Abstract
The Par-1 protein kinases are conserved from yeast to humans, where they function as key polarity determinants. The mammalian Par-1 family is comprised of 4 members (Par-1a, -b, -c, and -d). Previously, we demonstrated that atypical protein kinase C (aPKC) phosphorylates the Par-1 kinases on a conserved threonine residue (T595) to regulate localization and kinase activity. Here, we demonstrate that Par-1b is also regulated by another arm of the PKC pathway, one that involves novel PKCs (nPKC) and protein kinase D. Treatment of cells with the PKC activator phorbol-12-myristate-13-acetate (PMA) potently stimulated phosphorylation of Par-1b on serine 400 (S400), a residue that is conserved in all 4 mammalian Par-1 kinases as well as the fly ortholog. We demonstrate that PMA stimulates nPKC to activate PKD, which in turn directly phosphorylates Par-1b on S400 to positively regulate 14-3-3 binding and to negatively regulate membrane association. Thus, 2 arms of the PKC pathway regulate interactions between Par-1b and 14-3-3 proteins: one involving aPKC and the other nPKC/PKD.
Keywords: atypical protein kinase C, cell polarity, MARK2, EMK
Establishing and maintaining cellular polarity is critical for the homeostasis of unicellular and multicellular organisms alike. The PAR (partitioning-defective) genes (PAR 1–6) were identified in Caenorhabditis elegans as essential determinants of asymmetric cell division and polarized cell growth (1, 2). Par-1 is a serine/threonine protein kinase, and Par-1 homologues have been identified and studied in a number of organisms, including yeast, fruitflies, frogs, and mammals (3, 4). These studies have revealed disparate roles for Par-1 not only as a regulator of cell polarity but also as a component of mitogenic and Wnt signaling (4, 5). In mammals, there are 4 Par-1 family members named Par-1a (C-TAK1/MARK3), Par-1b (EMK/MARK2), Par-1c (MARK1), and Par-1d (MARKL1, MARK4).
Several Par-1 substrates have been identified, including Par-3 (6, 7). An antagonistic relationship between Par-1 and the Par-3/Par-6/aPKC complex has been revealed. In C. elegans embryos Par-1 is located at the posterior cortex whereas the Par-3/Par-6/atypical protein kinase C (aPKC) complex is located at the anterior cortex. In epithelial cells, the Par-3/Par-6/aPKC complex is found at tight junctions, whereas Par1 is located laterally beneath tight junctions. Par-1 phosphorylates Par-3 to exclude it from lateral membranes of epithelial cells (6, 7), whereas aPKC in complex with Par-3/Par-6 phosphorylates Par-1 to dislodge it from plasma membranes (8, 9). Thus, the establishment and/or maintenance of cell polarity likely require that Par-1 be physically sequestered from the Par-3/Par-6/aPKC complex, and phosphorylation of Par-1 by aPKC may enforce the mutual exclusion of Par-1 and Par-3/Par-6/aPKC. Negative regulation of Par-1b by the Par-3/Par-6/aPKC complex is also observed in hippocampal neurons (10). Here, we identify another protein kinase pathway that regulates Par-1 localization. We demonstrate that treatment of cells with phorbol-12-myristate-13-acetate (PMA) activates novel (n)PKCs to activate PKD and that PKD directly phosphorylates Par-1b on S400. Phosphorylation of S400, like phosphorylation of T595, regulates Par-1b/14-3-3 interactions and the ability of Par-1b to associate with cellular membranes.
Results
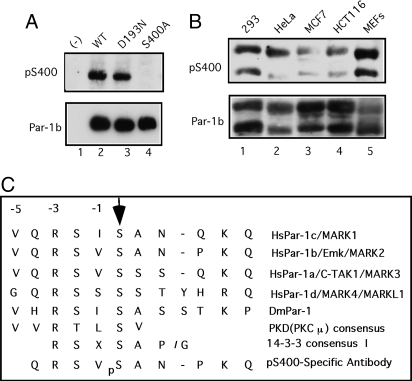
By using a combination of site-directed mutagenesis and tryptic phosphopeptide mapping, we identified serine 400 (S400) as a potential site of Par-1b phosphorylation in vivo (data not shown). To verify that Par-1b is indeed phosphorylated on S400 in vivo, a phosphospecific antibody was generated and used in Western blotting experiments (Fig. 1A Upper). Whereas the pS400 antibody recognized Par-1b (lane 2), mutation of S400 eliminated its recognition by the antibody (lane 4). Thus, the antibody is specific for Par-1b when it is phosphorylated on S400 and ectopically expressed Par-1b is phosphorylated on S400 in cultured cells. Also, phosphorylation of S400 did not require the kinase activity of Par-1b, because a kinase-inactive mutant of Par-1b was also phosphorylated on S400 in vivo (lane 3). The status of endogenous Par-1b phosphorylation was interrogated in several mammalian cell lines by using the pS400 antibody (Fig. 1B). Two electrophoretic forms of Par-1b that arise by alternative splicing (8, 11) were detected in each cell line and both splice variants reacted with the phosphospecific antibody. Thus, endogenous Par-1b is phosphorylated on S400 in vivo.
