Abstract
Ticks are ectoparasitic blood-feeders and important vectors for pathogens including arboviruses, rickettsiae, spirochetes and protozoa. As obligate blood-feeders, one possible strategy to retard disease transmission is disruption of the parasite’s ability to digest host proteins. However, the constituent peptidases in the parasite gut and their potential interplay in the digestion of the blood meal are poorly understood. We have characterized a novel asparaginyl endopeptidase (legumain) from the hard tick Ixodes ricinus (termed IrAE), which is the first such characterization of a clan CD family C13 cysteine peptidase (protease) in arthropods. By RT-PCR of different tissues, IrAE mRNA was only expressed in the tick gut. Indirect immunofluorescence and electron microscopy localized IrAE in the digestive vesicles of gut cells and within the peritrophic matrix. IrAE was functionally expressed in Pichia pastoris and reacted with a specific peptidyl fluorogenic substrate, and acyloxymethyl ketone and aza-asparagine Michael acceptor inhibitors. IrAE activity was unstable at pH ≥ 6.0 and was shown to have a strict specificity for asparagine at P1 using a positional scanning synthetic combinatorial library. The enzyme hydrolyzed protein substrates with a pH optimum of 4.5, consistent with the pH of gut cell digestive vesicles. Thus, IrAE cleaved the major protein of the blood meal, hemoglobin, to a predominant peptide of 4 kDa. Also, IrAE trans-processed and activated the zymogen form of Schistosoma mansoni cathepsin B1 – an enzyme contributing to hemoglobin digestion in the gut of that bloodfluke. The possible functions of IrAE in the gut digestive processes of I. ricinus are compared with those suggested for other hematophagous parasites.
Keywords: Asparaginyl endopeptidase, Legumain, protease, Tick, Midgut, Hemoglobin digestion, Ixodes ricinus
1. INTRODUCTION
Ticks are blood-feeding ectoparasites that transmit a wide variety of pathogens including arboviruses, rickettsiae, spirochetes and protozoa to humans and domestic animals (Kaufman, 1989). The hard tick Ixodes ricinus is a serious disease vector in Europe especially for transmitting tick borne-encephalitis virus and the spirochete, Borrelia burgdorferi, the causative agent of Lyme disease (Nutall, 1999). Digestion of host blood proteins, including hemoglobin, by gut-associated peptidases is an essential event for ticks that provides energy and nutrition for parasite molting and vitellogenesis (Grandjean, 1984). Also, hemoglobin hydrolysis derives antimicrobial peptides vital for the survival of both the hard and soft ticks Boophilus microplus (Fogaca et al. 1999) and Ornithodoros moubata (Nakajima et al., 2003), respectively. Therefore, preventing the parasite’s ability to digest host proteins might provide a useful strategy to interfere with disease transmission by decreasing the fecundity of ticks. In contrast to insect blood feeders (Terra and Ferreira, 1994), the contents of the tick gut are believed to be free of extracellular digestive enzymes. Rather, host blood proteins are gradually endocytosed with digestion occurring within gut cells (Sonenshine, 1991). Thus far, both cysteine (Clan CA) and aspartic (Clan AA) peptidases are believed to contribute to hemoglobinolysis (Coons et al. 1986; Mendiola et al., 1996), however, few reports characterizing individual peptidases exist (Renard et al., 2000, 2002; Boldbaatar et al., 2006). Here, we identify and characterize an asparaginyl endopeptidase (IrAE) from the hard tick Ixodes ricinus. The EST encoding a C-terminal portion of this enzyme was first identified among those genes induced by blood feeding, as determined by a subtractive cDNA hybridization approach (Rudenko et al., 2005).
AEs were first described in leguminous plants (Ishii et al., 1992) and are also known as legumains (clan CD, family C13; EC 3.4.22.34; Ishii, 1994). Since then, several orthologs have been described in both protozoa and metazoa (Müntz and Shutov, 2002; León-Félix et al., 2004; Caffrey et al., 2004, Watts et al., 2005), but, to date, not in arthropods. AEs are generally lysosomal enzymes, active at acid pH with a strict cleavage specificity for the carboxyl side of an asparagine residue at the P1 position (Chen et al., 2000). Given this discrete specificity, therefore, functions demonstrated for or ascribed to AEs have focused on their ability to process other proteins (rather than indiscriminate hydrolysis), and include, processing of peptide antigens prior to MHC II-loading (Manoury et al., 1998), processing of plant poly-proteins during seed germination in legumes (the vacuolar processing enzyme - Hara-Nishimura et al., 1993) and activation of clan CA cathepsin zymogens in the gut of hematophagous helminths such as Schistosoma mansoni (Sajid et al., 2003; Caffrey et al., 2004) and Haemonchus contortus (Oliver et al., 2006). Preliminary data indicate that I. ricinus posseses orthologous peptidases in the parasite gut. With this in mind, therefore, we have characterized the physico-chemical properties of IrAE, localized its expression to the parasite gut, and demonstrated its ability to process biologically relevant protein substrates.
2. MATERIALS AND METHODS
2.1. Animals
Ixodes ricinus was collected by flagging in woodlands around České Budějovice in the Czech Republic. Adult females were fed on laboratory guinea pigs and all animals were treated in accordance with the Animal Protection Law of the Czech Republic no. 246/1992 Sb.
2.2. Preparation and screening of cDNA library
Ten adult tick females were allowed to feed naturally on guinea pigs for five days, then removed and dissected. Total RNA was isolated from dissected guts using the TRI Reagent® solution (Sigma) and its quality checked by 1×TBE agarose gel electrophoresis according to Kevil et al. (1997). The gut-specific cDNA libary was constructed using the SMART™ cDNA Library Construction Kit (Clontech, BD Biosciences) and Gigapack® III Gold Packaging Extract (Stratagene), following protocols provided by the manufacturers. Using gut-specific first strand cDNA as a template, gene-specific primers, forward 5'-TCGGTGACGCTGAGAAGACTGAA-3' and reverse 5'-TAGATTATGCCCGA TGACTGTTGG-3' were designed to PCR amplify an EST of 239 bp encoding the C-terminal part of IrAE (Rudenko et al., 2005; AY339983). The product was radio-labeled with - [P32]dATP using the Deca Label DNA Labeling System (MBI Fermentas) and used as a probe in hybridization screening of plaques in 2 × SSC, 0.5% SDS at 63 °C overnight. Positive clones were picked, PCR tested with IrAE specific primers, and a secondary screening of the positive plaques was performed. Single positive plaques were excised into E. coli plasmids and sequenced using an automated sequencer model CEQ 2000 and the CEQ Dye Terminator Cycle Sequencing kit (Beckman Coulter) with appropriate sequencing primers.
2.3. Phylogenetic analysis
The primary sequences used for phylogenetic analysis comprised the mature catalytic domain and C- terminal extension of IrAE (394 amino acids residues). Sequences were obtained from GenBank and aligned in the program ClustalX 1.81 (Thompson et al., 1997). The alignment was manually checked using the BioEdit program (Hall, 1999). Tree reconstruction employed the Neighbor Joining (NJ) method (Saitou and Nei, 1987) in the program MEGA 2.1 (Kumar et al., 2001). Nodal supports were calculated with 500 replications.
2.4. Semi-quantitative RT-PCR
Total RNA was isolated from a variety of tissues dissected from adult female ticks fed for five days. First strand cDNA synthesis and the following PCR reactions were performed using the Enhanced Avian HS RT-PCR Kit (Sigma) following the provided instructions. Gene specific primers, forward 5'-GAGGCAGCGGGAAGGTAATC-3' and reverse 5'-AGCGCCACAAACGACACG-3', were used to amplify an IrAE product of 688 bp using an annealing temperature of 56°C. The identity of resulting PCR products was confirmed by DNA sequencing. Amplification of the ferritin mRNA was used as a loading control (Kopáček at al., 2003).
2.5. Expression and purification of recombinant IrAE proenzyme and production of antibodies
To recombinantly express pro-IrAE, the E. coli bacterial expression system, Champion™ pET directional expression kit (Invitrogen), was used. N-terminal (6×His) tagged fusion IrAE was prepared using the pET100/D-TOPO® expression vector and the following primers, forward 5′-CACCCTGGCGCTAGCTTCTCTTTTTc 3′ and reverse 5′-GTACAACACTAAACAATGCCG-3′. The resulting expression constructs were transformed into TOP10 cells (Invitrogen) and sequenced using the T7 forward and T7 reverse sequencing primers. The correct constructs were transformed into BL21 Star™ (DE3) E. coli. Expression of recombinant proteins was undertaken in 100 ml bacterial culture by induction with 1 mM IPTG according to the Champion™ pET Expression System protocol. The histidine-tagged fusion IrAE was purified from isolated inclusion bodies using Ni2+-chelating chromatography in the presence of 8 M urea. The purified protein was renatured by dialysis against 0.4 M L-arginine, 0.15 M NaCl, 1 mM mercapthoethanol, pH 7.5 and gradually decreasing the concentration of urea (from 8 to 0 M) with a final dialysis against 25 mM Tris-HCl, pH 7.5. Rabbits were immunized with purified recombinant IrAE using a standard immunization protocol (Kopáček et al., 2003). The quality of immune sera, referred to as Ra×IrAE, was checked by dot blot and Western blot analysis. Reducing SDS-PAGE and Western blotting were performed as already described (Kopáček et al., 1995).
