SUMMARY
Neuroligins are postsynaptic cell adhesion molecules that promote synaptic maturation and stabilization upon binding with presynaptic partners, the α- and β-neurexins. Using a combination of analytical ultracentrifugation, small angle x-ray, and neutron scattering, we have characterized the low-resolution three-dimensional structure of the extracellular domain of the neuroligins, free in solution, and in complex with β-neurexin. The globular extracellular domain of the neuroligins forms stable homodimers through a four-helix bundle typical of the cholinesterases and other members of the α/β-hydrolase fold family. The presence of the stalk region adds to the extracellular domain of neuroligin-1 an elongated structure, suggesting a rod-like nature of the stalk domain. Sedimentation equilibrium coupled with solution scattering data of the β-neurexin/neuroligin-1 complex indicated a 2:2 stoichiometry where two β-neurexin molecules bind to a neuroligin-1 dimer. Deuteration of neurexin allowed us to collect neutron scattering data that, in combination with other biochemical techniques, provide a basis for optimizing the positioning of each component in a higher resolution computational model of the neuroligin/neurexin complex. As several mutations of both neurexin and neuroligin genes have been linked to autism spectrum disorders and mental retardation, these new structures provide an important framework for the study of altered structure and function of these synaptic proteins.
INTRODUCTION
Autism is a heterogeneous spectrum of disorders, and research into its biological and genetic basis is still in its infancy. The genetics underlying autism spectrum disorders (ASD) are complex, involving multiple genetic variants, and multigenic interactions. Two major limitations prevail in understanding the pathogenesis of the ASD. First, the absence of a defined quantitative marker or gene product ascribed to the disorder makes the diagnosis of the disorder subjective, relying on rating scales and diagnostic checklists. Second, owing to the complicated nature of the human social behavior, animal models may not always be suitable surrogates for studying the disease. Therefore, much of what we know about the neurobiology of autism has been derived from clinical research on affected children or by postmortem analysis of brains of autistic individuals. Given these constraints, basic neuroscience research may ultimately provide crucial information to understand etiologic and pathophysiologic mechanisms.
Since the first study showing a linkage in a twin sets between the incidence of the ASD and mutations in the NLGN3 and 4 genes, several structural variants (including point mutations, truncations, and exon deletions) of the coding regions of neurexin (NX) and neuroligin (NL) were found to be associated with ASD and mental retardation(1–7). Emerging evidence also indicates that rare variations in copy number and common variations within the genes encoding NX-1 and NX-3 contribute to ASD susceptibility (8,9). Although the frequency of these individual mutations in the overall autistic population is probably low, these findings point to adhesion proteins or their associating partners as being important contributory gene products to autism and mental retardation. Predictably, associations are now emerging between autism and proteins known to associate intracellularly with NL, such as Shank3(10).
In humans, the neuroligins compose a family of transmembrane proteins composed of an N-linked glycosylated extracellular domain with strong sequence homology to acetylcholinesterase containing two sites of alternative splicing and a Ser-Thr rich stalk domain that carries N- and O-linked oligosaccharides. This domain connects the above globular domain to a single transmembrane spanning domain and a small intracellular C-terminal domain lacking a defined structure, but containing a PDZ recognition domain. The neuroligins bind in vitro to both NXα and NXβ in a Ca2+ dependent fashion. Neurexins compose a family of neuronal proteins with three genes (neurexins 1 through 3) driven by two promoters (α and β), resulting in the expression of at least six principal NX forms. Extensive alternative mRNA splicing confers additional complexity to the possible gene products (11). The atomic structures of the second and sixth LNS domain of the NXα have recently been solved(12,13); however, no structural information on the arrangement of the entire extracellular domain is available. The inhibitory role of the glycosylation carried by splice insert B in NL1 was extensively studied in both NXα and NXβ (14,15). More recently it was established that splice insert B has an even larger effect on the association than glycosylation alone(16–18). Viewed together, these data show a three-component hierarchical mechanism that could be specifically modulated by cell type and developmental stage: 1) NX and NL gene selection, 2) the splicing options afforded by both NXs and NLs, and 3) post-translational N-linked glycosylation of splice insert B in NL1 that negatively regulates both NXα and NXβ affinities.