Fig. 1.
Par-1b is phosphorylated on S400 in vivo. (A) HeLa cells were transfected with plasmids encoding the indicated proteins by using HeLa MONSTER reagent for 48 h. Flag-tagged Par-1b proteins were resolved by SDS/PAGE and Western blotting was performed with an antibody specific for Par-1b phosphorylated on S400 (Upper). Blots were stripped and reprobed with an antibody specific for the flag epitope (Lower). (B) Lysates prepared from the indicated cell lines were resolved by SDS/PAGE, and Western blotting was performed with an antibody specific for Par-1b phosphorylated on S400 (Upper). Blots were stripped and reprobed with an antibody specific for Par-1b (Lower). (C) Sequence alignment of Par-1 orthologs indicates conservation of the S400 phosphorylation site. The mode I 14-3-3 binding motif, the PKD phosphorylation motif and the phosphopeptide used to generate the pS400-specific Par-1b antibody are also indicated.
Members of the Par-1 family share a conserved amino-terminal kinase domain, followed by a ubiquitin-associated (UBA) domain, a divergent region of unknown function and a conserved C-terminal region of ≈100 aa [supporting information (SI) Fig. S1]. S400 resides within the divergent region and is conserved in all 4 human Par-1 kinases as well as the fly, but not worm ortholog (Fig. 1C). Par-1a also reacted with the pS400 antibody demonstrating that Par-1a is also phosphorylated on S410 (S400 equivalent) in vivo (Fig. S1B). The pS400-specific antibody did not recognize Par-1c and Par-1d, and this is likely because the phosphopeptide used to generate the pS400-specific antibody varies significantly in sequence from residues surrounding the equivalent phosphorylation sites in Par-1c and Par-1d (Fig. 1C).
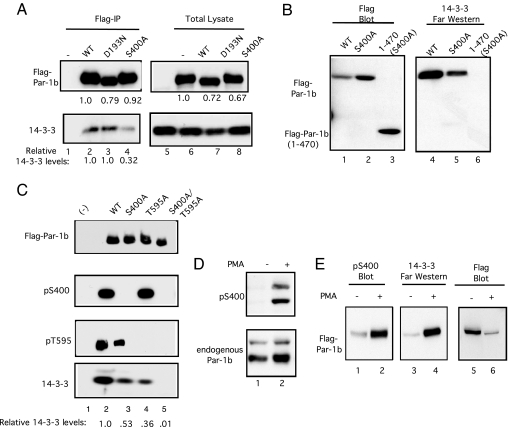
Interestingly, sequences inclusive of and surrounding S400 resemble a mode I 14-3-3 binding motif (12, 13) and Par-1b binds 14-3-3 proteins (6, 9, 14, 15). We tested whether S400 regulated interactions between Par-1b and 14-3-3 proteins in 2 ways. First, coprecipitation of 14-3-3 proteins with wild type and mutant forms of Par-1b were examined, and second, Far Western analysis with purified 14-3-3 proteins was used. As seen in Fig. 2A, coprecipitation of 14-3-3 proteins with WT (lane 2) and kinase-inactive Par-1b (lane 3) was observed. Substitution of S400 with alanine diminished, but did not eliminate, 14-3-3 binding to Par-1b (lane 4). Far Western analysis confirmed that mutation of S400 reduced interactions between Par-1b and 14-3-3 proteins (Fig. 2B, lane 5). A truncation mutant of Par-1b consisting of amino acids 1–470 but lacking S400 failed to bind 14-3-3 proteins (lane 6), indicating that phosphorylation of S400 regulated interactions between 14-3-3 proteins and the amino terminus of Par-1b. Additional 14-3-3 binding site(s) must reside within the C terminus of Par-1b, given that mutation of S400 diminished but did not eliminate interactions between 14-3-3 proteins with full length Par-1b (Fig. 2A). A previous study reported that 14-3-3 binding is facilitated by T595 phosphorylation (9). We monitored WT and mutant forms of Par-1b for their ability to bind to 14-3-3 proteins and to be phosphorylated on S400 and T595 in vivo (Fig. 2C and Fig. S1C). We observed that substitution of T595 with alanine reduced interactions between Par-1b and 14-3-3 proteins as did substitution of S400 with alanine. Simultaneous mutation of both phosphorylation sites severely compromised binding of 14-3-3 proteins to Par-1b.
Fig. 2.