2.6. Immunolocalization of IrAE by indirect immunofluorescent and electron microscopy
Semi-thin and ultra-thin sections of dissected tick guts were prepared according to a standard protocol (Grunclová et al., 2006). The semi-thin sections (0.5 µm) were blocked with 1% BSA and 5% non-fat dry milk solution in PBS-Tween [0.3% (v/v) Tween 20] and incubated with Ra×IrAE or pre-immune serum at a dilution of 1:25 in PBS-Tween in a wet chamber overnight at 4°C. Goat anti-rabbit IgG conjugated with FITC (Sigma) diluted 1:100 in PBS-Tween) was used as secondary antibody. Sections were than counterstained with Höchst-33 258 (Molecular Probes), mounted in 2.5% DABCO (Sigma) and examined with a fluorescence microscope Olympus BX-51 equipped with the Olympus DP-70 CCD camera. Ultra-thin sections of tick guts were blocked in 0.2 % glycine, 0.1 % NH4Cl, 0.05 % Tween-20, 10 % goat serum, 1 % fish skin gelatin (Sigma) in PBS, incubated with Ra×IrAE or pre-immune serum diluted 1:10 in 1 % fish skin gelatin/PBS for 2 h at room temperature and washed in PBS containing 0.05 % Tween 20. Protein A conjugated with 10 nm gold particles (Aurion) diluted 1:30 in PBS was used to detect primary antibody. Finally, labeled and washed ultra-thin sections were counterstained with uranyl acetate and lead citrate. Specimens were examined in a TEM JEOL 1010 electron microscope at accelerating voltage of 80 kV equipped with a MegaView III camera (SIS GMBh).
2.7. Expression of IrAE in Pichia pastoris
The starting position of the IrAE pro-region was predicted with the SignalP 3.0 server (http://www.cbs.dtu.dk/services/SignalP/) according to Bendtsen et al. (2004). IrAE zymogen was amplified using gut-specific first strand cDNA, Platinum™ Pfx polymerase (Invitrogen) and the following set of primers, forward 5′-TCTCTCGAGAAAAGAACCTCCGTACCTACCTCC-3′, containing an XhoI restriction site (underlined) and a Kex2 peptidase cleavage site, and reverse 5′-ATTCTAGATCAATGATGATGATGATGATGAACAATGCCG-3′, containing an XbaI restriction site (underlined), termination codon (bold underlined) and poly-histidine tag (in italics). Purified PCR products were ligated into the expression vector, pPICZαB (Invitrogen). The construct with correct sequence was linearized with BstXI and the plasmid electroporated into the P. pastoris X33 strain. Expression was induced following protocols provided by the manufacturer (Invitrogen). Activity in medium was determined by fluorometric assay with the peptidyl substrate, benzyloxy carbonyl (Z)-Ala-Ala-Asn-7-amido-4-methyl-coumarin (Z-AAN-AMC) (see below). Expression medium was filtered (0.45µm) and freeze-dried. One gram of the lyophilized medium was dissolved in 7 ml of distilled water and concentrated to a final volume of 500 µl in citrate-phosphate-salt (CPS) buffer pH 4.5 (50 mM citric acid and 100 mM Na2HPO4 mixed in appropriate ratio, 100 mM NaCl) by repeated ultrafiltration using a 15 ml Ultrafree™ unit (30 kDa cut-off, Millipore). The IrAE was activated by addition of 4 mM DTT and incubation at room temperature (25 °C) overnight. This preparation was stored at −20 °C.
2.8. Protease activity assays
Protease activity was measured with Z-AAN-AMC and Z-FR-AMC (Bachem) for legumain (IrAE) and S. mansoni cathepsin B (SmCB1), respectively. Assays were performed in black 96-well plates using an Infinite M200 fluorometer (TECAN) with excitation and emission wavelengths of 360 nm and 465 nm, respectively. Typical assay conditions were as follows: a 20 µl aliquot of adequately diluted activated IrAE was pre-incubated for 10 min at room temperature in 80 µl of CPS buffer adjusted to pH 5.5, and containing 4 mM DTT. The enzyme reaction was started by addition of 100 µl of 40 µM fluorescent substrate (diluted from 10 mM stock solutions in DMSO) in the CPS buffer of the same pH. For controls, the enzyme was replaced by the same volume of CPS buffer. Activity was expressed relative to that of recombinant human legumain (R&D Systems, Minneapolis, MN) where 20 µl of activated IrAE at dilution 1:64, was equivalent to 100 ng of human legumain, sufficient to yield 2000 relative fluorescent units (RFU)/min. Inhibition of IrAE was tested with an aza-peptide Michael acceptor [CBz-Ala-Ala-(aza-Asn)-CH=CH-COOEt) previously described to inhibit mammalian AE (Ekici et al., 2004). This inhibitor, referred to as Aza-Asn-11a, was kindly provided by Drs. James C. Powers and Marion Götz of the School of Chemistry and Biochemistry, Georgia Institute of Technology, Atlanta, Georgia.
2.9. IrAE specifity profiling using a positional scanning synthetic combinatorial library (PS-SCL)
Synthesis of the PS-SCL has been previously described (Choe et al., 2006). Randomized positions were incorporated by addition of the isokinetic mixture of 20 amino acids (cysteine was omitted and norleucine included). Enzyme assays were carried out in black 96-well microtiter plates (Dynex Technologies) and initiated by addition of activated recombinant IrAE (30µl per well) in 70µl CPS buffer, pH 6.0 or pH 4.0. Release of 7-amino-4-carbamoylmethylcoumarin (ACC) was measured in a Perkin–Elmer LS50B luminescence spectrometer with excitation and emission wavelengths set to 380 and 460 nm, respectively.
2.10. Activity-based labeling of yeast-recombinant IrAE
The peptide portion of fluorescein-hexanoic acid-Pro-Asp-acyloxymethyl ketone (Fhx-PD-AOMK) was synthesized as described (Kato et al., 2005). Following deprotection of the hexanoic acid, 4 eq. of fluorescein-5-isothiocyanate (FITC Isomer 1 from Invitrogen) and 7 eq. of DIEA were added in DMF and shaken overnight. Resin was then washed and the compound was cleaved from the resin by treatment with TFA/TIS/H2O (95/2.5/2.5) for 2 h. The cleavage solution was collected and concentrated in vacuo. The compound was then purified by HPLC and verified by LC/MS, with a final yield of 1.2%. This probe was used to specifically label the IrAE active site.
Aliquots of 20 µl of activated IrAE were mixed with 20 µl of CPS buffer pH 6.0 containing 4 mM DTT, and 10 µl of 25 µM Fhx-PD-AOMK in distilled water. Prior inhibition of IrAE was assessed with samples containing 50 µM Aza-Asn-11a and incubated for 20 minutes prior to labeling with Fhx-PD-AOMK for 2 hrs at room temperature. Samples were then heated to 70 °C in the reducing sample buffer (1 % DTT) and separated by SDS-PAGE on 5 – 17.5 % gradient gels. Gels were washed 3×15 min in water and then placed in a solution of 30% methanol, 10% acetic acid for 1 h. After washing for 5×5 min in water, fluorescence was detected in a Typhoon 8600 Variable Mode Imager (GE Amersham) using excitation at γ = 532 nm (green laser) and a cut-off filter set to γ = 526 nm.
2.11. Trans-activation and labeling of S. mansoni pro-cathepsin B1
The Clan CA peptidase active site-directed probe, DCG-04 (Greenbaum et al., 2000), was radio-iodinated as previously described (Xing at al., 1998). Activated IrAE (5 µl) was mixed with 5 µl (1 µg) of yeast recombinant pro-SmCB1 (Sajid et al., 2003) and 40 µl of CPS buffer of pH 4.5, 6.0 and 7.5 containing 4 mM DTT. Samples were incubated at room temperature for 4 hours in the presence or absence of Aza-Asn-11a inhibitor (50 µM, final concentration). In order to visualize SmCB1, samples were then incubated for 1 h with approximately 100 nM radio-iodinaed DCG-04 probe in CPS buffer, 4 mM DTT, pH 5.5 and resolved by reducing SDS-PAGE on a 7% gel. Gels were then vacuum-dried and labeled proteins visualized in a Typhoon 8600 Variable Mode Imager in phosphor imaging mode.
2.12. Hydrolysis of human hemoglobin by IrAE
Digestion of hemoglobin (Sigma) by IrAE was assayed overnight at room temperature (25°C) by incubating 5 µl of hemoglobin (20 mg/ml) and 5 µl of activated IrAE in 50 µL CPS buffer, pH 4.5, 6.0 or 7.5, containing 4mM DTT. Inhibition of IrAE was assessed with 50 µM of Aza-Asn-11a. The reaction mixture was resolved by reducing SDS-PAGE and staining with Coomassie blue.
