergen may differ among years and locations.
Cry j 1 cross-reacts with Jun a 1, and the two allergens share some linear and probable conformational epitopes (21). Moreover, cross-reactivity between Cry j 3 and Jun a 3 has also been predicted. Indeed, in the present study the anti-serum raised against Jun a 3 recognized Cry j 3, and it has been reported that 33% (12/36) of a group of patients allergic to Japanese cedar pollen showed cross-reaction with Jun a 3 (22). In Japanese cedar pollinosis patients, the IgE-binding frequency against Jun a 3 is comparable with that against Cry j 3 (33% and 27%, respectively). Furthermore, three sequential IgE epitopes of Jun a 3 (amino acids 120–131, 132–145 and 152–165) are highly conserved in Cry j 3.8, at 92% (120– 130), 100% (132–145) and 86% (152–153, 155–162 and 164–165) (23). These data strongly imply cross-reactivity between Cry j 3 and Jun a 3.
Cross-reactivity between allergens from Japanese cedar pollen and tomato have been reported in dogs and humans (24, 25), although the causative allergens remain unknown. Because two isoforms of TLP have been isolated from tomato fruit and their amounts shown to increase with ripening (26), Cry j 3 may be one of the causative allergens of the cross-reactivity. TLPs are reported to be contained in many fruits, as is chitinase. TLPs have significant amino acid sequence identity with thaumatin and are categorized in the PR-5 family (27). The PR-5 family comprises thaumatin, osmotin and zeamatin, and proteins highly homologous to them. PR-5 family proteins are well known as allergens from pollen and sweet fruits. Mal d 2 from apple (Malus domestica) was the first allergen described as a TLP (28). Following the identification of Mal d 2, several fruit allergens, including Pru av 2 from cherry (Prunus avium), Act c 2 and Act d 2 from kiwi (Actinidia chinensis and A. deliciosa, respectively), Cap a 1 from bell pepper (Capsicum annuum) and thaumatin from grape (Vitis vinifera), were reported as allergens belonging to the PR5 family (19, 29–32). Recently, PR-5 allergens from pollen have been identified as Jun a 3 from mountain cedar (J. ashei), Jun v 3 from eastern red cedar (J. virginiana) and Cup a 3 from C. arizonica (20, 22, 33). The PR proteins are considered important pan-allergens responsible for pollinosis and oral allergy syndrome (14). A recently identified allergen from Japanese cedar pollen, CJP-4, also belongs to the PR family. CJP-4 has been identified as a 34 kDa protein with endochitinase activity that cross-reacts with latex allergens (34). Therefore, both Cry j 3 and CJP-4 may act as causative allergens of the cross-reactivity in Japanese cedar pollinosis and oral allergy syndrome.
We purified native Cry j 3 from Japanese cedar pollen. The N-terminal sequence of purified Cry j 3 did not completely coincide with any previously reported sequences of Cry j 3, but did coincide with that of Jun a 3. Cry j 3.8 has the highest amino acid identity with Jun a 3 (85.8%) among the Cry j 3 isoforms (Cry j 3.1–3.6, 42.5– 54.8%; Cry j 3.7, 73.6%) (12, 13). However, it remains to be elucidated whether the other Cry j 3 isoforms exist in mature Japanese cedar pollen.
Acknowledgments
We thank Professor Kazuhisa Ono (Hiroshima University, Hiroshima, Japan) for providing the Japanese cedar pollen. This study was supported by a Grant-in-Aid for Scientific Research from the Ministry of Health, Labor and Welfare of Japan and by an EPA STAR research grant (no. 83-0399) (to R.M.G).