Phosphorylation of Par-1b on S400 regulates 14-3-3 binding and is stimulated by PMA. (A) HEK293 cells were transfected with plasmids encoding WT Par-1b (WT), kinase-inactive Par-1b (D193N), or a phosphorylation-site mutant of Par-1b (S400A) by using Superfect reagent for 48 h. Lysates were resolved directly by SDS/PAGE (lanes 5–8) or were incubated with anti-flag agarose before SDS/PAGE (lanes 1–4). Western blotting was performed with an antibody specific for the flag epitope to detect Par-1b (Upper) or with an antibody specific for 14-3-3 proteins (Lower). Relative levels of 14-3-3 in each precipitate were determined from Western blotting by using the ImageJ program and are indicated below the blot. (B) HEK293 cells were transfected with plasmids encoding the indicated fusion proteins by using Superfect reagent for 48 h. Lysates were incubated with anti-flag agarose and precipitates were resolved by SDS/PAGE. Precipitates were subjected to Western blotting to monitor Par-1b levels (lanes 1–3) or to Far Western analysis to monitor 14-3-3 binding (lanes 4–6). (C) HeLa cells were transfected with plasmids encoding the indicated flag-tagged proteins by using Lipofectamine 2000 for 24 h. Lysates were incubated with flag agarose. Precipitates were resolved by SDS/PAGE and analyzed by Western blotting by using the indicated antibodies. Relative levels of 14-3-3 in each precipitate were determined from Western blotting by using the ImageJ program and are indicated below the blot (Fig. S1C). A representative experiment is shown in C. (D) Lysates prepared from HeLa cells that had been cultured in the absence of serum for 16 h and then treated with vehicle (lane 1) or with 400 ng/mL of PMA for 10 min (lane 2) were resolved by SDS/PAGE and Western blotting was performed with pS400 antibody (Upper). Blots were stripped and reprobed with a Par-1b specific antibody (Lower). (E) HEK293 cells were transfected with plasmid encoding Flag-Par-1b by using Superfect reagent for 48 h. Cells were cultured in the absence of serum for 3 h and then treated with vehicle (lanes 1 and 3) or with 400 ng/mL of PMA for 20 min (lanes 2 and 4). Flag-tagged Par-1b was precipitated by using anti-flag agarose and resolved by SDS/PAGE. Flag-Par-1b was subjected to Western blotting to monitor S400 phosphorylation (lanes 1 and 2) or to Far Western analysis to monitor 14-3-3 binding (lanes 3 and 4). The pS400 blot was stripped and probed with antibody specific to the flag epitope (lanes 5 and 6).
In our search to identify signaling pathways that regulate Par-1b in vivo, we observed that treatment of cells with the PKC activator PMA potently stimulated phosphorylation of endogenous (Fig. 2D) and ectopically-produced (Fig. 2E, lane 2) Par-1b on S400 in vivo. Also, Far Western analysis demonstrated that PMA-treatment enhanced the binding of 14-3-3 proteins to Par-1b (Fig. 2E, lane 4). Although PMA enhanced the binding of 14-3-3 proteins to Par-1b, its relative binding to WT and mutant forms of Par-1b was not significantly altered in PMA-treated cells (Fig. S1D). Enhanced Par-1b phosphorylation on S400 by PMA implicated members of the PKC family as upstream regulators of Par-1b. In particular, both the conventional (c)PKCs and nPKCs are activated by diacylglycerol (DAG) or phorbol esters (16). Sequences surrounding and inclusive of S400 (KVQRSVpSA) do not form a consensus PKC phosphorylation site but rather more closely resemble that of a PKD consensus site (Fig. 1C) (17). A major pathway for the activation of PKD is translocation to membrane compartments by means of binding to DAG or phorbol esters followed by phosphorylation of activation loop residues by nPKCs (δ, ε, η, and θ), which are directly activated by PMA (18). As seen in Fig. S1E, PMA treatment resulted in activation loop phosphorylation of both PKD1 (lane 2) and PKD2 (lane 4) indicative of PKD activation.