We report here the results of small-angle solution scattering experiments on the extracellular domains of the neuroligins and the complex formed between NL1 and NXβ. X-ray scattering data provide overall shape information on the extracellular domains of the NLs and the initial structural definition of the O-linked glycosylated domain linking the extracellular domain to the transmembrane span. We also use the X-ray scattering data to refine our homology model for the NL1 dimer. A combination of X-ray scattering and neutron contrast variation data with ab initio and rigid body modeling has yielded a structural model of the extracellular domain of NL1 in complex with NXβ (19). Our model is presented in relation to the synaptic disposition of the complex, and while of inherently low resolution, it provides an important structural framework for linking genetic information on mutated neurexins and neuroligins with neuro-developmental disorders.
RESULTS
The extracellular domain of the neuroligins belongs to the α/β-hydrolase fold family of proteins
It was previously shown that the NLs behave as dimers(14,16,20). The extracellular domain of NL1, lacking both splice inserts A and B, was truncated following the α/β-hydrolase fold domain at residue 638 (NL1-638-Δ (A&B)). Using small-angle X-ray scattering data in combination with homology modeling, we first determined a three-dimensional structural model of the dimeric acetylcholinesterase-like domain for NL1 in solution. The scattering data can provide accurate and precise structural parameters that relate to the overall shape of the scattering object, as well as information on the domain or subunit dispositions within that shape. Under dilute solution conditions in which the scattering molecules are mono-disperse and identical, one can determine the molecular volume (which for proteins is generally proportional to molecular weight), radius of gyration, Rg, vector length distribution function, P(r), and the maximum linear dimension Dmax. Once these basic structural parameters are obtained, three-dimensional shape reconstruction of the protein of interest can be performed. If there are reliable homology models, crystal structure or NMR data available on domains or components of the molecule or complex, rigid body modeling can be used to develop a structural model that best fits the scattering data and incorporates the know structural components (Grishaev et al., 2005).
The distribution of the distances between scattering centers (i.e. atoms) in the protein molecule (P(r)) that is obtained as a Fourier transform of the scattering data reflects the molecular shape and allows calculation of the global descriptors such as the radius of gyration, Rg, and the maximum dimension of the protein, Dmax. The P(r) profile for NL1-638-Δ (A&B) gives values for Rg and Dmax of 42.5+0.4Å and ~130Å respectively, confirming that the NLs exist as dimers in solution, free of larger aggregates under our experimental conditions(19). These values show a close correspondence to the values estimated from the partial specific volume and the molecular weights determined from sequences and by mass spectrometry, respectively(16,21). The solution scattering data further indicate that the stalk region connecting the globular domain of the NLs with their transmembrane domain is elongated and projects away from the globular domain(19). The presence of oligosaccharides, combined with an abundance of Pro residues, presumably maintains the peptide chain in a semi-rigid, bottlebrush-like structure, as demonstrated in the P-selectin molecule and the T-cell co-receptor CD8 protein(22,23). Shape restoration and rigid body modeling show that the dimerization surface of the NLs is located at a characteristic 4-helix bundle, similarly to what was found in the crystal structure of Torpedo Californica AChE(24) and in the crystal structures of mouse and eel AChE lacking that C-terminal cysteine(25). As expected from the high sequence identities among the NLs, our scattering data show that the extracellular domains of NL2, 3 and 4 are similar in overall shape and dimensions, supporting the notion that they share a common fold and dimerize at similar surfaces(19).