3. RESULTS
3.1. Isolation and characterization of full-length IrAE
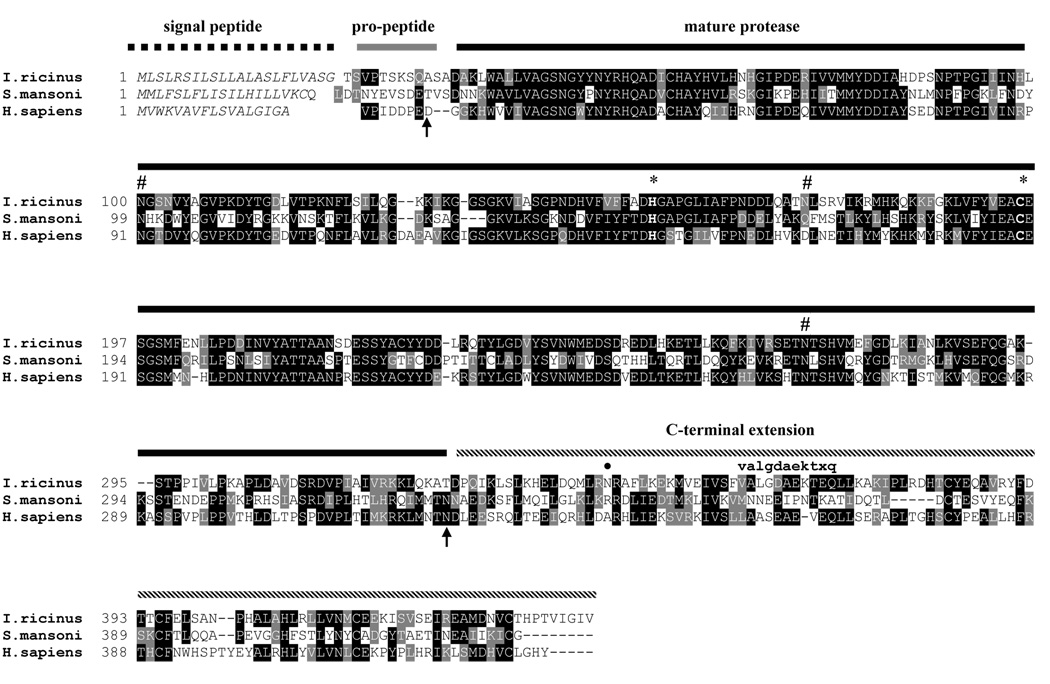
A full clone of IrAE cDNA was isolated from a gut-specific cDNA libary using a radiolabeled probe synthesized based on the EST sequence published by Rudenko et al. (2005). The complete cDNA is 1461 bp long and contains one open reading frame encoding a polypeptide of 441 amino-acid residues. The amino-acid sequence alignment of IrAE with the S. mansoni AE (Caffrey et al., 2000) and human AE (Chen et al., 1997) is shown in Fig. 1. The predicted signal peptide of IrAE is formed by the first 22 residues. The pro-enzyme has a theoretical mass of 46778.39 Da and an isoelectric point of 6.18. The conserved residues His154 and Cys195 forming the catalytic dyad are depicted with asterisks. Three Asn-linked glycosylation motifs, centering on residues 100, 171 and 272, are present. The schematic depiction of the pro-peptide, mature protease and C-terminal domain in Fig. 1 is derived from the experimental results with human AE (Chen et al., 2000; Li et al., 2003).
Figure 1. Multiple alignment of IrAE with S. mansoni and human asparaginyl-endopeptidases.
I. ricinus: the hard tick Ixodes ricinus, this work (GenBank AY584752); S. mansoni: the blood fluke Schistosoma mansoni (Caffrey et al., 2000; GenBank AJ250582); H. sapiens: human (Chen et al., 1997; GenBank Y09862). The conserved His and Cys residues forming the catalytic dyad of AE are marked with an asterisk. Space symbol indicates the possible N-glycosylation sites. The arrows depict the cleavage sites of the N- and C-terminal pro-domains experimentally determined for human AE (Chen et al., 2000; Li et al., 2003). The bold dot marks the predicted Asn residue cleavage of the IrAE C-terminal domain. The lower-case partial sequence shows the experimental N-terminal sequence of an enriched IrAE fragment.
3.2. IrAE is related to mammalian and amphibian AEs
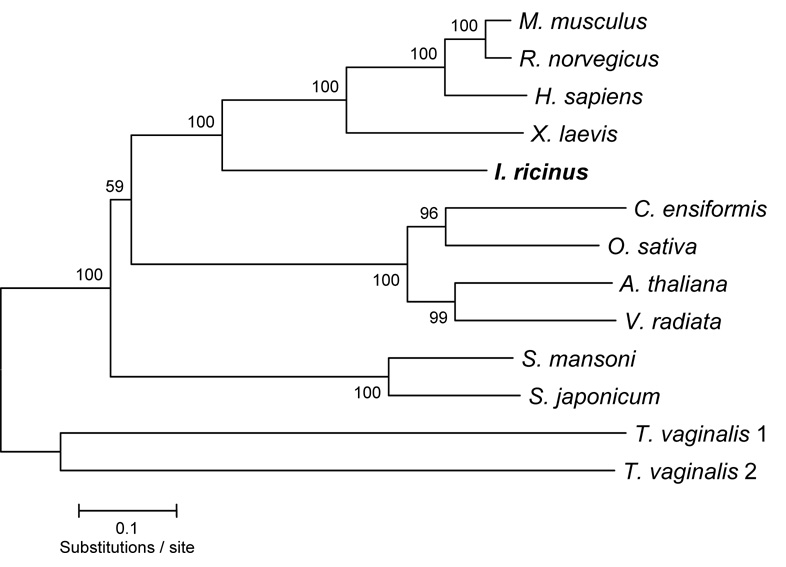
The phylogenetic analysis of IrAE with representative AEs from plants, mammals, helminths, amphibians and protists was performed using the catalytic and C-terminal domains. As shown in the Fig. 2, IrAE groups together with mammalian and amphibian orthologues, being more distant from helminth and plant AEs. An identical phylogeny was obtained using only the primary sequences of the mature domains (data not shown). The multiple alignments are available upon request, or at http://www.paru.cas.cz/fig/sojka1.zip.
Figure 2. Phylogenetic comparison of IrAE with selected AEs.
Trees were reconstructed using the Neighbor Joining method (Saitou and Nei, 1987) using amino-acid sequences spanning across the putative mature enzymes and C-terminal extensions. Mammal - Mus musculus (O89017), Rattus norvegicus (Q9R0J8); Homo sapiens (Q99538); amphibian - Xenopus laevis (AAH56842); tick - Ixodes ricinus (AAS94231); plant - Canavalia ensiformis (P49046), Oryza sativa (Q8GS39); Arabidopsis thaliana (P49047), Vigna radiata (Q9AUD9); helminth - Haemonchus contortus (CAJ45481), Schistosoma mansoni (Q9NFY9), Schistosoma japonicum (CAA50304), Fasciola hepatica (CAC85636); protist - Trichomonas vaginalis 1 and 2 (GenBank AAQ93039, AAQ93040, respectively). The horizontal bar represents a distance of 0.1 substitutions per site. Numbers at the branches represent bootstrap support. The sequence obtained in this study is marked in boldface.
3.3. IrAE is expressed solely in the gut
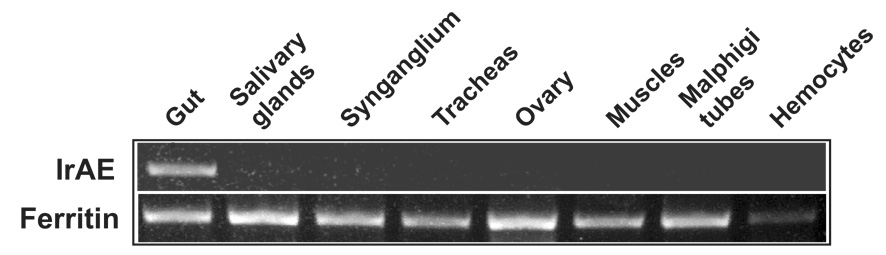
Semi-quantitative RT-PCR profiling of IrAE mRNA levels in different tissues of semi-engorged I. ricinus females revealed that enzyme message was present specifically in the gut (Fig. 3). Ferritin mRNA, levels of which are independent of blood-feeding (Kopáček et al., 2003), was used as a loading control.
Figure 3. Tissue expression profile of IrAE in female I. ricinus.
Messenger RNA levels were determined by semi-quantitative two-step RT PCR. Tissues were dissected and pooled from 10 semi-engorged females fed for five days on guinea pigs. I. ricinus ferritin mRNA was used as a loading control. For details, see Material and Methods.
3.4. IrAE is localized to the digestive vesicles and peritrophic matrix of the gut
N-terminal histidine-tagged IrAE was expressed in E. coli, purified from inclusion bodies using Ni2+ chelating sepharose in the presence of 8 M urea and refolded as described in the Material and Methods. The protein remained soluble and migrated at 50 kDa by reducing SDS PAGE (data not shown), thus agreeing with the predicted mass of 52 114 Da. By immunoblotting, the resulting rabbit anti-serum reacted with the corresponding antigen whereas no cross-reactivity was found with the pre-immune serum (data not shown). This recombinant protein did not display any hydrolytic activity of Z-AAN-AMC substrate even after extensive pre-incubation at pH 4.5 in the presence of DTT.