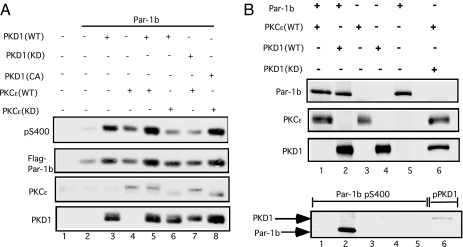
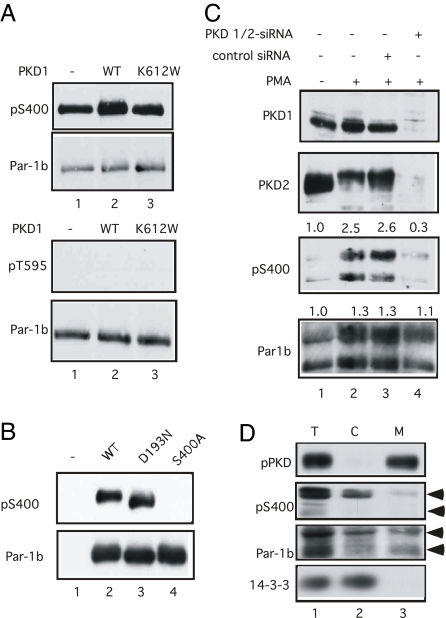
Several experiments were performed to determine whether Par-1b is directly regulated by PKD. First, the phosphorylation status of Par-1b was monitored after coproduction with wild type and mutant forms of PKD1 and PKCε (Fig. 3A). Enhanced S400-phosphorylation was observed when Par-1b was coproduced with either WT PKD1 (lane 3), WT PKCε (lane 4), or both PKD1 and PKCε (lane 5). Also, the stimulatory effects of PKD1 and PKCε on Par-1b S400-phosphorylation were blocked by kinase-inactive PKCε (lane 6) and kinase-inactive PKD1 (lane 7), respectively. Importantly, kinase-inactive PKCε was unable to block the stimulatory effects of constitutively active PKD1 on Par-1b S400 phosphorylation (lane 8). These findings suggest that PKCε functions upstream of PKD to regulate phosphorylation of Par-1b on S400. Next, kinase assays were performed in vitro to determine whether Par-1b was a direct substrate of either PKCε or PKD1 (Fig. 3B). PKD1 phosphorylated a kinase-inactive mutant of Par-1b on S400 in vitro (Lower, lane 2). Although PKCε phosphorylated activation loop residues of PKD1 (Lower, lane 6), it was incapable of phosphorylating Par-1b on S400 in vitro (Lower, lane 1). Also, PKD1 did not phosphorylate Par-1b on T595, the aPKC site, in vitro (Fig. 4A Lower). Interestingly, S400 (Fig. 4A Upper), but not T595 (Fig. 4A Lower), was a site of Par-1b autophosphorylation (lane 1). To determine whether auto/trans phosphorylation contributed significantly to S400 phosphorylation in vivo, WT and mutant forms of Par-1b were expressed in Par-1b null mouse embryo fibroblasts (MEFs) (11). Note that kinase-active (Fig. 4B, lane 2) and kinase-inactive (lane 3) forms of Par-1b were similarly phosphorylated on S400. Thus, although Par-1b is capable of phosphorylating itself on S400, additional cellular kinase(s) also serve this function in vivo.
Fig. 3.
Phosphorylation of Par-1b on S400 is regulated by nPKC/PKD pathway. (A) HEK293 cells were transfected with the indicated plasmids by using Superfect reagent for 24 h. Lysates were resolved by SDS/PAGE and subjected to Western blotting. (B) Kinase assays were performed in vitro with a kinase-inactive mutant of Par-1b and the indicated purified protein kinases. Western blotting was performed to monitor levels of each protein in the assay (Upper 3 panels) and to monitor the phosphorylation status of Par-1b (Lower, lanes 1–5) and PKD1 (Lower, lane 6).
Fig. 4.
Phosphorylation and localization of Par-1b regulated by PKD in vivo. (A) Kinase assays were performed in vitro with kinase-active Par-1b alone (lane 1) or in the presence of kinase-active (lane 2) or kinase-inactive (K612W, lane 3) PKD1. Reactions were resolved by SDS/PAGE and subjected to Western blotting. The phosphorylation status of Par-1b was monitored by using the indicated phospho-specific antibodies. (B) Par-1b null MEFs (32) were transfected with plasmids expressing the indicated proteins by using Lipofectamine 2000 for 24 h. Lysates were incubated with anti-flag agarose and precipitates were resolved by SDS/PAGE. Precipitates were subjected to Western blotting with antibodies specific for Par-1b phosphorylated on S400 (Upper). Blots were stripped and reprobed with Flag-specific antibodies (Lower). (C) HeLa cells were untreated, incubated with control siRNAs or with siRNAs specific for PKD1 and PKD2 as described in the methods section. Cells were incubated for 1 min with 200 ng/mL of PMA, lysed and subjected to Western blotting with the indicated antibodies. Relative levels of S400 phoshorylation and Par-1b protein are indicated above each blot. (D) Lysates from PMA-treated HeLa cells (T) were fractionated into soluble (S) and membrane (M) compartments. Fractions were probed for the indicated proteins by Western blotting. Arrows denote 2 isoforms of Par-1b.
Next, siRNA was used to knockdown expression of PKD1 and PKD2 in HeLa cells. As seen in Fig. 4C, stimulation of S400 phosphorylation was not observed in PMA-treated cells deficient for PKD1 and PKD2 (lane 4), whereas control cells (lane 2) or cells incubated with scrambled siRNA (lane 3) showed a robust stimulation of S400 in response to PMA-treatment. Together, these results suggest that PMA stimulates a signaling pathway from nPKCs to PKD to Par-1b to regulate S400-phosphorylation and 14-3-3 binding.
Phosphorylation of Par-1b by aPKC induces dissociation of Par-1b from the plasma membrane into soluble fractions in a T595-dependent manner (9). To test the consequences of S400 phosphorylation, PMA-treated HeLa cells were fractionated by sequential centrifugation and membrane (Fig. 4D, M) and soluble (Fig. 4D, S) fractions were analyzed for endogenous Par-1b by Western blotting (Fig. 4D). As expected, phosphorylated PKD was found in membrane fractions, whereas 14-3-3 proteins were observed in soluble fractions. Importantly, endogenous Par-1b phosphorylated on S400 was present predominantly in soluble fractions.
