Abstract
DNA-dependent ATPases participate in a broad range of biological processes that include transcription, DNA repair, and chromatin dynamics. Mutations in the HARP ATPase are responsible for Schimke immuno-osseous dysplasia (SIOD), but the function of the protein is unknown. Here we report that HARP is an ATP-dependent annealing helicase. HARP rewinds single-stranded DNA bubbles that are stably bound by replication protein A. Other related ATPases, including the DNA translocase Rad54, do not exhibit annealing helicase activity. Analysis of mutant HARP proteins suggests that SIOD is caused by a deficiency in annealing helicase activity. Moreover, the pleiotropy of HARP mutations is consistent with the function of HARP as an annealing helicase that acts throughout the genome to oppose the action of DNA-unwinding activities in the nucleus.
HARP (HepA-related protein; also known as SMARCAL1 and DNA-dependent ATPase A) is a member of the SNF2 family of ATP-driven molecular motor proteins (1–4). The biological importance of HARP was revealed by the discovery that mutations in HARP are responsible for a pleiotropic disorder known as Schimke immuno-osseous dysplasia (SIOD) (5). However, the molecular function of the HARP ATPase activity is unknown.
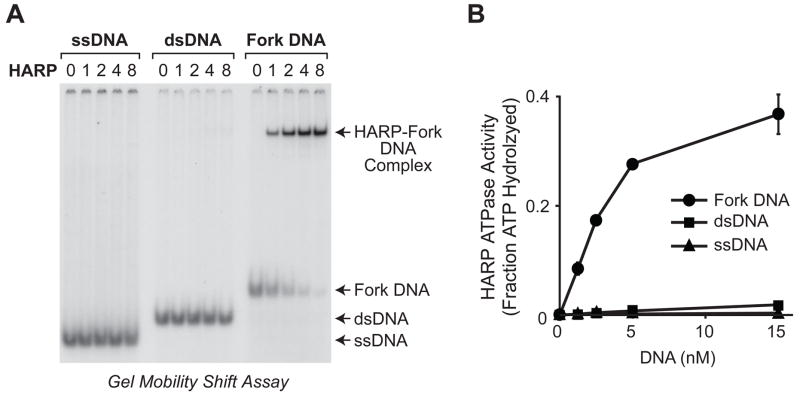
We investigated how human HARP functions as an ATP-dependent molecular motor by synthesizing and purifying human HARP protein (fig. S1). By using the gel mobility shift assay, we determined that HARP binds with higher affinity to fork DNA than to single-stranded DNA or to double-stranded DNA (Fig. 1A). In addition, the ATPase activity of HARP is stimulated to a much greater extent by fork DNA than by single- or double-stranded DNA (Fig. 1B) (6). These results are consistent with the finding that the HARP ATPase is activated by M13 single-stranded DNA (4), which probably contains hairpin structures, as well as the observation that the HARP ATPase domain is stimulated by DNA structures that contain both single- and double-stranded DNA (7,8).
Fig. 1.
HARP protein binds selectively to fork DNA. (A) HARP protein binds with higher affinity to fork DNA than to single-stranded DNA (ssDNA) or double-stranded DNA (dsDNA). Gel mobility shift experiments were performed with a 30 nt ssDNA, a 30 bp dsDNA, and a 30 nt fork DNA that is identical to the double-stranded DNA except for a 9 nt mismatch at one end. The relative concentrations of HARP are shown. The actual concentrations of HARP are 0, 0.05, 0.1, 0.2, and 0.4 nM. (B) HARP ATPase activity is stimulated to a greater extent by fork DNA than by single- or double-stranded DNA. The DNA substrates used in the ATPase assays are identical to those used in A, except that the DNA samples were not radiolabelled. Error bars represent SD (N = 3).
The stimulation of the HARP ATPase activity upon binding to fork DNA suggested that HARP may be an ATP-driven helicase that unwinds DNA. Helicases generate single-stranded DNA regions that can be bound by single-stranded DNA-binding proteins, such as replication protein A (RPA) in eukaryotes (see, for example, 9). However, we tested the ability of HARP to function as a helicase with several different assays and substrates, but did not observe any detectable helicase activity (for example, see fig. S2). Thus, HARP does not appear to be a helicase.