References
- 1.Okuda M. Epidemiology of Japanese cedar pollinosis throughout Japan. Ann Allergy Asthma Immunol. 2003;91:288–296. doi: 10.1016/S1081-1206(10)63532-6. [DOI] [PubMed] [Google Scholar]
- 2.Yasueda H, Yui Y, Shimizu T, Shida T. Isolation and partial characterization of the major allergen from Japanese cedar (Cryptomeria japonica) pollen. J Allergy Clin Immunol. 1983;71(1 Pt 1):77–86. doi: 10.1016/0091-6749(83)90550-x. [DOI] [PubMed] [Google Scholar]
- 3.Sakaguchi M, Inouye S, Taniai M, Ando S, Usui M, Matuhasi T. Identification of the second major allergen of Japanese cedar pollen. Allergy. 1990;45:309–312. doi: 10.1111/j.1398-9995.1990.tb00501.x. [DOI] [PubMed] [Google Scholar]
- 4.Sone T, Komiyama N, Shimizu K, Kusakabe T, Morikubo K, Kino K. Cloning and sequencing of cDNA coding for Cry j I, a major allergen of Japanese cedar pollen. Biochem Biophys Res Commun. 1994;199:619–625. doi: 10.1006/bbrc.1994.1273. [DOI] [PubMed] [Google Scholar]
- 5.Taniguchi Y, Ono A, Sawatani M, Nanba M, Kohno K, Usui M, et al. Cry j I, a major allergen of Japanese cedar pollen, has pectate lyase enzyme activity. Allergy. 1995;50:90–93. doi: 10.1111/j.1398-9995.1995.tb02489.x. [DOI] [PubMed] [Google Scholar]
- 6.Komiyama N, Sone T, Shimizu K, Morikubo K, Kino K. cDNA cloning and expression of Cry j II the second major allergen of Japanese cedar pollen. Biochem Biophys Res Commun. 1994;201:1021–1028. doi: 10.1006/bbrc.1994.1804. [DOI] [PubMed] [Google Scholar]
- 7.Namba M, Kurose M, Torigoe K, Hino K, Taniguchi Y, Fukuda S, et al. Molecular cloning of the second major allergen, Cry j II, from Japanese cedar pollen. FEBS Lett. 1994;353:124–128. doi: 10.1016/0014-5793(94)01022-6. [DOI] [PubMed] [Google Scholar]
- 8.Ohtsuki T, Taniguchi Y, Kohno K, Fukuda S, Usui M, Kurimoto M. Cry j 2, a major allergen of Japanese cedar pollen, shows polymethylgalacturonase activity. Allergy. 1995;50:483–488. doi: 10.1111/j.1398-9995.1995.tb01183.x. [DOI] [PubMed] [Google Scholar]
- 9.Fujimura T, Shigeta S, Kawamoto S, Aki T, Masubuchi M, Hayashi T, et al. Two-dimensional IgE-binding spectrum of Japanese cedar (Cryptomeria japonica) pollen allergens. Int Arch Allergy Immunol. 2004;133:125–135. doi: 10.1159/000076438. [DOI] [PubMed] [Google Scholar]
- 10.Maeda M, Kamamoto M, Hino K, Yamamoto S, Kimura M, Okano M, et al. Glycoform analysis of Japanese cedar pollen allergen, Cry j 1. Biosci Biotechnol Biochem. 2005;69:1700–1705. doi: 10.1271/bbb.69.1700. [DOI] [PubMed] [Google Scholar]
- 11.Goto Y, Kondo T, Ide T, Yasueda H, Kuramoto N, Yamamoto K. Cry j 1 isoforms derived from Cryptomeria japonica trees have different binding properties to monoclonal antibodies. Clin Exp Allergy. 2004;34:1754–1761. doi: 10.1111/j.1365-2222.2004.02108.x. [DOI] [PubMed] [Google Scholar]
- 12.Futamura N, Mukai Y, Sakaguchi M, Yasueda H, Inouye S, Midoro-Horiuti T, et al. Isolation and characterization of cDNAs that encode homologs of a pathogenesis-related protein allergen from Cryptomeria japonica. Biosci Biotechnol Biochem. 2002;66:2495–2500. doi: 10.1271/bbb.66.2495. [DOI] [PubMed] [Google Scholar]
- 13.Futamura N, Tani N, Tsumura Y, Nakajima N, Sakaguchi M, Shinohara K. Characterization of genes for novel thaumatin-like proteins in Cryptomeria japonica. Tree Physiol. 2006;26:51–62. doi: 10.1093/treephys/26.1.51. [DOI] [PubMed] [Google Scholar]
- 14.Midoro-Horiuti T, Brooks EG, Goldblum RM. Pathogenesis-related proteins of plants as allergens. Ann Allergy Asthma Immunol. 2001;87:261–271. doi: 10.1016/S1081-1206(10)62238-7. [DOI] [PubMed] [Google Scholar]
- 15.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 16.Yasueda H, Mita H, Yui Y, Shida T. Isolation and characterization of two allergens from Dermatophagoides farinae. Int Arch Allergy Appl Immunol. 1986;81:214–223. doi: 10.1159/000234137. [DOI] [PubMed] [Google Scholar]
- 17.Siraganian RP. An automated continuous-flow system for the extraction and fluorometric analysis of histamine. Anal Biochem. 1974;57:383–394. doi: 10.1016/0003-2697(74)90093-1. [DOI] [PubMed] [Google Scholar]
- 18.Krebitz M, Wagner B, Ferreira F, Peterbauer C, Campillo N, Witty M, et al. Plant-based heterologous expression of Mal d 2, a thaumatin-like protein and allergen of apple (Malus domestica), and its characterization as an antifungal protein. J Mol Biol. 2003;329:721–730. doi: 10.1016/s0022-2836(03)00403-0. [DOI] [PubMed] [Google Scholar]