Discussion
In this study, we identified S400 as a nPar-1b phosphorylation site. Phosphorylation of S400 was shown to regulate interactions between Par-1b and 14-3-3 proteins and to be mediated by PKD. Par-1b is also phosphorylated on T595, and this phosphorylation is catalyzed by atypical PKC (8, 9). Thus, Par-1b is regulated by 2 arms of the PKC pathway: one arm is indirect, involving activation of nPKCs, which, in turn, activate PKD to phosphorylate Par-1b on S400; the second arm is direct and involves phosphorylation of T595 by aPKCs.
Previous studies reported that 14-3-3 binding to Par-1 either does not involve Par-1 phosphorylation (19) or is facilitated by T595 phosphorylation (9). Another study reported 17 new Par-1b phosphorylation sites and mutation of all 17 of these residues eliminated 14-3-3 binding. Surprisingly, S400 was not identified as a site of phosphorylation in this study (14). Here, we report that mutation of the 2 PKC-regulated sites (S400 and T595) is sufficient to ablate 14-3-3 binding to Par-1b. It may be that mutation of 17 residues of Par-1b indirectly effected phosphorylation at S400 and this phosphorylation, in turn, explains the loss of 14-3-3 binding in the Goransson et al. study (14).
The PKC family of protein kinases are subdivided into conventional PKCs that are activated by calcium, acidic phospholipids, and DAG; nPKCs activated by DAG and acidic phospholipids but insensitive to calcium and aPKCs that are activated, in part, by PKD1 (16). The PKD family consists of 3 members. PKD1 was originally reported to be a nPKC and was given the name PKCμ. However, it was later appreciated that the PKD family distinguished itself from the PKC family in amino acid composition, domain structure, regulation, and substrate specificity (20). A major pathway for the activation of PKD is translocation to membrane compartments by means of binding to DAG or phorbol esters followed by phosphorylation of activation loop residues by the nPKCs, which are directly activated by PMA (18). Stimulation of S400 phosphorylation by PMA implicated members of the PKC family as regulators of Par-1b. However, sequences inclusive of and surrounding S400 do not conform to a typical PKC consensus sequence but rather more closely resembles that of a PKD phoshorylation site (17). The experimental evidence leading to the conclusion that PKD directly phosphorylates Par-1b on S400 in a nPKC-dependent manner is as follows: enhanced S400-phosphorylation was observed when Par-1b was coproduced with PKD1 and/or PKCε (Fig. 3A); the stimulatory effects of PKD1 and PKCε on Par-1b S400-phosphorylation were blocked by kinase-inactive forms of each kinase (Fig. 3A); importantly, kinase-inactive PKCε was unable to block the stimulatory effects of constitutively-active PKD1 on Par-1b S400 phosphorylation (Fig. 3A); PKD, but not nPKC, directly phosphorylated Par-1b on S400 in vitro (Fig. 3B); and last, stimulation of S400 phosphorylation by PMA was not observed in cells deficient for PKD1 and PKD2 (Fig. 4C). Together, these data argue that PMA stimulates nPKCs to activate PKD and PKD, in turn, directly phosphorylates Par-1b on S400.
PKD functions downstream of activated G protein coupled receptors and receptor tyrosine kinases that signal through phospholipase C. PKD is activated by any agent that activates nPKCs and, as such PKD functions to regulate a diverse set of cellular processes including proliferation, apoptosis, stress responses, vesicle trafficking from golgi, neuronal development, and immune cell signaling (21–23). If Par-1b is an important downstream target of PKD in each of these pathways, it may explain the diverse phenotypes observed in mice disrupted for Par-1b (5). It has been demonstrated that the regulated cycling of Par-1b on and off lateral membranes is critical for maintenance of apicobasal polarity in mammalian cells (9). Our study provides mechanistic insight into how Par-1b shuttling is regulated. Phoshorylation of Par-1b on S400 by PKD, like phosphorylation of T595 by aPKC, not only promotes the binding of 14-3-3 proteins to Par-1b but also promotes the release of Par-1b off of cellular membranes. Also, these results implicate a role for PKD in regulating cellular polarity through phosphorylation of Par-1b.
Materials and Methods
Cell Culture.
HeLa, HEK-293, and HCT116 cells were routinely maintained in DMEM (Gibco-BRL) supplemented with 10% bovine growth serum (HyClone), 100 units per mL of penicillin and streptomycin, and 1 mM l-glutamine. MCF7 cells were grown in the presence of 10% FBS (HyClone). Par-1b null MEFs (11) were cultured in DMEM, 10% FBS (HyClone), and 1 mM l-glutamine, 0.2 mM nonessential amino acids, 140 mM 2-mercaptoethanol, 100 units per mL of penicillin G, and 100 μg/mL streptomycin.
Plasmids and Reagents.