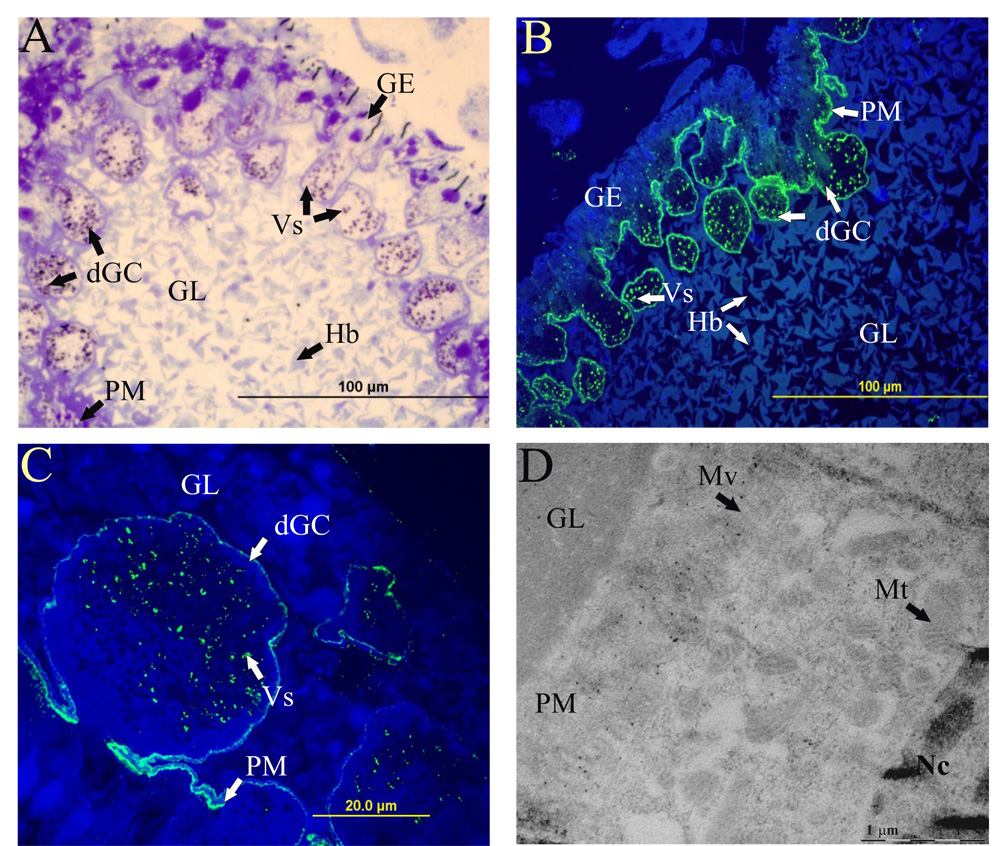
The general structure of the sections from the semi-engorged female (five days of feeding) is shown in Fig 4A. By immuno-fluorescence microscopy with specific anti-serum, IrAE was localized in the digestive vesicles of the gut epithelium cells and within the peritrophic matrix (Fig. 4B, C). No staining was observed with pre-immune sera (data not shown). The presence of IrAE within the peritrophic matrix (boundary between the gut epithelium and lumen) was further confirmed by immuno-gold electron microscopy, where the IrAE-specific gold particles were found to be clearly associated with the microvilli (Fig. 4D).
Figure 4. Localization of IrAE in the gut of female I. ricinus by indirect immunofluorescence microscopy and immunogold electron microscopy.
Sections were prepared from guts dissected from semi-engorged I. ricinus females (5 days of feeding). Panel A – semi-thin sections stained with toluidine blue - general structure of the tick gut showing the boundary between the gut epithelium (GE) digestive vesicles and the gut lumen (GL), containing large hemoglobin crystals (Hb) and digestive gut cells (dGC); Nc–nuclei; Vs – digestive vesicles. Panel B – semi-thin section labeled with Ra×IrAE serum (1:25) and FITC-conjugated anti-rabbit antibody merged with Höchst 33–258 staining (blue). Note, IrAE-specific signal was markedly enriched within the peritrophic matrix (PM) and also present intracellularly in the digestive vesicles. Panel C – a detailed image of a digestive gut cell in the phase of detachment from the gut epithelium, same staining as in Panel B. Panel D – electron microscopy of ultrathin-sections labeled with Ra×IrAE serum (1:25) and protein A conjugated with immunogold particles. IrAE-specific labeling within the peritrophic matrix showing the association of IrAE with microvilli (Mv).
3.5. Functional expression and characterization of yeast-recombinant IrAE
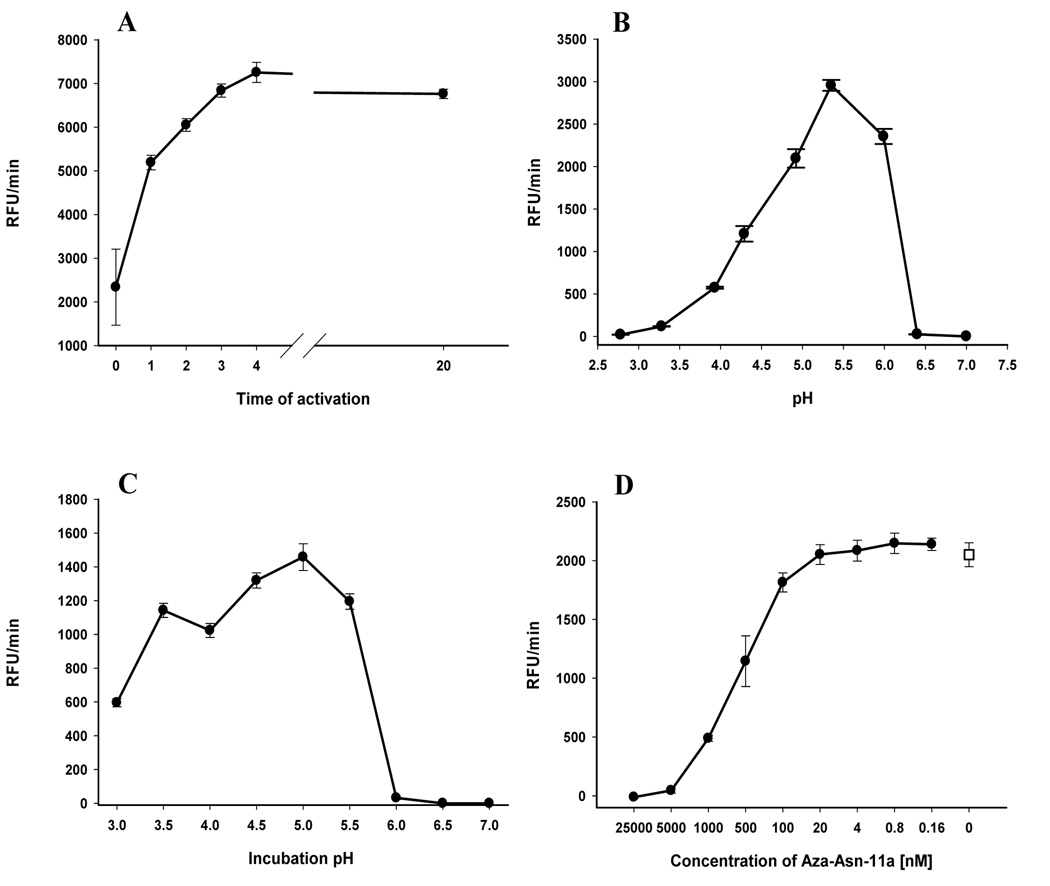
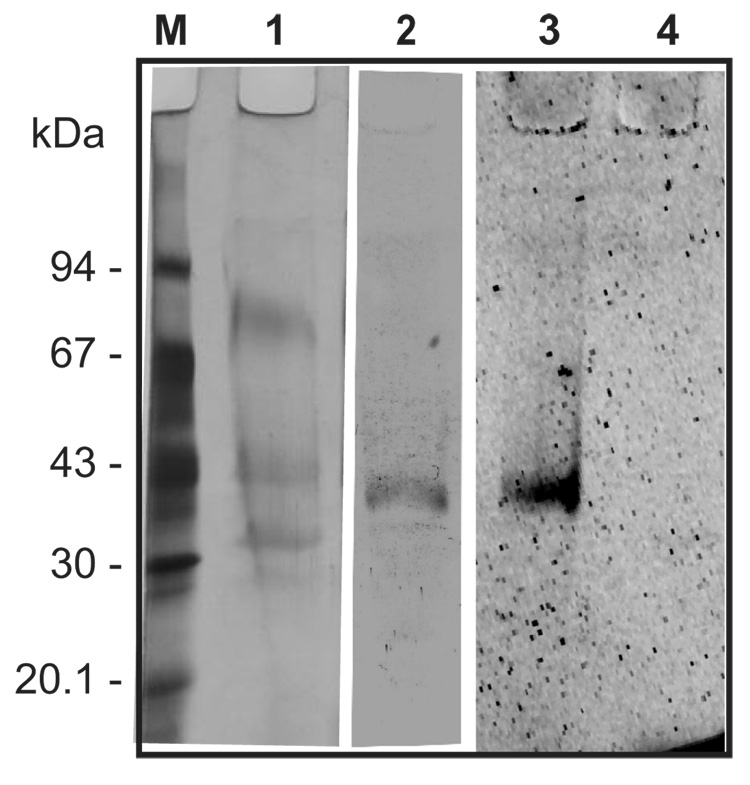
IrAE, including a C-terminal histidine tag, was expressed in Pichia pastoris and the expression medium concentrated in the by ultrafiltration. Endopeptidase activity in this concentrated medium was detectable using the peptidyl substrate Z-AAN-AMC prior to activation with 4 mM DTT. AE activity was never detected in non-transformed yeast clones similarly processed (data not shown). However, activity could be increased 3–4-fold in the presence of DTT reaching a maximum after 3–4 h and remaining stable for at least 20 hours at room temperature (Fig. 5A). The pH optimum of the activated IrAE in the concentrated yeast expression medium was approximately 5.5 and decreased sharply above pH 6.0 (Fig. 5B). The steep decline in activity was likely due to the irreversible inactivation of IrAE at pH ≥ 6.0 as determined by pre-incubating activated IrAE in CPS buffers over a pH range of 3.0 to 7.0 and subsequently measuring residual activity at pH 5.5 (Fig. 5C). Activated IrAE was effectively inhibited by the specific azapeptidyl inhibitor, Aza-Asn-11a, with an IC50 of approximately 500 nM (Fig. 5D). Despite being able to measure its activity, recombinant IrAE could not be detected as a distinct band by SDS-PAGE and silver staining on the background of proteins from the concentrated yeast expression medium (Fig. 6, lane 1). However, specific rabbit antiserum in immunoblots reacted with a protein band of approximately 38 – 40 kDa consistent with the Mr of fully activated IrAE (Fig. 6, lane 2). Also, a protein band of about 40 kDa was also detected in the concentrated yeast medium by the fluorescent activity-based probe, Fhx-PD-AOMK (Fig. 6, lane 3). Labeling of IrAE by Fhx-PD-AOMK was completely inhibited by prior reaction with the Aza-Asn-11a inhibitor (Fig. 6, lane 4). Thus, we interpret that this band likely represents the glycosylated mature form of IrAE. No alteration in the migration of the 40 kDa Fhx-PD-AOMK-labeled protein was observed after activation in the presence of DTT (see above).