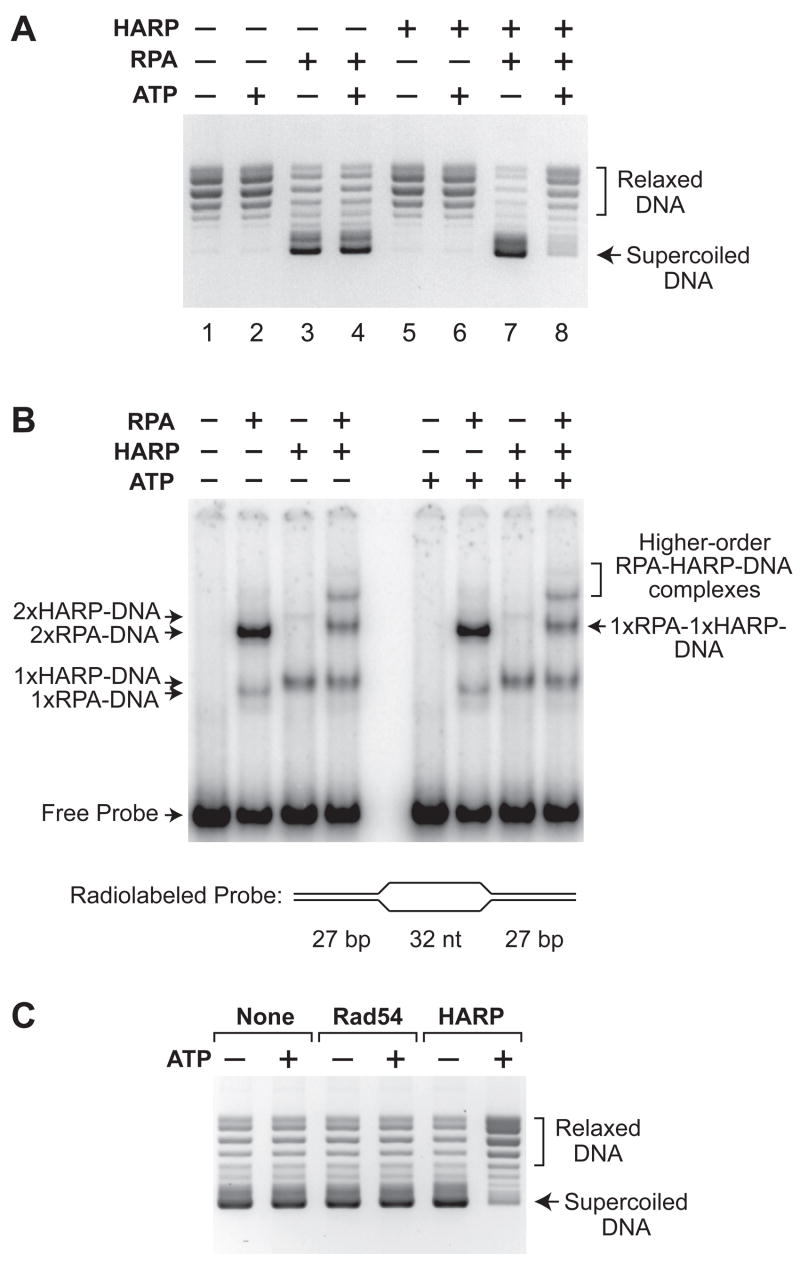
We therefore considered the possibility that HARP is an ATP-driven annealing helicase that anneals complementary RPA-bound single-stranded DNA. To test this hypothesis, we devised an assay for annealing helicase activity (fig. S3). We generated a stable, partially unwound DNA substrate by adding RPA to plasmid DNA in the presence of topoisomerase I (10,11). Under these conditions, RPA binds to small transient bubbles and then wedges the DNA strands apart to form stable single-stranded bubbles in which RPA is bound to the unwound DNA (12). Upon addition of SDS (to inactivate enzymes such as topoisomerase I) and subsequent deproteinization, the RPA-unwound DNA yields negatively supercoiled DNA (Fig. 2A, lanes 3 and 4). As a control, when plasmid DNA is treated in an identical manner in the absence of RPA, the resulting DNA is relaxed, as expected (Fig. 2A, lanes 1 and 2). In addition, HARP protein does not alter DNA supercoiling in the presence or absence of ATP (Fig. 2A, lanes 5 and 6).
Fig. 2.