Sedimentation equilibrium allows the determination of the oligomeric state of the neuroligins and the NXβ/NL complex stoichiometry
Analytical ultracentrifugation involves the combination of sedimentation velocity (SV) and sedimentation equilibrium (SE) analyses. As SV can give information on the mass and shape of the macromolecular species, and SE is sensitive only to the mass of the particle, these measurements provide complementary information. SE analysis was used to determine that whereas the extracellular domain of NXβ is monomeric in solution at concentrations < 0.5 mg/ml, NL1 is an extremely stable dimer virtually devoid of free monomer even at low concentrations (Figure 1)(16). To further characterize the association of NXβ with neuroligins, we conducted sedimentation equilibrium experiments of NL1 mixed with NXβ in the presence of 2 mM Ca2+. These experiments enabled us to calculate a molecular mass consistent with the formation of a tight complex in a 2:2 stoichiometry (Figure 1)(16).
Figure 1. Equilibrium Ultracentrifugation Analysis of NL1 and NL1/NXβ complex.
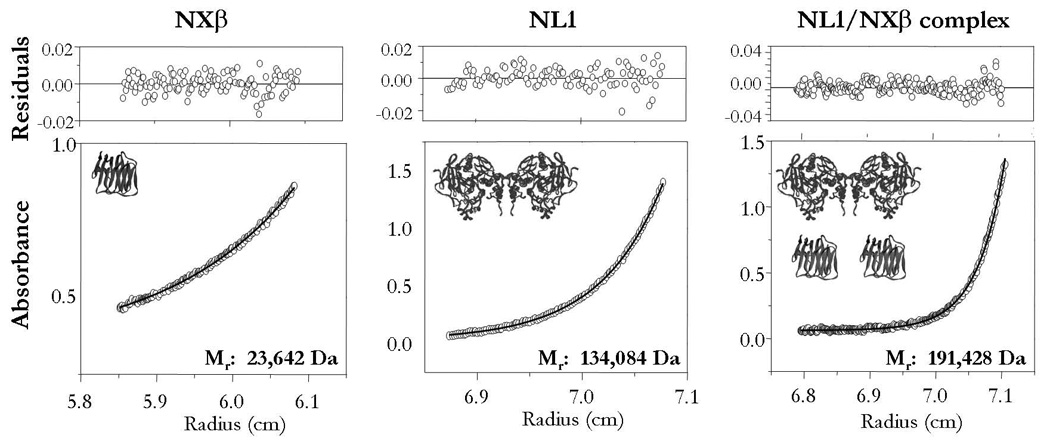
Left panel, the extracellular domain of NXβ appears monomeric in solution (sequence based MW= 24,770 Da). Center panel, the glycosylated extracellular domain of NL1 is a dimer in solution (sequence based MW= ~135,000 Da). Right panel, a 2:2 molar mixture of NXβ and NL1 was ran at equilibrium (sequence based MW of the complex= ~184,000 Da). The extracted molecular mass indicates that the stoichiometry of the complex is compatible with a 2:2 assembly. Raw data are shown as circles; solid line is the best fit. The distribution of the residuals of the best fit is shown above.
Two NXβ monomers associates with NL1 at opposite sides of the long axis of the NL1 dimer
Surface plasmon resonance refractivity measurements were used to quantify the association between NL1 and NXβ (16). Removal of splice insert B in NL1 led to a ~9 fold increase in affinity with respect to the wild-type NL1 protein. These data indicate that splice insert B exerts a strong inhibitory influence on the NXβ binding suggesting that the focal point of the NL surface binding for NXβ is in the vicinity of this alternative splice sequence B(16).
Small angle X-ray scattering was employed to characterize complex formation between NL1-638-Δ (A&B) and NXβ. The P(r) analysis of these data yielded Rg and Dmax values of 48.7±.0.9 Å and ~150Å, respectively, an increase of ~20 Å in maximum dimension compared to the free NL1-638-Δ (A&B). This increase, in combination with a marked change in the shape of the P(r) profile also indicates that the added mass of NXβ sits at the periphery of each NL1 monomer, distant from both the center of mass and the dimer axis of symmetry(19). To determine the positions of the NXβ subunits relative to the NL1 dimer, deuterated NXβ was prepared and stoichiometric amounts of deuterated NXβ were combined with protonated NL1-638-Δ (A&B) in the presence of 2mM Ca2+. By acquiring neutron scattering data from the complex in 42% D2O solution, the protonated NL1 subunits were effectively “solvent-matched”. In these conditions, the scattering from the deuterated NXβ dominates the signal, allowing for direct measurement of the relative position of the NXβ units(19). The combined X-ray and neutron scattering data provide the approximate binding locations for the NXβ subunits and the separation between their centers of mass; a powerful set of restraints for refining a low resolution structure of the complex.