Figure 5. Enzymatic characteristics of recombinant IrAE expressed in Pichia pastoris.
IrAE was expressed in P. pastoris, desalted and concentrated by ultrafiltration (30 kDa cutoff). All activity assays were performed at room temperature in triplicate using the fluorogenic substrate Z-AAN-AMC. Panel A – Activation of IrAE in the presence of DTT. IrAE was transferred to citrate-phosphate-salt buffer (CPS) pH 4.5 and the activation started by addition of 4 mM DTT. IrAE was fully activated in 3 hrs and the activity was stable for at least 20 hrs. Panel B – pH optimum of activated IrAE. Activated IrAE (20 h activation) was transferred to CPS of specified pH and the rate of AMC production was measured. Notice the loss of activity at pH > 6.0. Panel C – IrAE stability at different pH values. An aliquot of activated IrAE was pre-incubated for 4 h in CPS buffer of specified pH, then transferred to the CPS buffer of pH 5.5 with 4 mM DTT and the rate of AMC production was measured. The activated IrAE was stable only at pH < 6.0. Panel D – Inhibition of activated IrAE with a legumain-specific inhibitor. The activated IrAE was pre-incubated with two-fold serial dilutions of the inhibitor Aza-Asn-11A in CPS buffer, pH 5.5 for 30 minutes and then the rate of AMC production was measured. The IC50 concentration of the inhibitor was in the range of about 500 nM. The open symbol represents the IrAE activity without inhibitor.
Figure 6. Visualization of active recombinant IrAE by specific antiserum and the activity-based probe, Fhx-PD-AOMK.
Desalted and concentrated IrAE in P. pastoris medium (see above) was resolved by reducing, gradient SDS-PAGE. Parts of the gel were silver stained or electroblotted onto PVDF membrane and visualized with either Ra×IrAE antibodies or the activity-based fluorescent probe, Fhx-PD-AOMK. Lane 1 – Silver stain of desalted and concentrated IrAE in P. pastoris medium; Lane 2 – Western blot using Ra×IrAE serum (1 :100), swine×Ra-IgG – peroxidase conjugate and diaminobenzidine as substrate. Lanes 3, 4 – IrAE in P. pastoris medium pre-incubated with Fhx-PD-AOMK in the absence and presence of a legumain-specific inhibitor Aza-Asn-11a, respectively. For details, see Material and Methods.
Attempts to purify intact recombinant IrAE zymogen from the concentrated yeast medium through use of a C-terminal polyhistidine tag and Ni2+ sepharose chromatography were unsuccessful. By SDS-PAGE and silver staining, the only protein detected in the column eluate migrated approximately as a 17 kDa band (data not shown). Edman degradation determined the N-terminus to be a mixture of two proteins; one unknown and the other IrAE, with the sequence VALGDAE(K)TXQ (Fig. 1). This sequence commences 15 residues downstream from the theoretical cleavage site (Asn345) between the catalytic and C-terminal domains, as predicted using the server ‘NetAEP: Predicting Asparaginyl Endopeptidase specificity’ (http://theory.bio.uu.nl/kesmir/AEP/). Thus, it seems that the IrAE zymogen had already auto-activated, perhaps during expression in the yeast or subsequent handling prior to Ni2+ sepharose chromatography.
3.6. IrAE has a strict specificity for Asn at P1
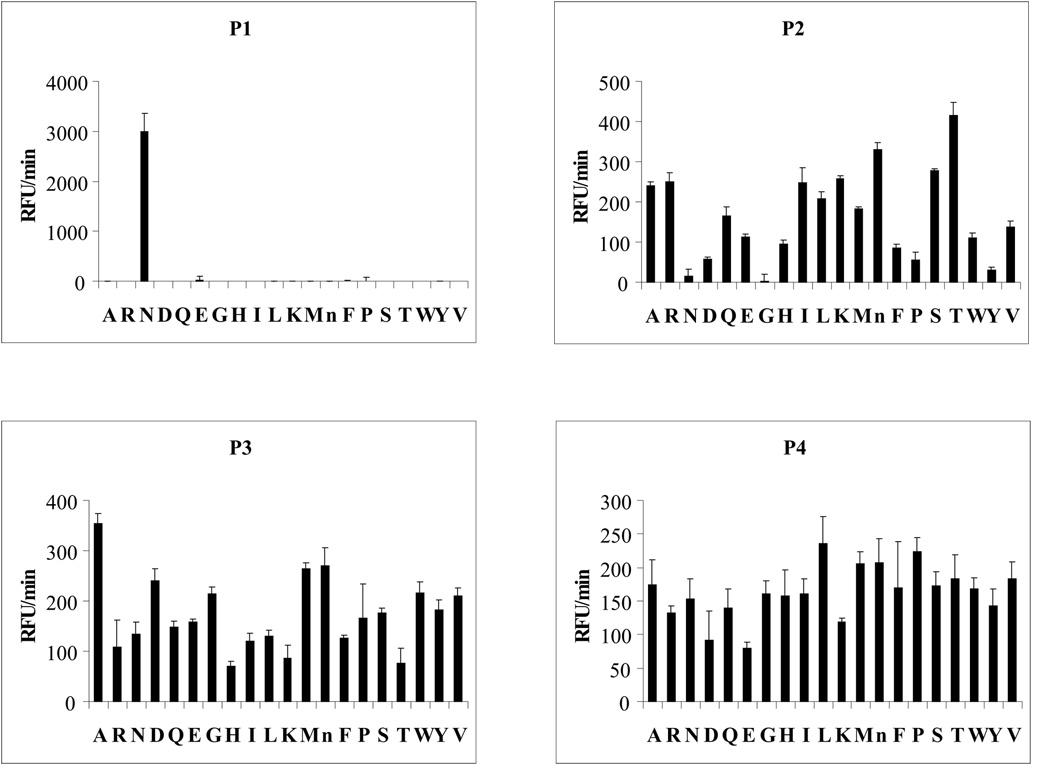
The mapping of IrAE specificity using the PS-SCL at pH 6.0 defined the specificity of IrAE for Asn at P1 position (Fig. 7). At P2, partial preferences for Thr at P2 and Ala at P3 were revealed and no preference for any residue was noted at P4 (Fig. 7). Use of the PS-SCL at pH 4.0 yielded similar results (not shown).
Figure 7. Specificity profile of IrAE using a positional scanning synthetic combinatorial library.
P1-P4 specificities were determined with a synthetic peptidyl library in which randomized positions were incorporated by addition of the isokinetic mixture of 20 amino acids (Choe et al., 2006).