HARP is an ATP-dependent annealing helicase. (A) HARP catalyzes the rewinding of DNA in an ATP-dependent manner. Annealing helicase assays, as depicted in fig. S3, were carried out in the presence or absence of the indicated factors, and the resulting DNA species were resolved by agarose gel electrophoresis. An equimolar concentration of UTP was used as a control for the absence of ATP. (B) HARP does not catalyze the ATP-dependent displacement of RPA from DNA. Gel mobility shift experiments were performed with radiolabeled bubble DNA that contains two high affinity sites for RPA (the two 32 nt single-stranded DNA segments) and for HARP (the two DNA forks). HARP (2 nM), RPA (3 nM), and ATP (1.5 mM) were included, as indicated. The apparent compositions of the shifted complexes are specified. Quantitation of the bands is shown in fig. S5. (C) Rad54, a member of the SNF2 family that translocates along DNA, does not exhibit annealing helicase activity. Annealing helicase assays were performed as in A with RPA and an equimolar concentration of Rad54 or HARP, where indicated.
The addition of an annealing helicase to the partially unwound DNA substrate should yield circular DNA that is relaxed by topoisomerase I due to the elimination of the stable single-stranded regions within the double-stranded DNA (fig. S3). In this assay, HARP catalyzes the ATP-dependent relaxation of the RPA-unwound DNA (Fig. 2A, compare lanes 7 and 8). This effect could be due to an ATP-dependent annealing helicase activity, as depicted in fig. S4, or to the ATP-dependent removal of RPA from single-stranded DNA by HARP. To distinguish between these two possibilities, we carried out gel mobility shift analyses with RPA-bound DNA and found that HARP does not catalyze the ATP-dependent displacement of RPA from DNA (Fig. 2B; figs. S5 and S6). These results thus reveal that HARP is an annealing helicase. In addition, we found that RPA does not directly stimulate the ATPase activity of HARP (fig. S7). These findings indicate that the properties of HARP are distinct from those of Mot1, a SNF2 family member protein that removes TATA box-binding protein (TBP) from DNA and whose ATPase activity is stimulated by TBP (13,14).
We also investigated the possibility that the basis for the annealing helicase activity is translocation along double-stranded DNA, which has been observed in some SNF2 family proteins such as Sth1 and Rad54 (15–17). To test this idea, we compared the properties of Rad54 and HARP. Rad54 is an SNF2 family protein involved in homologous recombination that been shown to translocate along DNA in both triple-helix strand displacement (16) and single-molecule assays (17). In triple-helix strand-displacement assays, we found that Rad54 has a higher DNA translocation activity than HARP (fig. S8). In contrast, Rad54 does not exhibit any detectable annealing helicase activity (Fig. 2C). Thus, the ability of a factor to translocate along double-stranded DNA is not sufficient for the removal of RPA from the unwound DNA. We also tested two other SNF2 family proteins, ACF (which contains the ISWI ATPase) and Brg1, and found that neither protein exhibits annealing helicase activity (fig. S9). These experiments further reveal that the annealing helicase activity of HARP is not a general property of SNF2 ATPases.
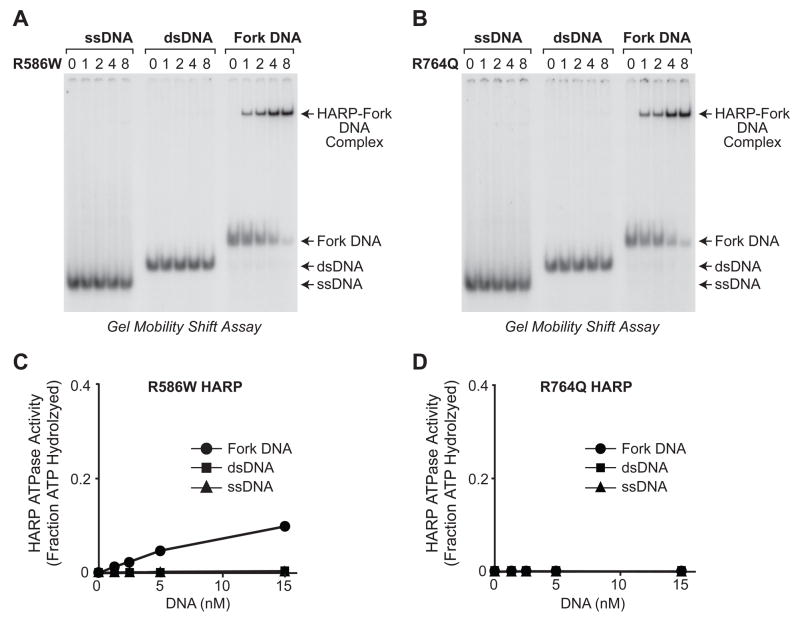
To gain insight into the molecular basis of SIOD, we purified two mutant versions of HARP that are associated with the disease (fig. S10). The R764Q mutation causes a severe form of SIOD, and the R586W mutation results in a milder form of SIOD (5). Both of these mutations are in the conserved ATPase region of HARP. As seen with wild-type HARP (Fig. 1A), both mutant proteins bind selectively to fork DNA relative to single- or double-stranded DNA (Fig. 3A,B). In addition, the wild-type and mutant HARP proteins bind with nearly the same affinity to fork DNA (fig. S11). However, in ATPase assays, the R586W HARP protein exhibits partial activity, whereas the R764Q HARP protein has no detectable activity (Fig. 3C,D).