Ab initio shape restoration from the X-ray scattering data form the complex gives a shape that shows two clearly recognizable protrusions from the basic NL1 dimer shape with their approximate separation corresponding to the values determined for the bound NXβ molecules in the neutron solvent matching experiment(19). In laminin-neurexin-sex hormone binding globulin (LNS) domains, the metal binding pocket is located at the rim of the β-sheet sandwich opposite the N- and C-termini(12) and alanine mutations of D137 and N238, residing on this rim, lack synaptogenic activity(17), possibly participating in the binding of Ca2+. On the NL1 surface, mapping NXβ binding determinants revealed that the region flanking splice insert B (including E297 and K306)(14–16) is the focal point for NXβ binding. Therefore, initial placement of the NXβ subunits relative to the NL1-638-Δ (A&B) dimer was done to best-fit the low-resolution density reconstruction of the complex and according to known biochemical constraints(14–17). The final calculation produced a structure in which the NXβ monomers are positioned in agreement with the available mutagenesis data(16,17) and the C- termini of the NL1 and NXβ subunits facing opposite directions, consistent with the complex bridging pre- and post-synaptic surfaces(19). High resolution crystal structures of NL4, free in solution and in complex with NXβ, have recently been solved(26). Comparative analysis of the low resolution structures obtained by SAXS with the high resolution structure of NL4 confirms that the α/β-hydrolase fold and the reciprocal orientation of the two NL monomers were accurately predicted. Moreover, the crystal structure of the NL/NXβ complex reveals that the binding surface of NXβ and the relative positioning of the NX monomer (with its β- strands sandwich oriented perpendicularly to the NL dimer) on the NL dimer are virtually identical in the high and low resolution structures. In the crystal structure only the orientation of NX differs slightly from the assignment obtained with solution scattering techniques. The solution scattering data reflect the ensemble average solution of a structure, while crystallization would select the most stable interaction consistent with maintaining crystal contacts. The relatively small interaction area between the neuroligin and neurexin components raises the question as to whether the different orientation deduced from the solution scattering experiments versus the crystallographic result may be due to some dynamic variability in the relative orientations of neurexin in the complex. The formation of a highly dynamic complex could be crucial for the signaling mechanism at the synapse.
CONCLUSIONS
Using small angle solution scattering we described the first low resolution, three-dimensional structure of the AChE-like domain of the NLs, showing that all four NLs have similar overall shapes and dimensions. The stalk region connecting the globular domain of the NLs with their transmembrane domain is elongated and projects away from the globular domain. Using the newly determined NL1 structure, and complementing our X-ray data with neutron contrast variation data from a complex of NL1 and a deuterated NXβ, we determined the first three dimensional structural model of NL1 in complex with NXβ. The combination of X-ray scattering with neutron contrast variation, sequence analysis, modeling, biochemical and mutagenesis data has allowed us to put forward a model of the NX and NL complex in the synaptic space (figure 2)(19).
Figure 2. Model of the NL1/NXβ Assembly in the Synapse.