- 19.Inschlag C, Hoffmann-Sommergruber K, O’Riordain G, Ahorn H, Ebner C, Scheiner O, et al. Biochemical characterization of Pru a 2, a 23-kD thaumatin-like protein representing a potential major allergen in cherry (Prunus avium) Int Arch Allergy Immunol. 1998;116:22–28. doi: 10.1159/000023920. [DOI] [PubMed] [Google Scholar]
- 20.Cortegano I, Civantos E, Aceituno E, del Moral A, Lopez E, Lombardero M, et al. Cloning and expression of a major allergen from Cupressus arizonica pollen, Cup a 3, a PR-5 protein expressed under polluted environment. Allergy. 2004;59:485–490. doi: 10.1046/j.1398-9995.2003.00363.x. [DOI] [PubMed] [Google Scholar]
- 21.Midoro-Horiuti T, Schein CH, Mathura V, Braun W, Czerwinski EW, Togawa A, et al. Structural basis for epitope sharing between group 1 allergens of cedar pollen. Mol Immunol. 2006;43:509–518. doi: 10.1016/j.molimm.2005.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Midoro-Horiuti T, Goldblum RM, Kurosky A, Wood TG, Brooks EG. Variable expression of pathogenesis-related protein allergen in mountain cedar (Juniperus ashei) pollen. J Immunol. 2000;164:2188–2192. doi: 10.4049/jimmunol.164.4.2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soman KV, Midoro-Horiuti T, Ferreon JC, Goldblum RM, Brooks EG, Kurosky A, et al. Homology modeling and characterization of IgE binding epitopes of mountain cedar allergen Jun a 3. Biophys J. 2000;79:1601–1609. doi: 10.1016/S0006-3495(00)76410-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kondo Y, Tokuda R, Urisu A, Matsuda T. Assessment of cross-reactivity between Japanese cedar (Cryptomeria japonica) pollen and tomato fruit extracts by RAST inhibition and immunoblot inhibition. Clin Exp Allergy. 2002;32:590–594. doi: 10.1046/j.0954-7894.2002.01337.x. [DOI] [PubMed] [Google Scholar]
- 25.Fujimura M, Ohmori K, Masuda K, Tsujimoto H, Sakaguchi M. Oral allergy syndrome induced by tomato in a dog with Japanese cedar (Cryptomeria japonica) pollinosis. J Vet Med Sci. 2002;64:1069–1070. doi: 10.1292/jvms.64.1069. [DOI] [PubMed] [Google Scholar]
- 26.Pressey R. Two isoforms of NP24: a thaumatin-like protein in tomato fruit. Phytochemistry. 1997;44:1241–1245. doi: 10.1016/s0031-9422(96)00667-x. [DOI] [PubMed] [Google Scholar]
- 27.Breiteneder H. Thaumatin-like proteins – a new family of pollen and fruit allergens. Allergy. 2004;59:479–481. doi: 10.1046/j.1398-9995.2003.00421.x. [DOI] [PubMed] [Google Scholar]
- 28.Hsieh LS, Moos M, Jr, Lin Y. Characterization of apple 18 and 31 kd allergens by microsequencing and evaluation of their content during storage and ripening. J Allergy Clin Immunol. 1995;96 (6 Pt 1):960–970. doi: 10.1016/s0091-6749(95)70234-2. [DOI] [PubMed] [Google Scholar]
- 29.Gavrovic-Jankulovic M, Cirkovic T, Vuckovic O, Atanaskovic-Markovic M, Petersen A, Gojgic G, et al. Isolation and biochemical characterization of a thaumatin- like kiwi allergen. J Allergy Clin Immunol. 2002;110:805–810. doi: 10.1067/mai.2002.128947. [DOI] [PubMed] [Google Scholar]
- 30.Bublin M, Mari A, Ebner C, Knulst A, Scheiner O, Hoffmann-Sommergruber K, et al. IgE sensitization profiles toward green and gold kiwifruits differ among patients allergic to kiwifruit from 3 European countries. J Allergy Clin Immunol. 2004;114:1169–1175. doi: 10.1016/j.jaci.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 31.Jensen-Jarolim E, Santner B, Leitner A, Grimm R, Scheiner O, Ebner C, et al. Bell peppers (Capsicum annuum) express allergens (profilin, pathogenesis-related protein P23 and Bet v 1) depending on the horticultural strain. Int Arch Allergy Immunol. 1998;116:103–109. doi: 10.1159/000023932. [DOI] [PubMed] [Google Scholar]
- 32.Pastorello EA, Farioli L, Pravettoni V, Ortolani C, Fortunato D, Giuffrida MG, et al. Identification of grape and wine allergens as an endochitinase 4, a lipid-transfer protein, and a thaumatin. J Allergy Clin Immunol. 2003;111:350–359. doi: 10.1067/mai.2003.35. [DOI] [PubMed] [Google Scholar]
- 33.Midoro-Horiuti T, Goldblum RM, Brooks EG. Identification of mutations in the genes for the pollen allergens of eastern red cedar (Juniperus virginiana) Clin Exp Allergy. 2001;31:771–778. doi: 10.1046/j.1365-2222.2001.01079.x. [DOI] [PubMed] [Google Scholar]
- 34.Fujimura T, Shigeta S, Suwa T, Kawamoto S, Aki T, Masubuchi M, et al. Molecular cloning of a class IV chitinase allergen from Japanese cedar (Cryptomeria japonica) pollen and competitive inhibition of its immunoglobulin E-binding capacity by latex C-serum. Clin Exp Allergy. 2005;35:234–243. doi: 10.1111/j.1365-2222.2005.02167.x. [DOI] [PubMed] [Google Scholar]