Fig. 3.
Mutant HARP proteins bind selectively to fork DNA, but exhibit less ATPase activity than wild-type HARP. The R764Q mutation results in a stronger SIOD phenotype than the R586W mutation (5). (A, B) The mutant HARP proteins bind with higher affinity to fork DNA than to single-stranded DNA (ssDNA) or double-stranded DNA (dsDNA). These gel mobility shift experiments were performed as in Fig. 1A. (C, D) The R586W HARP protein has low ATPase activity, whereas the R764Q HARP protein has no detectable ATPase activity. ATPase assays were carried out as in Fig. 1B. Error bars represent SD (N = 3).
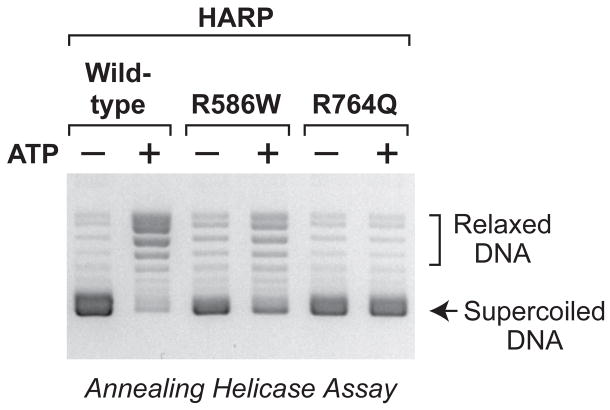
In the annealing helicase assay, R586W HARP exhibits less activity than wild-type HARP, whereas R764Q HARP has no detectable activity (Fig. 4). Therefore, the two SIOD-associated mutations have little or no effect on DNA-binding by HARP; instead, the mutations reduce the ATPase and annealing helicase activities of HARP in a manner that correlates with the severity of the disease with which they occur. In addition, the studies of the mutant HARP proteins further reveal that the DNA-binding activity of HARP is not sufficient for annealing helicase activity, because R764Q HARP is fully active for selective binding to fork DNA (Fig. 3B and fig. S11) yet is deficient in annealing helicase activity (Fig. 4).
Fig. 4.
Mutant HARP proteins are defective in annealing helicase activity. The R586W HARP protein has less annealing helicase activity than wild-type HARP, whereas the R764Q HARP protein has no detectable annealing helicase activity. Annealing helicase assays were performed as in Fig. 2. All reactions contained plasmid DNA, RPA, and topoisomerase I. UTP was used as a control for the absence of ATP.
HARP is an ATP-driven molecular zipper of complementary RPA-bound single-stranded DNA (fig. S4). Whereas many helicases convert double-stranded DNA into RPA-bound unwound DNA, HARP performs the opposite reaction. The annealing helicase activity of HARP is also distinct from fork regression activity (see, for example, 18–20), which involves the dissociation and annealing of four strands of DNA without any involvement of RPA (fig. S12).
The biological importance of HARP is revealed by its causal role in SIOD. The defects in the ATPase and annealing helicase activities of two SIOD mutant proteins correlate with the severity of disease. It is thus likely that SIOD is caused by a deficiency in annealing helicase activity. Moreover, SIOD patients exhibit a diverse range of symptoms. The pleiotropic nature of HARP mutations is consistent with the ubiquitous expression of HARP in mammalian tissues (4,21) and the molecular function of HARP as an annealing helicase that acts throughout the genome to reanneal stably unwound DNA. There are many enzymes, such as helicases and polymerases, that unwind DNA. In addition, single-stranded DNA bubbles could arise spontaneously, such as in A/T-rich sequences. Hence, there is considerable potential for the incomplete reannealing of DNA and formation of stable, RPA-bound DNA bubbles, which could be deleterious to the transcription of genes or may interfere with replication or repair processes. In this manner, HARP would be able to promote the proper functioning of the cell by catalyzing the rewinding of the stably unwound DNA. More generally, HARP would serve as an opposing force to the numerous DNA-unwinding activities in the nucleus.