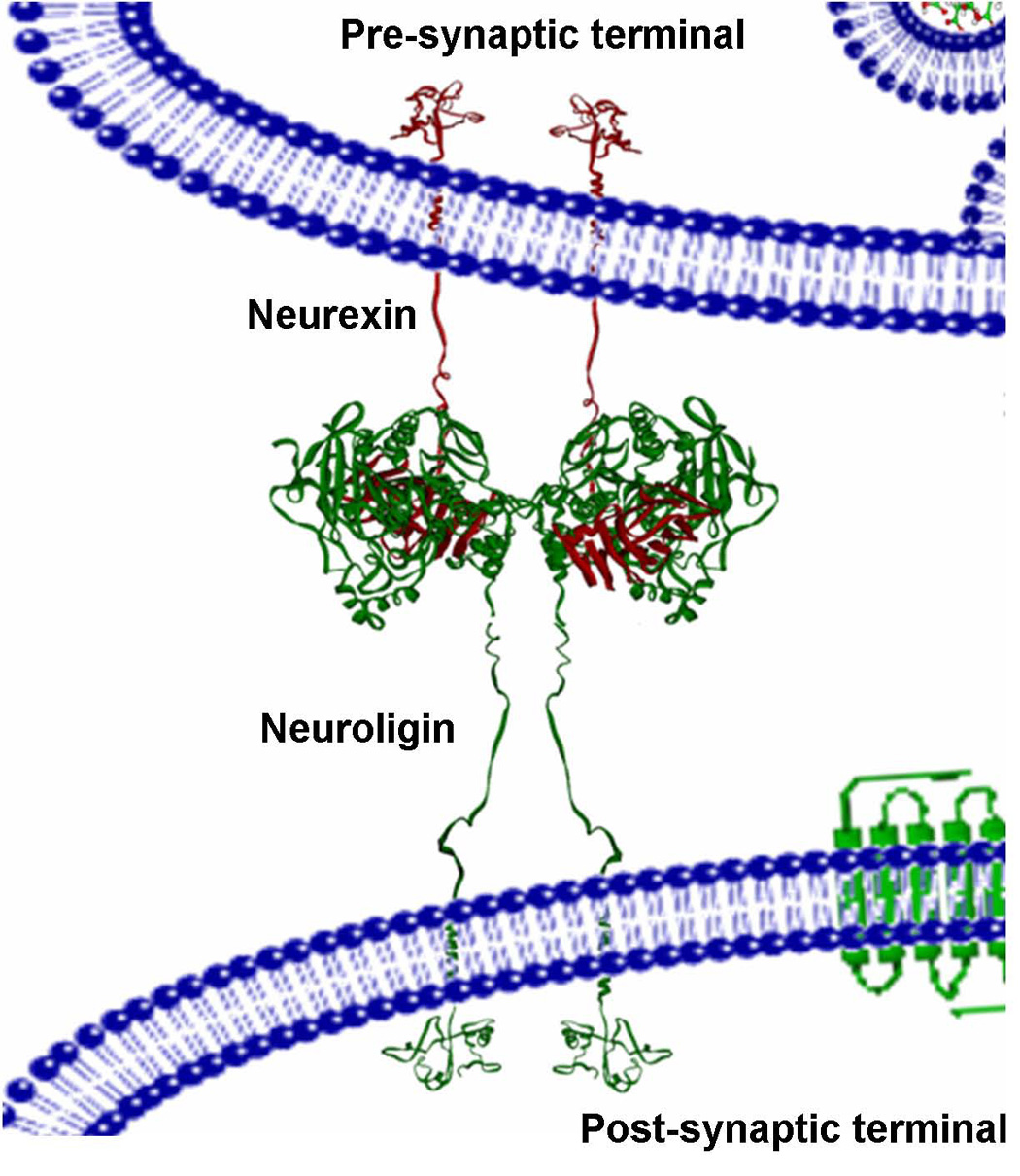
The NX/NL complex is tethered at the synaptic membranes through their respective O-linked glycosylated regions. NL1 is displayed as green ribbon and is tethered to the post-synaptic membrane, whereas NXβ is displayed as red ribbon and is tethered to the pre-synapse.
Our study provides an important framework for analyzing disposition of these molecules within the synaptic space and for linking genetic information on mutated NXs and NLs with neurodevelopmental disorders such as autism.
EXPERIMENTAL PROCEDURES
Expression Vectors and Mutagenesis
Soluble FLAG-NL1 to NL4 plasmids devoid of both transmembrane and intracellular domains were engineered as described previously(16). NL1-638 was also engineered with (NL1-638) and without splice inserts A and B (NL1-638-Δ (A&B)). All mutations were obtained using the Quickchange Mutagenesis Kit (Stratagene, San Diego, CA) and verified by both restriction digests and DNA sequencing. Preparation of the GST-cleavable NXβ construct was described elsewhere(16).