3.7. Processing of macromolecular substrates by the activated IrAE
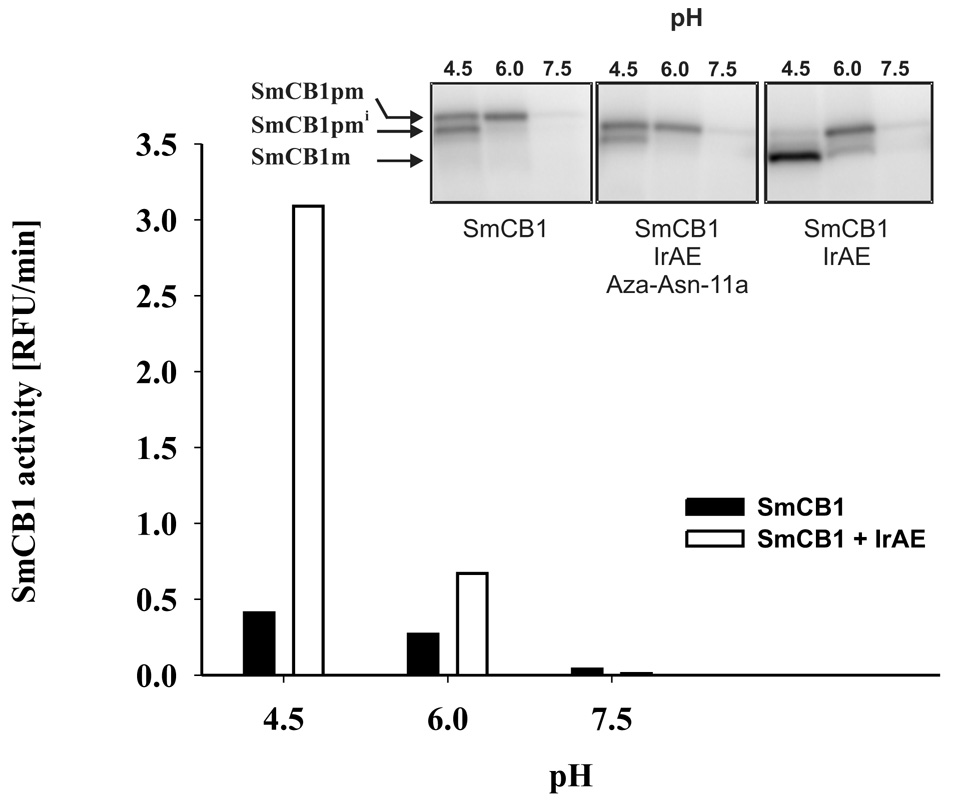
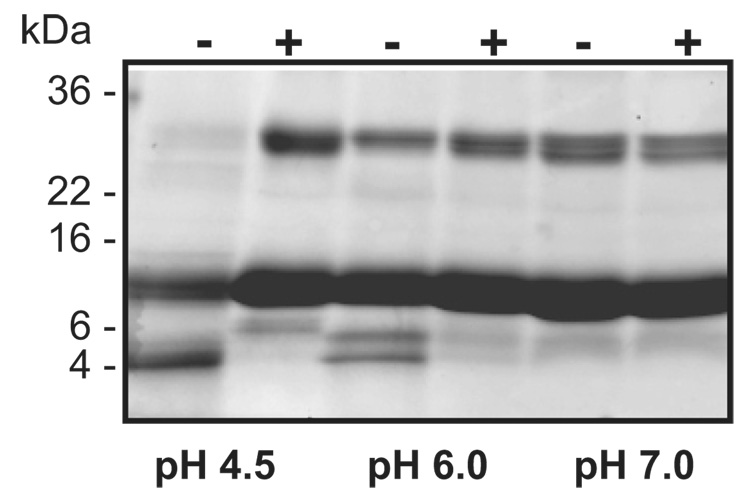
IrAE trans-processed both the full-length and intermediate forms of recombinant pro-SmCB1 (Sajid et al., 2003) to the active mature form as visualized by the Clan CA activity-based probe, DCG-04 (Figure 8 and inset, right panel). Processing was maximal at pH 4.5, being progressively less efficient at pH 6.0 and 7.5. Trans-processing was inhibited by pre-incubation of IrAE with the Aza-Asn-11a inhibitor (Fig. 8 inset, middle panel). IrAE digested human hemoglobin to a major product of 4 kDa at pH 4.5 (Fig. 9). Hemoglobinolysis was less efficient at pH 6.0, yielding two products at 6 and 4 kDa, and did not occur at pH 7.0. Edman N-terminal sequence analysis of both products resulted in a mixed signal due to the presence of at least two peptides. Nevertheless, it was possible to identify two fragments (in bold face) of the appropriate molecular mass resulting from the cleavage of the α-hemoglobin by IrAE: DKTN10/VKAA…ALTN69/ (6332.16 Da) and DPVN98/FKLL…SKYR (4797.7 Da) for the 6 and 4 kDa bands, respectively (data not shown). No hemoglobin digestion occurred when IrAE was pre-incubated with the Aza-Asn-11a inhibitor (Fig. 9).
Figure 8. Trans-processing of S. mansoni pro-cathepsin B1 by IrAE.
Trans-processing of the SmCB1 zymogen (expressed in P. pastoris) was performed as described using the endogenous S. mansoni AE (Sajid et al., 2003). Graph – pro-SmCB1 was pre-incubated with or without activated IrAE at the specified pH and the SmCB1 activity assayed with the fluorogenic substrate Z-Phe-Arg-AMC. Inset – processing of pro-SmCB1. Pro-SmCB1 was visualized using a radio-iodinated version of the DCG-04 activity-based probe (Sajid et al., 2003). Processing could be inhibited by prior incubation with the legumain-specific inhibitor, Aza-Asn-11a. SmCB1pm, SmCB1pmi, SmCB1m refer to the zymogen, partially processed SmCB1 zymogen and the mature form of SmCB1, respectively (Sajid et al., 2003).
Figure 9. Digestion of hemoglobin by activated IrAE.
Human hemoglobin (10 µg) was incubated with activated IrAE in CPS buffers of specified pH values in the presence (+) or absence (−) of the legumain-specific inhibitor, Aza-Asn-11a. The reaction mixture was resolved on reducing, gradient SDS-PAGE and stained with Coomassie Brilliant Blue. Digestion of hemoglobin was most efficient at pH 4.5 and was inhibited by Aza-Asn-11a. The scale on right of panel indicates the positions of pre-stained molecular weight standards.
4. DISCUSSION
The hard tick I. ricinus is a major vector of tick borne-encephalitis virus and Lyme disease and, consequently, of concern to public health in Europe and northern Asia (Nutall, 1999). An improved understanding of the molecular physiology I. ricinus should conceivably offer novel opportunities for interrupting the transmission of these serious diseases. With this in mind and because hard ticks are obligate blood-feeders, we have focused on characterizing digestive peptidases in the gut, with which host proteins, particularly hemoglobin, are degraded to absorbable peptides and amino acids for parasite molting and egg-production (Grandjean, 1984; Coons et al.,1986). Hematophagy in endo- and ecto-parasites, such as in the bloodfluke S. mansoni (Caffrey et al., 2004; Delcroix et al., 2006), the intestinal nematodes Ancylostoma caninum and Haemonchus contortus, (Williamson et al., 2003) and other ticks (Renard et al., 2000, 2002; Boldbaatar et al., 2006), involves a number of aspartic and cysteine peptidases to completely degrade ingested proteins. Thus far for I. ricinus, however, nothing is known regarding component alimentary peptidases, their substrate specificities, or potential redundancy of action.
Here, we report the full sequence, isolation and characterization of an AE (a Clan CD, Family C13 cysteine protease), found in the gut of I. ricinus. To our knowledge this is the first characterization of an AE from an arthropod. An EST encoding a C-terminal portion of IrAE was identified by subtractive hybridization designed to highlight those genes up-regulated after blood feeding (Rudenko et al., 2005). The full primary structure of IrAE described herein displays features in common with those in humans (Chen et al., 1997) and schistosomes (Caffrey et al., 2000). IrAE has a predicted signal sequence and a short N-terminal pro-domain. The mature enzyme contains conserved His and Cys residues forming the catalytic dyad. Within the putative C-terminal domain, the sequence similarity of aligned AEs is much lower but still it contains four conserved cysteine residues likely forming two intrachain disulphide bridges. Maturation of AEs to the catalytic form requires autocatalytic removal of the short N-terminal and longer C-terminal peptide domains (Caffrey et al., 2000; Ishi et al., 1994). However, the C-terminal cleavage motif T322N/N(D) (numbering for human AE) shared by mammalian, schistosomal and amphibian AEs is missing in the IrAE and the nearest theoretical candidate for cleavage in the tick enzyme is at Asn345 located 18 residues downstream from the respective human AE site. It was not possible to experimentally determine the cleavage site for pro-IrAE as the enzyme had already converted to the mature form (38 – 40 kDa) during expression in Pichia and/or subsequent handling of the yeast expression medium. This processing also explains why only part of the C-terminal propeptide was captured on the Ni2+ -chelating column via the engineered polyhistidine tag. For the same reason, the precise processing site for cleavage of the N-terminal propeptide can only be hypothesized as being between Asp28 and Ala29, based on the experimentally demonstrated site at Asp25/Gly26 for human AE (Chen et al., 2000; Li et al., 2003).
The enzymatic properties of IrAE expressed in P. pastoris accord with the data reported for purified or recombinant mammalian (Chen et al., 1997; 1998; Li et al., 2003) and S. mansoni AEs (Caffrey et al., 2000). IrAE has a pH optimum for hydrolysis of Z-AAN-AMC at pH 5.5 and the activity declines sharply above pH 6.0. This phenomenon is most-likely due to the instability of activated IrAE at neutral pH reported also for plant (Ishi S., 1994) and mammalian legumains (Chen et al., 1997). The activity of IrAE was effectively inhibited with a legumain-specific Aza-peptidyl Michael acceptor inhibitor (Ekici et al., 2004) and active IrAE could be clearly visualized using the fluorescent activity-based probe, Fhx-PD-AOMK.
Mapping of the IrAE hydrolytic specificity by the PS-SCL revealed a strict specificity for N at P1, partial preferences for T and A at P2 and P3, respectively, and a broad specificity at P4. The preferred sequence at positions P4 through P1 of X-A-T-N reveals a specificity similar to those mapped for the human (P-T-N; P3 through P1) and S. mansoni AEs (A/T-A-N; P3 through P1) using similar libraries (Mathieu et al., 2002). The identified α-hemoglobin fragments revealed cleavage sites that were in reasonable agreement with the preferred sequence determined by PS-SCL. Two of the three cleavage sites contain T at P2 along with the required N at P1. One site with N at P1 (position 79) on the α-chain seemed not to be cleaved by IrAE, possibly due to the presence of an unfavorable P at P2, as judged by the PS-SCL. Although the β-chain contains six N residues, it is interesting to note that no fragments have thus far been identified. Many of the potential cleavage sites contain unfavorable P2 residues, including G which appears twice. A favorable A at P2 appears once near the C-terminus, but cleavage would result in a fragment only seven residues in length, too short to identify by SDS-PAGE.