Supplementary Material
References and Notes
- 1.Gorbalenya AE, Koonin EV. Curr Opin Struct Biol. 1993;3:419. [Google Scholar]
- 2.Eisen JA, Sweder KS, Hanawalt PC. Nucl Acids Res. 1995;23:2715. doi: 10.1093/nar/23.14.2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Flaus A, Martin DM, Barton GJ, Owen-Hughes T. Nucl Acids Res. 2006;34:2887. doi: 10.1093/nar/gkl295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coleman MA, Eisen JA, Mohrenweiser HW. Genomics. 2000;65:274. doi: 10.1006/geno.2000.6174. [DOI] [PubMed] [Google Scholar]
- 5.Boerkoel CF, et al. Nat Genet. 2002;30:215. doi: 10.1038/ng821. [DOI] [PubMed] [Google Scholar]
- 6.The kcat values (mean +/− SD; N = 4) for HARP ATPase activity with fork DNA and double-stranded DNA are approximately 1200 +/− 270 min−1 and 81 +/− 29 min−1, respectively. ATPase activity was not detectable in the absence of DNA or in the presence of single-stranded DNA.
- 7.Muthuswami R, Truman PA, Mesner LD, Hockensmith JW. J Biol Chem. 2000;275:7648. doi: 10.1074/jbc.275.11.7648. [DOI] [PubMed] [Google Scholar]
- 8.Hockensmith JW, Wahl AF, Kowalski S, Bambara RA. Biochemistry. 1986;25:7812. doi: 10.1021/bi00372a005. [DOI] [PubMed] [Google Scholar]
- 9.Wu L, Hickson ID. Annu Rev Genet. 2006;40:279. doi: 10.1146/annurev.genet.40.110405.090636. [DOI] [PubMed] [Google Scholar]
- 10.Georgaki A, Strack B, Podust V, Hübscher U. FEBS Lett. 1992;308:240. doi: 10.1016/0014-5793(92)81283-r. [DOI] [PubMed] [Google Scholar]
- 11.Treuner K, Ramsperger U, Knippers R. J Mol Biol. 1996;259:104. doi: 10.1006/jmbi.1996.0305. [DOI] [PubMed] [Google Scholar]
- 12.Lao Y, Lee CG, Wold MS. Biochemistry. 1999;38:3974. doi: 10.1021/bi982371m. [DOI] [PubMed] [Google Scholar]
- 13.Auble DT, Wang D, Post KW, Hahn S. Mol Cell Biol. 1997;17:4842. doi: 10.1128/mcb.17.8.4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chicca JJ, 2nd, Auble DT, Pugh BF. Mol Cell Biol. 1998;18:1701. doi: 10.1128/mcb.18.3.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saha A, Wittmeyer J, Cairns BR. Genes Dev. 2002;16:2120. doi: 10.1101/gad.995002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jaskelioff M, Van Komen S, Krebs JE, Sung P, Peterson CL. J Biol Chem. 2003;278:9212. doi: 10.1074/jbc.M211545200. [DOI] [PubMed] [Google Scholar]
- 17.Amitani I, Baskin RJ, Kowalczykowski SC. Mol Cell. 2006;23:143. doi: 10.1016/j.molcel.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 18.McGlynn P, Lloyd RG. Cell. 2000;101:35. doi: 10.1016/S0092-8674(00)80621-2. [DOI] [PubMed] [Google Scholar]
- 19.Ralf C, Hickson ID, Wu L. J Biol Chem. 2006;281:22839. doi: 10.1074/jbc.M604268200. [DOI] [PubMed] [Google Scholar]
- 20.Blastyák A, et al. Mol Cell. 2007;28:167. doi: 10.1016/j.molcel.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Elizondo LI, et al. Am J Med Genet. 2006;140A:340. [Google Scholar]
- 22.We thank Barbara Rattner, Jer-Yuan Arthur Hsu, Tammy Juven-Gershon, Debra Urwin, and Joshua Theisen for critical reading of the manuscript. We thank Matthew Coleman (Lawrence Livermore National Laboratories) for the gift of HARP cDNAs in the early stages of this study; Marc Wold (University of Iowa) for the human RPA expression vector; Bradley Cairns for the triple-helix plasmid; and Robert Kingston (Harvard Medical School) for the BRG1 expression vector. We also thank Martin Gellert for a helpful discussion on this work prior to publication. This work was supported by a grant from the NIH (GM058272) to J. T.K. T.Y. was supported in part by an NRSA fellowship from the NIH (F32 GM76936).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.