PMA was purchased from Sigma. Purified PKCε and the PKC lipid activator were purchased from Upstate Cell Signaling. Sf9 cell purified HA-PKD and HA-PKD (K612W) as well as plasmids pcDNA3.HA-PKD1, pcDNA3.HA-PKD1(K612W), and pcDNA3.HA-PKD1 (S738E, S742E) have been described (24, 25). Plasmids encoding PKCε(WT) and PKCε(KD) and Flag tagged Par-1b, Par-1b(D193N) and Par-1b(T595A) have been described (8, 26). Substitution of alanine for serine at position 400 and/or threonine at position 595 was accomplished by using the QuikChange XL Site-Directed Mutagenesis Kit (Stratagene). DNA sequencing was performed to verify the mutant sequence; siRNAs specific for PKD1 and PKD2 were purchased from Dharmacon as well as SiGenome Smart Pool reagent to Human PRKCM (PKD1) and Smart Pool reagent to Human PRKD2 (PKD2). The control siRNA sequence was: 5′UAAGGCUAUGAAGAGAUACUU. Transient transfections were performed by using TransIT HeLa MONSTER reagent (Mirus Bio), with Superfect reagent (Qiagen) or with Lipofectamine 2000 reagent (Invitrogen). Par-1b null MEFs have been described (11).
Antibodies and Western Blotting.
Antibodies against 14-3-3 (K19) and glutathione S-transferase (GST) were purchased from Santa Cruz. Other antibodies used in this study were specific for PKD1 (Abcam or Cell Signaling Technology), PKD2 (Abcam), phosphoPKD (Cell Signaling Technology) and PKCε (Upstate Cell Signaling). Antibodies used for experiments shown in Fig. S1E include a rabbit polyclonal antibody specific for PKD1 raised against a NH2-MAECQNDSGEMQDP-amide peptide (amino acids 372–385 in human PKD1), a rabbit polyclonal antibody specific for PKD2 was from Upstate Cell Signaling, and a rabbit polyclonal antibody specific for PKD3 from Bethyl Laboratories. These antibodies are specific for the respective PKD isoenzyme and do not cross react with other PKD family members. Antibodies specific for Par-1b have been described (11). Antibodies specific for Par-1b phosphorylated on S400 were generated by immunizing rabbits with the phosphopeptide CQRSV-pS-ANPKQ coupled to keyhole limpet hemocyanin (KLH). Antibodies specific for Par-1b phosphorylated on T595 have been described (8). Flag-fusion proteins were precipitated with anti-Flag M2 antibody-agarose affinity gel (Sigma) and detected by Western blotting with anti-Flag M2 monoclonal antibody (Sigma). For Western blotting, antibodies were diluted in 5% milk in 50 mM Tris·HCl (pH 8.0), 0.15 M NaCl, and 0.02% Tween-20 (TBST), and membranes were washed 4 times in TBST after application of both primary and secondary antibody. Bound primary antibodies were detected with horseradish peroxidase-conjugated goat anti-mouse (Jackson ImmunoResearch), goat anti-rabbit (Invitrogen), or donkey anti-goat (Santa Cruz) secondary antibodies, and visualized by using the ECL reagent (GE Healthcare) or Super Signal West Femto reagent (Pierce).
Immunoprecipitations.
Cells were lysed in 50 mM Tris·HCl (pH 8.0), 0.1 M NaCl, 5 mM EDTA, 0.5% Nonidet P-40, and 1 mM DTT (MCLB) supplemented with 1 μM microcystin-LR and protease inhibitors (2 mM PMSF, 0.15 units/mL aprotinin, 20 μM leupeptin, and 20 μM pepstatin.). Lysates were clarified by centrifugation at 16, 000 × g. Cell lysates were precleared with protein A agarose for 1 h at 4 °C, then incubated with anti-FLAG agarose (Sigma) for 1 h at 4 °C. Beads were washed 4 times in MCLB, and bound proteins were eluted by boiling in SDS/PAGE sample buffer.
14-3-3 Far Western Analysis.
Samples were subjected to SDS/PAGE and transferred to nitrocellulose membranes as for Western blotting. Proteins bound to the membrane were denatured by incubating for at least 1 h in denaturation buffer [50 mM Tris·HCl (pH 8.0), 6 M guanidine-HCl, 6.25 mM EDTA, 1 mM DTT, 10% glycerol, 0.05% Tween-20], then renatured by incubating for at least 1 h in renaturation buffer (denaturation buffer without guanidine-HCl or DTT). Membranes were blocked for 1 h in 5% milk in TBST, rinsed in TBST, and then incubated for 2 h at room temperature or overnight at 4 °C in TBST containing 0.1 μg/mL of GST-14-3-3ζ and σ, and 1 mg/mL BSA. GST-14-3-3 proteins were purified as described (15). Membranes were washed 4 times in TBST, then incubated for 1 h with anti-GST primary antibody in 5% milk/TBST. Bound primary antibody was detected with horseradish peroxidase-conjugated secondary antibody, and visualized by using ECL reagent, as for Western blotting.