Neuroligin and Neurexin Expression and Purification
To purify each soluble NL, M2 anti-FLAG-affinity column (Sigma) was used(14). Samples were concentrated by micro filtration and separated in analytical gel filtration and SDS-PAGE gels and stained with Coomassie to check for purity and absence of degradation products. For neutron scattering experiments, deuterated NXβ was expressed in BL21 E. coli (Invitrogen, Carlsbad, CA) using Spectra 9 deuterated media (Spectragases, Branchburg, NJ) and purified as described previously(16,19).
Initial Topology Modeling of the Neuroligins
Structural models of the extracellular domain of the NLs were generated with the programs Homology and InsightII (Accelrys, Inc., 2002) using the crystal structure of mouse AChE (1MAH in the PDB database) as a template.
Sedimentation equilibrium analysis
Analytical ultracentrifugation was conducted in a Beckman/Coulter XL-I centrifuge using an An 60 Ti rotor. Protein solutions of 30, 100 and 300 µg/mL of NL1 were used in a six-channel charcoal-filled epon centerpiece loaded with 120 µL of sample and 125 µL of reference buffer, pH 7.4. Samples were spun at 20 °C for 16 hours at 8,000, 10,000 and 12,000 rpm. Equilibrium was attained as judged by the overlay of the last three scans. The partial specific volume was calculated using Sednterp software (ver. 1.06) accounting for total sugars. Molar masses of the proteins were calculated using XL-A/XL-I Data Analysis Software version 4.0 based on OriginTM program.
Small-angle scattering
X-ray scattering data were acquired using Beam Line 4.2 at SSRL(19) and the instrument at the University of Utah that is described in Heidorn and Trewhella(27). Neutron scattering data were acquired using the 30-m SANS instrument NG3 at the National Institutes of Standards and Technology(19). Standard data reduction protocols were used to obtain scattering due to the proteins in solution and scattering data analysis used the ATSAS suite of programs(28). Refinement of the NL dimer and NL/NXβ complex used the protocols described in Grishaev et al., 2005 (29).
Acknowledgments
This work was supported by: USPHS Grant R37 GM-18360 and NIEHS to PT; ARC Federation Fellowship (FF0457488) to JT. The scattering experiments were performed at the University of Utah supported by DOE Grant No. DE-FG02-05ER64026 to JT.
List of abbreviations
- ASD
Autism spectral disorders
- NL
Neuroligin
- NXβ
β-neurexin
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Cited references
- 1.Jamain S, Quach H, Betancur C, Rastam M, Colineaux C, Gillberg IC, Soderstrom H, Giros B, Leboyer M, Gillberg C, Bourgeron T. Paris Autism Research International Sibpair Study. (2003) Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34:27–29. doi: 10.1038/ng1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Laumonnier F, Bonnet-Brilhault F, Gomot M, Blanc R, David A, Moizard MP, Raynaud M, Ronce N, Lemonnier E, Calvas P, Laudier B, Chelly J, Fryns JP, Ropers HH, Hamel BC, Andres C, Barthelemy C, Moraine C, Briault S. X-linked mental retardation and autism are associated with a mutation in the NLGN4 gene, a member of the neuroligin family. Am J Hum Genet. 2004;74:552–557. doi: 10.1086/382137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yan J, Oliveira G, Coutinho A, Yang C, Feng J, Katz C, Sram J, Bockholt A, Jones IR, Craddock N, Cook EH, Vicente A, Sommer SS. Analysis of the neuroligin 3 and 4 genes in autism and other neuropsychiatric patients. Mol Psychiatry. 2005;10:329–332. doi: 10.1038/sj.mp.4001629. [DOI] [PubMed] [Google Scholar]