In contrast to the optimal hydrolysis of the peptidyl AMC substrate at pH 5.5, the cleavage of protein substrates including IrAE self-activation, the trans-processing of S. mansoni pro-cathepsin B1, and the hydrolysis of hemoglobin was most efficient at pH 4.0 – 4.5, which is consistent with the acidic environment within the digestive vesicles of tick gut cells (Coons, et al., 1986; Lara, et al., 2005). The trans-processing of pro-SmCB1 by IrAE is similar to that demonstrated in vitro by the endogenous S. mansoni AE (Sajid et al., 2003) whereby fully activated and mature SmCB1 only appeared in the presence of IrAE. As has been hypothesized for Schistosoma (Sajid et al., 2003), it is possible that IrAE may contribute to the activation of putative Clan CA (cysteine) and AA (aspartic) in the tick gut via trans-processing. Preliminary evidence suggests that, like parasitic helminths, I. ricinus expresses a number of gut Clan CA and AA peptidases (Sojka, unpublished). Likewise, the discrete hydrolysis of hemoglobin to a major fragment of 4 kDa by IrAE at pH 4.5 is remarkably similar to that demonstrated for the S. mansoni ortholog in worm regurgitant (Delcroix et al., 2006). SmAE is postulated to synergize with a network of Clan CA and AA proteases to complete the breakdown of hemoglobin to absorbable peptides and amino acids (Delcroix et al., 2006) and it is possible that a similar system operates in the tick gut. The discrete processing activity of hemoglobin by IrAE is consistent with its restricted P1-N specificity and contrasts with the complete hydrolysis of the same substrate by Clan CA peptidases from B. microplus (Mendiola et al., 1996) or with the Clan AA peptidase, longepsin, from H. longicornis (Boldbaatar et al., 2006). Importantly, the present data suggest that IrAE is not responsible for the generation of anti-bacterial peptides from hemoglobin (Fogaca et al., 1999; Nakajima et al., 2003) as none of the peptides described resulted from hydrolysis with N at P1.
Of interest was the finding that IrAE was extracellularly localized within the peritrophic matrix of partially engorged females (after the 5th day of feeding). Our data support previous results of morpho-functional (Coons et al., 1986; Grigor'eva 2003, 2004) and histochemical studies (Agyei, 1992) showing that gut cells grow, differentiate and finally detach from the gut lumen during the slow feeding period (up to 6 days after tick attachment to the host). Released contents of the detached cells apparently become constituents of the peritrophic matrix on the gut epithelial surface. Entrapment and immobilization of secreted enzymes at the surface of the peritrophic membrane has been demonstrated in insects (Ferreira et al., 1994). Thus, it is conceivable that IrAE is exported from the lysosomal vesicles onto the peritrophic matrix. Whether IrAE retains catalytic activity on the peritrophic matrix is as yet unclear. With respect to the remarkable instability of IrAE above pH 6.0, we conclude that IrAE is unlikely to be active in the gut lumen as the pH of gut contents of several tick species has been reported to be at or above 7.0 (Podboronov and Berdyev, 1991).
To conclude, the demonstration of an AE in the gut of I. ricinus suggests a function for this enzyme associated with the degradation of host hemoglobin. That orthologous AEs are also found in the gut of other hematophagous parasites such as S. mansoni (Caffrey et al., 2000) and H. contortus (Oliver et al., 2006) suggests a conserved function(s) for this discrete specificity of action, perhaps by contributing directly to cleavage of hemoglobin and/or the processing of other gut peptidase zymogens (Clan CA and AA) that then act to complete the degradation of the substrate (Dalton and Brindley, 1996; Caffrey et al., 2004; Delcroix et al., 2006). Overall, the conservation of Clan CA, CD and AA gut peptidases across diverse hematophagic invertebrate parasites is remarkable and it is possible that one or more of these enzyme components represent targets for chemo- or immuno-therapeutic intervention (Sajid and McKerrow, 2002; Caffrey et al., 2004), thereby decreasing the incidence and prevalence of tick-borne diseases. Characterization of other I. ricinus peptidases will be the subject of future reports.
Acknowledgements
This work was supported by grants to P.K.: No. A6022307 from the Grant Agency of the Academy of Sciences of the Czech Republic; No. 206/06/0865 from the Grant Agency of the Czech Republic and the Research Centre No. LC06009. We thank the Sandler Family Supporting Foundation for financial support of research visits by D.S. to the San Francisco laboratory. J.H.M., C.S.C. and E.L.S. were supported by the NIH grant AI35707. D.S. was supported by project FRG3/3646/2005 and PhD program No. 524/03/H133 from Ministry of Education, Youth and Sports of the Czech Republic. Research at the Institute of Parasitology, BC ASCR and the Faculty of Biological Sciences, USB is covered by research plans Z60220518 and MSMT 6007665801, respectively. We thank James C. Powers and Marion Götz, School of Chemistry and Biochemistry, Georgia Institute of Technology, GA, for the gift of Aza-Asn-11a.
Footnotes
Note: The nucleotide sequence of the IrAE has been deposited in the GenBank database with accession number AY584752.
REFERENCES
- Agyei AD, Runham NW, Blackstock N. Histochemical changes in the midgut of two ixodid tick species Boophilus microplus and Rhipicephalus appendiculatus during digestion of the blood meal. Exp. Appl. Acarol. 1992;13:187–212. doi: 10.1007/BF01194936. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Boldbaatar D, Sikalizyo Sikasunge C, Battsetseg B, Xuan X, Fujisaki K. Molecular cloning and functional characterization of an aspartic protease from the hard tick Haemaphysalis longicornis. Insect Biochem. Mol. Biol. 2006;36:25–36. doi: 10.1016/j.ibmb.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Caffrey CR, Mathieu MA, Gaffney AM, Salter JP, Sajid M, Lucas KD, Franklin C, Bogyo M, McKerrow JH. Identification of a cDNA encoding an active asparaginyl endopeptidase of Schistosoma mansoni and its expression in Pichia pastoris. FEBS Lett. 2000;466:244–248. doi: 10.1016/s0014-5793(99)01798-6. [DOI] [PubMed] [Google Scholar]
- Caffrey CR, McKerrow JH, Salter JP, Sajid M. Blood ‘n’ guts: an update on schistosome digestive peptidases. Trends Parasitol. 2004;20:241–248. doi: 10.1016/j.pt.2004.03.004. [DOI] [PubMed] [Google Scholar]
- Chen JM, Dando PM, Rawlings ND, Brown MA, Young NE, Stevens RA, Hewitt E, Watts C, Barrett AJ. Cloning, isolation, and characterization of mammalian legumain, an asparaginyl endopeptidase. J. Biol. Chem. 1997;272:8090–8098. doi: 10.1074/jbc.272.12.8090. [DOI] [PubMed] [Google Scholar]
- Chen JM, Dando PM, Stevens RA, Fortunato M, Barrett AJ. Cloning and expression of mouse legumain, a lysosomal endopeptidase. Biochem. J. 1998;335:111–117. doi: 10.1042/bj3350111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JM, Fortunato M, Barrett AJ. Activation of human prolegumain by cleavage at a C-terminal asparagine residue. Biochem J. 2000;352:327–334. [PMC free article] [PubMed] [Google Scholar]
- Choe Y, Leonetti F, Greenbaum DC, Lecaille F, Bogyo M, Bromme D, Ellman JA, Craik CS. Substrate profiling of cysteine proteases using a combinatorial peptide library identifies functionally unique specificities. J. Biol. Chem. 2006;281:12824–12832. doi: 10.1074/jbc.M513331200. [DOI] [PubMed] [Google Scholar]
- Coons LB, Rosell-Davis R, Tarnowski BI. Bloodmeal digestion in ticks. In: Sauer JR, Hair JA, editors. Morphology, physiology, and behavioral biology of ticks. Chichester, England: Ellis Horwood, Ltd.; 1986. pp. 248–279. [Google Scholar]
- Dalton JP, Brindley PJ. Schistosome asparaginyl endopeptidase SM32 in hemoglobin digestion. Parasitol Today. 1996;12:125. doi: 10.1016/0169-4758(96)80676-4. [DOI] [PubMed] [Google Scholar]
- Delcroix M, Sajid M, Caffrey CR, Lim KC, Dvořák J, Hsieh I, Bahgat M, Dissous C, McKerrow JH. A multienzyme network functions in intestinal protein digestion by a platyhelminth parasite. J. Biol. Chem. 2006 doi: 10.1074/jbc.M607128200. in press. [DOI] [PubMed] [Google Scholar]
- Ekici ÖD, Götz MG, James KE, Li ZZ, Rukamp BJ, Asgian JL, Caffrey CR, Hansell E, Dvořák J, McKerrow JH, Potempa J, Travis J, Mikolajczyk J, Salvesen GS, Powers JC. Aza-peptide Michael acceptors: a new class of inhibitors specific for caspases and other clan CD cysteine proteases. J. Med. Chem. 2004;47:1889–1892. doi: 10.1021/jm049938j. [DOI] [PubMed] [Google Scholar]