Coprecipitation of 14-3-3 Proteins with WT and Mutant Forms of Par-1b.
HeLa cells at 75% confluency were transfected by using Lipofectamine 2000 reagent for 24 h. Cells were lysed in MCLB supplemented with 1 μM microcystin and protease inhibitors. Lysates were clarified by centrifugation at 16,000 × g. Cell lysates were preincubated with protein A agarose 1 h at 4 °C, then incubated with anti-FLAG agarose for 1 h at 4 °C. Bound proteins were eluted by incubating 1 h with a 3× flag peptide (Sigma).
Kinase Assays.
Kinase reactions were carried out in the presence of 0.5 μg bacterially-purified GST-Par-1b, GST-Par-1b (S400A), or GST-Par-1b (D160N) with 250 ng Sf9 cell purified HA-PKD or HA-PKD (K612W) or with 5 ng active PKCε (Upstate Cell Signaling) and a buffer consisting of 12 μM ATP, 2 mM DTT, 10 mM MgCl2, and 50 mM Tris (pH 7.5) supplemented with sonicated PKC lipid activator containing phosphatidyl serine and diacylglycerol (Upstate Cell Signaling). Samples were incubated at room temperature for 30 min. Reactions were terminated by the addition of SDS sample buffer followed by incubation for 10 min at 37 °C.
RNAi Experiments.
RNAi was performed by using protocols supplied by Dharmacon; siRNA duplexes were transfected into HEK293 cells by using DharmaFECT 2 at a final concentration of 10 nM. After ≈30 h, cells were transfected a second time with siRNA at 10 nM final concentration. Cells were incubated an additional 48 h and then incubated in the absence or presence of 200 ng/mL of PMA for 1 min. Cells were lysed in MCLB and analyzed by Western blotting.
Cell Fractionation.
HeLa cells were grown to near confluency on 10 cm tissue culture dishes. Cells were treated with 200 ng/mL of PMA for 1 min before harvest. Cells were trypsinized and washed in ice-cold PBS. Cells were suspended in 800 μL of hypotonic buffer (HB: 12.5 mM Tris pH 7.5, 1 mM EDTA, 1 mM DTT) supplemented with 5 mM NaP4PO4, 1 μM microcystin, 1 mM sodium orthovanadate, 2 mM PMSF, 0.15 units/mL aprotinin, 20 μM leupeptin, and 20 μM pepstatin for ≈40 min. Cells were dounce homogenized, and, when the majority of cells were lysed, cell lysates were centrifuged at 1,000 × g for 5 min. The resulting supernatant was transferred to a fresh tube, and the pellet was washed once with 200 μL of HB. The washed supernatant was added to the first collected supernatant [ = total (T)]. A portion of this supernatant was reserved for Western blotting, and the remainder was centrifuged at 100,000 × g for 30 min. The supernatant was transferred to a fresh tube [ = soluble (S) fraction] and the pellet was vortexed in 100 μL of HB and recentrifuged at 100,000 × g for 15 min. The supernatant was added to the soluble fraction and the pellet [ = membrane (M) fraction] was resuspended in 300 μL of MCLB supplemented with 1 μM microcystin, 1 mM sodium orthovanadate, and protease inhibitors. Fractions were resolved by SDS/PAGE and proteins visualized by Western blotting by using Super Signal West Femto reagent.
Supplementary Material
Acknowledgments.
We thank Seong-Kyoun Kim, Paul Graves, Geoff Uy, Elizabeth Tank, Stephanie Willerth, and Jonathan Hurov for help in the early stages of this work; and Gwanghee Lee for help with data interpretation. K.T.L. is supported by Training Grant T32 CA113275 and the Alvin J. Siteman Cancer Center. This work was supported by National Institutes of Health Grant CA075134 (to A.T.). H.P.-W. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0809661105/DCSupplemental.