- 4.International Molecular Genetic Study of Autism Consortium (IMGSAC) Blasi F, Bacchelli E, Pesaresi G, Carone S, Bailey AJ, Maestrini E. Absence of coding mutations in the X-linked genes neuroligin 3 and neuroligin 4 in individuals with autism from the IMGSAC collection. Am J Med Genet B Neuropsychiatr Genet. 2006;141:220–221. doi: 10.1002/ajmg.b.30287. [DOI] [PubMed] [Google Scholar]
- 5.Talebizadeh Z, Lam DY, Theodoro MF, Bittel DC, Lushington GH, Butler MG. Novel splice isoforms for NLGN3 and NLGN4 with possible implications in autism. J Med Genet. 2006;43:e21. doi: 10.1136/jmg.2005.036897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feng J, Schroer R, Yan J, Song W, Yang C, Bockholt A, Cook EH, Jr, Skinner C, Schwartz CE, Sommer SS. High frequency of neurexin 1beta signal peptide structural variants in patients with autism. Neurosci Lett. 2006;409:10–13. doi: 10.1016/j.neulet.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 7.Chocholska S, Rossier E, Barbi G, Kehrer-Sawatzki H. Molecular cytogenetic analysis of a familial interstitial deletion Xp22.2-22.3 with a highly variable phenotype in female carriers. Am J Med Genet A. 2006;140:604–610. doi: 10.1002/ajmg.a.31145. [DOI] [PubMed] [Google Scholar]
- 8.Duvall JA, Stone JL, Cantor RM, Nelson SF, Geschwind DH. Independent replication of autism association in several neuronal connectivity genes [abstract 1652]. Presented at the annual meeting of The American Society of Human Genetics; New Orleans, Louisiana. October, 2006; 2006. http://www.ashg.org/genetics/ashg06s/ [Google Scholar]
- 9.The Autism Genome Project Consortium. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nature Genetics. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, Nygren G, Rastam M, Gillberg IC, Anckarsater H, Sponheim E, Goubran-Botros H, Delorme R, Chabane N, Mouren-Simeoni MC, de Mas P, Bieth E, Roge B, Heron D, Burglen L, Gillberg C, Leboyer M, Bourgeron T. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Missler M, Fernandez-Chacon R, Sudhof TC. The making of neurexins. J Neurochem. 1998;71:1339–1347. doi: 10.1046/j.1471-4159.1998.71041339.x. [DOI] [PubMed] [Google Scholar]
- 12.Rudenko G, Nguyen T, Chelliah Y, Sudhof TC, Deisenhofer J. The structure of the ligand-binding domain of neurexin-Ibeta: regulation of LNS domain function by alternative splicing. Cell. 1999;99:93–101. doi: 10.1016/s0092-8674(00)80065-3. [DOI] [PubMed] [Google Scholar]
- 13.Sheckler LR, Henry L, Sugita S, Sudhof TC, Rudenko G. Crystal structure of the second LNS/LG domain from neurexin 1alpha: Ca2+-binding and the effects of alternative splicing. J Biol Chem. 2006;281:22896–22905. doi: 10.1074/jbc.M603464200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Comoletti D, Flynn RE, Jennings LL, Chubykin A, Matsumura T, Hasegawa H, Südhof TC, Taylor P. Characterization of the interaction of a recombinant soluble neuroligin1 with neurexin-1β. J Biol. Chem. 2003;278:50497–50505. doi: 10.1074/jbc.M306803200. [DOI] [PubMed] [Google Scholar]
- 15.Boucard AA, Chubykin AA, Comoletti D, Taylor P, Südhof TC. A splice-code for trans-synaptic cell adhesion mediated by binding of neuroligins to α- and β-neurexins. Neuron. 2005;48:229–236. doi: 10.1016/j.neuron.2005.08.026. [DOI] [PubMed] [Google Scholar]
- 16.Comoletti D, Flynn RE, Boucard AA, Demeler B, Schirf V, Shi J, Jennings LL, Newlin HR, Sudhof TC, Taylor P. Gene selection, alternative splicing, and post-translational processing regulate neuroligin selectivity for β-neurexins. Biochemistry. 2006;45:12816–12827. doi: 10.1021/bi0614131. [DOI] [PubMed] [Google Scholar]