- Ferreira C, Capella AN, Sitnik R, Terra WR. Digestive enzymes in midgut cells, endo- and ectoperithrophic contents, and peritrophic membranes of Spodoptera frugiperda (Lepidoptera) larvae. Arch. Insect Biochem. Physiol. 1994;26:299–313. [Google Scholar]
- Fogaca AC, da Silva PI, Jr, Miranda MT, Bianchi AG, Miranda A, Ribolla PE, Daffre S. Antimicrobial activity of a bovine hemoglobin fragment in the tick Boophilus microplus. J. Biol. Chem. 1999;274:25330–25334. doi: 10.1074/jbc.274.36.25330. [DOI] [PubMed] [Google Scholar]
- Grandjean O. Blood digestion in Ornithodoros moubata Murray sensu stricto Walton (Ixodoidea: Argasidae) females. I. Biochemical changes in the midgut lumen and ultrastructure of the midgut cells, related to intracellular digestion. Acarologia. 1984;25:147–165. [Google Scholar]
- Greenbaum D, Medzihradszky KF, Burlingame A, Bogyo M. Epoxide electrophiles as activity-dependent cysteine protease profiling and discovery tools. Chem. Biol. 2000;7:569–581. doi: 10.1016/s1074-5521(00)00014-4. [DOI] [PubMed] [Google Scholar]
- Grigor'eva LA. Morphofunctional changes in the midgut of tick females of the genus Ixodes (Acari: Ixodidae) during and after feeding. Parazitologiia. 2003;37:169–176. In Russian. [PubMed] [Google Scholar]
- Grigor'eva LA, Amosova LI. Peritrophic matrix in the midgut of tick females of the genus Ixodes (Acari: Ixodidae) Parazitologiia. 2004;38:3–11. In Russian. [PubMed] [Google Scholar]
- Grunclová L, Horn M, Vancová M, Sojka D, Franta Z, Mareš M, Kopáček P. Two secreted cystatins of the soft tick Ornithodoros moubata: Differential expression pattern and inhibitory specificity. Biol. Chem. 2006;387 doi: 10.1515/BC.2006.204. in press. [DOI] [PubMed] [Google Scholar]
- Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl. Acids. Symp. Ser. 1999;41:95–98. [Google Scholar]
- Hara-Nishimura I, Takeuchi Y, Nishimura M. Molecular characterization of a vacuolar processing enzyme related to a putative cysteine proteinase of Schistosoma mansoni. Plant Cell. 1993;5:1651–1659. doi: 10.1105/tpc.5.11.1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii S, Abe Y, Mitta M, Matsushita H, Kato I. A novel protease from jack bean seeds: Asparaginyl endopeptidase. J. Protein Chem. 1992;11:367–368. [Google Scholar]
- Ishii S-I. Legumain: asparaginyl endopeptidases. Methods Enzymol. 1994;244:604–615. doi: 10.1016/0076-6879(94)44044-1. [DOI] [PubMed] [Google Scholar]
- Kato D, Boatright KM, Berger AB, Nazif T, Blum G, Ryan C, Chehade KA, Salvesen GS, Bogyo M. Activity-based probes that target diverse cysteine protease families. Nat. Chem. Biol. 2005;1:33–38. doi: 10.1038/nchembio707. [DOI] [PubMed] [Google Scholar]
- Kaufman WR. Tick-host interaction: a synthesis of current concepts. Parasitol. Today. 1989;13:165. doi: 10.1016/0169-4758(89)90191-9. [DOI] [PubMed] [Google Scholar]
- Kevil CG, Walsh L, Laroux FS, Kalogeris T, Grisham MB, Alexander JS. An improved, rapid Northern protocol. Biochem. Biophys. Res. Commun. 1997;238:277–279. doi: 10.1006/bbrc.1997.7284. [DOI] [PubMed] [Google Scholar]
- Kopáček P, Weise C, Götz P. The prophenoloxidase from the wax moth Galleria mellonella: purification and characterization of the proenzyme. Insect. Biochem.Mol. Biol. 1995;25:1081–1091. doi: 10.1016/0965-1748(95)00040-2. [DOI] [PubMed] [Google Scholar]
- Kopáček P, Ždychová J, Yoshiga T, Weise C, Rudenko N, Law JH. Molecular cloning, expression and isolation of ferritins from two tick species--Ornithodoros moubata and Ixodes ricinus. Insect Biochem. Mol. Biol. 2003;233:103–113. doi: 10.1016/s0965-1748(02)00181-9. [DOI] [PubMed] [Google Scholar]
- Kumar S, Tamura K, Jakobsen IB, Nei M. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics. 2001;17:1244–1245. doi: 10.1093/bioinformatics/17.12.1244. [DOI] [PubMed] [Google Scholar]
- Lara FA, Lins U, Bechara GH, Oliveira PL. Tracing heme in a living cell: hemoglobin degradation and heme traffic in digest cells of the cattle tick Boophilus microplus. J. Exp. Biol. 2005;208:3093–3101. doi: 10.1242/jeb.01749. [DOI] [PubMed] [Google Scholar]
- Leon-Felix J, Ortega-Lopez J, Orozco-Solis R, Arroyo R. Two novel asparaginyl endopeptidase-like cysteine proteinases from the protist Trichomonas vaginalis: their evolutionary relationship within the clan CD cysteine proteinases. Gene. 2004;335:25–35. doi: 10.1016/j.gene.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Li DN, Matthews SP, Antoniou AN, Mazzeo D, Watts C. Multistep autoactivation of asparaginyl endopeptidase in vitro and in vivo. J. Biol. Chem. 2003;278:38980–38990. doi: 10.1074/jbc.M305930200. [DOI] [PubMed] [Google Scholar]
- Manoury B, Hewitt EW, Morrice N, Dando PM, Barrett AJ, Watts C. An asparaginyl endopeptidase processes a microbial antigen for class II MHC presentation. Nature. 1998;396:695–699. doi: 10.1038/25379. [DOI] [PubMed] [Google Scholar]
- Mathieu MA, Bogyo M, Caffrey CR, Choe Y, Lee J, Chapman H, Sajid M, Craik CS, McKerrow JH. Substrate specificity of schistosome versus human legumain determined by P1-P3 peptide libraries. Mol. Biochem. Parasitol. 2002;121:99–105. doi: 10.1016/s0166-6851(02)00026-9. [DOI] [PubMed] [Google Scholar]
- Mendiola J, Alonso M, Marquetti MC, Finlay C. Boophilus microplus: multiple proteolytic activities in the midgut. Exp. Parasitol. 1996;82:27–33. doi: 10.1006/expr.1996.0004. [DOI] [PubMed] [Google Scholar]
- Müntz K, Shutov AD. Legumains and their functions in plants. Trends Plant Sci. 2002;7:340–344. doi: 10.1016/s1360-1385(02)02298-7. [DOI] [PubMed] [Google Scholar]
- Nakajima Y, Ogihara K, Taylor D, Yamakawa M. Antibacterial hemoglobin fragments from the midgut of the soft tick, Ornithodoros moubata (Acari: Argasidae) J. Med. Entomol. 2003;40:78–81. doi: 10.1603/0022-2585-40.1.78. [DOI] [PubMed] [Google Scholar]
- Nutall PA. Pathogen-tick-host interactions: Borrelia bugdorferi and TBE virus. Zentralbl. Bakteriol. 1999;289:492–505. doi: 10.1016/s0934-8840(99)80002-4. [DOI] [PubMed] [Google Scholar]
- Oliver EM, Skuce PJ, McNair CM, Knox DP. Identification and characterization of an asparaginyl proteinase (legumain) from the parasitic nematode, Haemonchus contortus. Parasitology. 2006;133:237–244. doi: 10.1017/S0031182006000229. [DOI] [PubMed] [Google Scholar]
- Podboronov VM, Berdyev A. Protective mechanisms of Ixodidea tick and their vertebrate hosts (host-parasite interface) (Ashgabad, Ylym) 1991 In Russian. [Google Scholar]
- Renard G, Garcia JF, Cardoso FC, Richter MF, Sakanari JA, Ozaki LS, Termignoni C, Masuda A. Cloning and functional expression of a Boophilus microplus cathepsin L-like enzyme. Insect Biochem. Mol. Biol. 2000;30:1017–1026. doi: 10.1016/s0965-1748(00)00070-9. [DOI] [PubMed] [Google Scholar]
- Renard G, Lara FA, de Cardoso FC, Miguens FC, Dansa-Petretski M, Termignoni C, Masuda A. Expression and immunolocalization of a Boophilus microplus cathepsin L-like enzyme. Insect Mol. Biol. 2002;11:325–328. doi: 10.1046/j.1365-2583.2002.00342.x. [DOI] [PubMed] [Google Scholar]
- Rudenko N, Golovchenko M, Edwards MJ, Grubhoffer L. Differential expression of Ixodes ricinus tick genes induced by blood feeding or Borrelia burgdorferi infection. J. Med. Entomol. 2005;42:36–41. doi: 10.1093/jmedent/42.1.36. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Sajid M, McKerrow JH. Cysteine proteases of parasitic organisms. Mol. Biochem Parasitol. 2002;120:1–21. doi: 10.1016/s0166-6851(01)00438-8. [DOI] [PubMed] [Google Scholar]
- Sajid M, McKerrow JH, Hansell E, Mathieu MA, Lucas KD, Hsieh I, Greenbaum D, Bogyo M, Salter JP, Lim KC, Franklin C, Kim JH, Caffrey CR. Functional expression and characterization of Schistosoma mansoni cathepsin B and its trans-activation by an endogenous asparaginyl endopeptidase. Mol. Biochem. Parasitol. 2003;131:65–75. doi: 10.1016/s0166-6851(03)00194-4. [DOI] [PubMed] [Google Scholar]
- Sonenshine DE. Biology of Ticks. vol. 1. New York: Oxford university Press; 1991. [Google Scholar]
- Terra WR, Ferreira C. Insect digestive enzymes: properties, compartmentalization and function. Comp. Biochem. Physiol. 1994;109B:1–62. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts C, Matthews SP, Mazzeo D, Manoury B, Moss CX. Asparaginyl endopeptidase: case history of a class II MHC compartment protease. Immunol. Rev. 2005;207:218–228. doi: 10.1111/j.0105-2896.2005.00312.x. [DOI] [PubMed] [Google Scholar]
- Williamson AL, Brindley PJ, Knox DP, Hotez PJ, Loukas A. Digestive proteases of blood-feeding nematodes. Trends Parasitol. 2003;9:417–423. doi: 10.1016/s1471-4922(03)00189-2. [DOI] [PubMed] [Google Scholar]
- Xing R, Addington AK, Mason RW. Quantification of cathepsins B and L in cells. Biochem J. 1998;332:499–505. doi: 10.1042/bj3320499. [DOI] [PMC free article] [PubMed] [Google Scholar]