References
- 1.Kemphues KJ, Priess JR, Morton DG, Cheng N. Identification of genes required for cytoplasmic localization in early embryos of C. elegans. Cell. 1988;52:311–320. doi: 10.1016/s0092-8674(88)80024-2. [DOI] [PubMed] [Google Scholar]
- 2.Kemphues K. PARsing embryonic polarity. Cell. 2000;101:345–348. doi: 10.1016/s0092-8674(00)80844-2. [DOI] [PubMed] [Google Scholar]
- 3.Goldstein B, Macara IG. The PAR proteins: Fundamental players in animal cell polarization. Dev Cell. 2007;13:609–622. doi: 10.1016/j.devcel.2007.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tassan J-P, Schultz SJ, Bartek J, Nigg EA. Cell cycle analysis of the activity, subcellular localization, and subunit composition of human CAK(CDK-activating kinase) J Cell Biol. 1994;127:467–478. doi: 10.1083/jcb.127.2.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hurov J, Piwnica-Worms H. The Par-1/MARK family of protein kinases: From polarity to metabolism. Cell Cycle. 2007;6:1966–1969. doi: 10.4161/cc.6.16.4576. [DOI] [PubMed] [Google Scholar]
- 6.Benton R, St Johnston D. Drosophila PAR-1 and 14–3-3 inhibit Bazooka/PAR-3 to establish complementary cortical domains in polarized cells. Cell. 2003;115:691–704. doi: 10.1016/s0092-8674(03)00938-3. [DOI] [PubMed] [Google Scholar]
- 7.Hurd TW, et al. Phosphorylation-dependent binding of 14-3-3 to the polarity protein Par3 regulates cell polarity in mammalian epithelia. Curr Biol. 2003;13:2082–2090. doi: 10.1016/j.cub.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 8.Hurov JB, Watkins JL, Piwnica-Worms H. Atypical PKC Phosphorylates PAR-1 Kinases to Regulate Localization and Activity. Curr Biol. 2004;14:736–741. doi: 10.1016/j.cub.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki A, et al. aPKC acts upstream of PAR-1b in both the establishment and maintenance of mammalian epithelial polarity. Curr Biol. 2004;14:1425–1435. doi: 10.1016/j.cub.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 10.Chen YM, et al. Microtubule affinity-regulating kinase 2 functions downstream of the PAR-3/PAR-6/atypical PKC complex in regulating hippocampal neuronal polarity. Proc Natl Acad Sci USA. 2006;103:8534–8539. doi: 10.1073/pnas.0509955103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hurov JB, et al. Immune cell dysfunction and autoimmune disease in mice lacking the EMK1/Par-l protein kinase. Mol Cell Biol. 2001;21:3853–3861. doi: 10.1128/MCB.21.9.3206-3219.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14-3-3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–897. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 13.Yaffe MB, et al. The structural basis for 14-3-3:phosphopeptide binding specificity. Cell. 1997;91:961–971. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- 14.Goransson O, et al. Regulation of the polarity kinases PAR-1/MARK by 14-3-3 interaction and phosphorylation. J Cell Sci. 2006;119:4059–4070. doi: 10.1242/jcs.03097. [DOI] [PubMed] [Google Scholar]
- 15.Meek SE, Lane WS, Piwnica-Worms H. Comprehensive proteomic analysis of interphase and mitotic 14-3-3-binding proteins. J Biol Chem. 2004;279:32046–32054. doi: 10.1074/jbc.M403044200. [DOI] [PubMed] [Google Scholar]
- 16.Newton AC. Regulation of protein kinase C. Curr Opin Cell Biol. 1997;9:161–167. doi: 10.1016/s0955-0674(97)80058-0. [DOI] [PubMed] [Google Scholar]
- 17.Nishikawa K, Toker A, Johannes FJ, Songyang Z, Cantley LC. Determination of the specific substrate sequence motifs of protein kinase C isozymes. J Biol Chem. 1997;272:952–960. doi: 10.1074/jbc.272.2.952. [DOI] [PubMed] [Google Scholar]
- 18.Rozengurt E, Rey O, Waldron RT. Protein kinase D signaling. J Biol Chem. 2005;280:13205–13208. doi: 10.1074/jbc.R500002200. [DOI] [PubMed] [Google Scholar]
- 19.Benton R, Palacios IM, St Johnston D. Drosophila 14-3-3/PAR-5 is an essential mediator of PAR-1 function in axis formation. Dev Cell. 2002;3:659–671. doi: 10.1016/s1534-5807(02)00320-9. [DOI] [PubMed] [Google Scholar]
- 20.Rykx A, et al. Protein kinase D: A family affair. FEBS Lett. 2003;546:81–86. doi: 10.1016/s0014-5793(03)00487-3. [DOI] [PubMed] [Google Scholar]
- 21.Wang QJ. PKD at the crossroads of DAG and PKC signaling. Trends Pharmacol Sci. 2006;27:317–323. doi: 10.1016/j.tips.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 22.Toker A. The biology and biochemistry of diacylglycerol signalling. Meeting on molecular advances in diacylglycerol signalling. EMBO Rep. 2005;6:310–314. doi: 10.1038/sj.embor.7400378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Van Lint J, et al. Protein kinase D: An intracellular traffic regulator on the move. Trends Cell Biol. 2002;12:193–200. doi: 10.1016/s0962-8924(02)02262-6. [DOI] [PubMed] [Google Scholar]
- 24.Storz P, Doppler H, Johannes FJ, Toker A. Tyrosine phosphorylation of protein kinase D in the pleckstrin homology domain leads to activation. J Biol Chem. 2003;278:17969–17976. doi: 10.1074/jbc.M213224200. [DOI] [PubMed] [Google Scholar]
- 25.Storz P, Doppler H, Toker A. Protein kinase Cdelta selectively regulates protein kinase D-dependent activation of NF-kappaB in oxidative stress signaling. Mol Cell Biol. 2004;24:2614–2626. doi: 10.1128/MCB.24.7.2614-2626.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cenni V, et al. Regulation of novel protein kinase C epsilon by phosphorylation. Biochem J. 2002;363:537–545. doi: 10.1042/0264-6021:3630537. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.