- 17.Graf ER, Kang Y, Hauner AM, Craig AM. Structure, function and splice site analysis of the synaptogenic activity of the neurexin-1 beta LNS domain. J Neurosci. 2006;26:4256–4265. doi: 10.1523/JNEUROSCI.1253-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chih B, Gollan L, Scheiffele P. Alternative splicing controls selective trans-synaptic interactions of the neuroligin-neurexin complex. Neuron. 2006;51:171–178. doi: 10.1016/j.neuron.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 19.Hoffman RC, Jennings LL, Tsigelny I, Comoletti D, Flynn R, Südhof TC, Taylor P. Structural characterization of recombinant soluble rat neuroligin 1: Mapping of secondary structure and glycosylation by mass spectrometry. Biochemistry. 2004;43:1496–1506. doi: 10.1021/bi035278t. [DOI] [PubMed] [Google Scholar]
- 20.Comoletti D, Grishaev A, Whitten A, Tsigelny I, Taylor P, Trewhella J. Synaptic Arrangement of the Neuroligin/β-Neurexin Complex Revealed by X-ray and Neutron Scattering. Structure. 2007;15:693–705. doi: 10.1016/j.str.2007.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dean C, Dresbach T. Neuroligins and neurexins: linking cell adhesion, synapse formation and cognitive function. Trends Neurosci. 2006;29:21–29. doi: 10.1016/j.tins.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 22.Li F, Erickson HP, James JA, Moore KL, Cummings RD, McEver RP. Visualization of P-selectin glycoprotein ligand-1 as a highly extended molecule and mapping of protein epitopes for monoclonal antibodies. J. Biol. Chem. 1996;271:6342–6348. doi: 10.1074/jbc.271.11.6342. [DOI] [PubMed] [Google Scholar]
- 23.Merry AH, Gilbert RJ, Shore DA, Royle L, Miroshnychenko O, Vuong M, Wormald MR, Harvey DJ, Dwek RA, Classon BJ, et al. O-glycan sialylation and the structure of the stalk-like region of the T cell co-receptor CD8. J Biol Chem. 2003;278:27119–27128. doi: 10.1074/jbc.M213056200. [DOI] [PubMed] [Google Scholar]
- 24.Sussman JL, Harel M, Frolow F, Oefner C, Goldman A, Toker L, Silman I. Atomic structure of acetylcholinesterase from Torpedo Californica: a prototypic acetylcholine-binding protein. Science. 1991;253:872–879. doi: 10.1126/science.1678899. [DOI] [PubMed] [Google Scholar]
- 25.Bourne Y, Taylor P, Marchot P. Acetylcholinesterase inhibition by fasciculin: crystal structure of the complex. Cell. 1995;83:503–512. doi: 10.1016/0092-8674(95)90128-0. [DOI] [PubMed] [Google Scholar]
- 26.Fabrichny IP, Leone P, Sulzenbacher G, Comoletti D, Miller MT, Taylor P, Bourne Y, Marchot P. Structural analysis of the synaptic protein neuroligin and its β-neurexin complex: determinants for folding and cell adhesion Neuron. 2007;56:1–13. doi: 10.1016/j.neuron.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heidorn DB, Trewhella J. Comparison of the crystal and solution structures of calmodulin and troponin C. Biochemistry. 1988;27:909–915. doi: 10.1021/bi00403a011. [DOI] [PubMed] [Google Scholar]
- 28.Konarev PV, Petoukhov MV, Volkov VV, Svergun D. ATSAS 2.1, a program package for small-angle scattering data analysis. Journal of Applied Crystallography Part 2. 2006;39:277–286. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grishaev A, Wu J, Trewhella J, Bax A. Refinement of multi-domain protein structures by combination of solution small angle X-ray scattering and NMR data. J. Am. Chem. Soc. 2005;127:16621–16628. doi: 10.1021/ja054342m. [DOI] [PubMed] [Google Scholar]


