Abstract
Acetylation of histone proteins by the yeast Spt-Ada-Gcn5-acetyltansferase (SAGA) complex has served as a paradigm for understanding how posttranslational modifications of chromatin regulate eukaryotic gene expression. Nonetheless, it has been unclear to what extent the structural complexity of the chromatin substrate modulates SAGA activity. By using chromatin model systems, we have found that SAGA-mediated histone acetylation is highly cooperative (cooperativity constant of 1.97 ± 0.15), employing the binding of multiple noncontiguous nucleosomes to facilitate maximal acetylation activity. Studies with various chromatin substrates, including those containing novel asymmetric histone octamers, indicate that this cooperativity occurs only when both H3 histone tails within a nucleosome are properly oriented and unacetylated. We propose that modulation of maximal SAGA activity through this dual-tail recognition could facilitate coregulation of spatially proximal genes by promoting cooperative nucleosome acetylation between genes.
Keywords: chromatin, enzymology, protein modification, transcription, S. cerevisiae
The 1.8-MDa Spt-Ada-Gcn5-acetyltansferase (SAGA) complex in the budding yeast Saccharomyces cerevisiae, and highly homologous complexes in higher eukaryotes, plays a major role in regulating gene expression (1, 2). In yeast, this regulation occurs predominantly by enhancing transcription of inducible genes (3) and occurs in part through the SAGA-mediated acetylation of chromatin at the promoter and transcribed regions of genes (4, 5).
The chromatin targets of SAGA-mediated acetylation are the amino-terminal portions of histone proteins, also called histone tails, which extend past the DNA of nucleosomes. In vitro studies have shown that the histone H3 tail is the primary target of SAGA-mediated acetylation, although the histone H2B tail can also be weakly acetylated (6). Within the H3 tail, the side chain of lysine-14 is the major site of acetylation, with H3 lysine-9 and lysine-18 being secondary sites, and H3 lysine-23 being a tertiary site (7). Interestingly, the histone H3 tail not only interacts with the catalytic domain of the SAGA complex subunit Gcn5 (8) but can also bind to a number of other domains contained within the 20-protein subunits that comprise the complex. Some of the currently identified domains include the SANT domain of Ada2 (9) and the bromo domains of both Gcn5 and Spt7, which recognize acetylated H3 tails (10).
The ability of the H3 tail to serve as both substrate and binding partner of the SAGA complex potentially allows for complex regulation of the action of SAGA by chromatin. This regulation could occur at many levels of chromatin structure. In the most basic structural unit of chromatin, the nucleosome, all 4 histones, H2A, H2B, H3, and H4, are present in 2 copies, and these histones are associated with 147 bp of nucleosomal DNA wrapped 1.67 times around the histone octamer (11). Thus, the core nucleosome component and the presence of 2 copies of the H3 tails have the potential to influence SAGA activity. In addition, H3 tails are presented at relatively regular intervals because nucleosomes in the genome occur frequently and in relatively close proximity. For example, in yeast ≈69% of chromosome III is sequestered by nucleosomes, with an average distance between nucleosome centers of 212 bp (12). Although these nucleosomes are arrayed in a linear fashion in genomic sequence, in many cases they adopt more complex higher-order structures. Short-range interactions between nucleosomes within a strand of chromatin allow chromatin to adopt a more compact 30-nm fiber structure (13, 14), and long-range interactions between distant nucleosomes provide a means for chromatin to potentially adopt 100- to 400-nm fiber structures (15). Moreover, a number of recent studies suggest that different functional forms of chromatin can adopt complex looping structures, where regions of the genome that are separated by large distances in sequence can be brought together in space (16). These complex forms of higher-order chromatin structure further complicate the presentation of the H3 tails to the SAGA complex.
We hypothesized that the rich structural complexity of chromatin would be exploited by the SAGA complex to both regulate and adjust its acetyltransferase activity. To test this possibility, we measured rates of nucleosome acetylation by the SAGA complex and found 2 aspects of chromatin structure that direct SAGA acetylation activity. Within a nucleosome the enzyme complex interacts with both copies of the histone H3 tail, and this interaction stimulates SAGA to bind to different, noncontiguous nucleosomes, generating cooperative acetylation.
Results
SAGA-Mediated Nucleosome Acetylation Is Cooperative.
To characterize the kinetics of chromatin acetylation by the SAGA complex, the enzyme complex was TAP-tagged on the carboxyl terminus of the Spt7 subunit and affinity-purified to a high degree of homogeneity, as described by Wu et al. (17). Nucleosomal arrays were assembled as a model of chromatin fibers by using a salt dialysis deposition strategy (18). In these nucleosomal arrays, 12 recombinant, wild-type, Xenopus laevis histone octamers are spaced approximately every 208 nt apart on a Lytechinus variegates 208-12 5S rDNA template. Initial rates of nucleosome acetylation were determined by measuring the time-dependent incorporation of the radiolabeled acetyl group transferred from acetyl-CoA to the histones (19). Initial velocities were determined over a large range of nucleosomal array concentrations and showed good linearity for early time points. In these time courses, the extent of acetyl incorporation per H3 histone was from 0.47% to 4.4%.
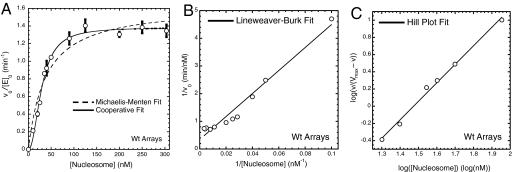
Originally, we sought to fit the extensive initial-velocity data with respect to nucleosome concentration according to the standard Michaelis–Menten model (Fig. 1A). However, the data were not well suited for such a model of hyperbolic saturation because few data points were within error of the best theoretical fit, and systematic deviation above and below the fit was observed. Instead, a sigmoidal, cooperative saturation kinetics model provided a fit that closely tracked the data (Fig. 1A). Fitting these data to linear formulations of the noncooperative and cooperative models reinforces the superiority of this approach. By using the Lineweaver–Burk form of the Michaelis–Menten model, it is clear the data shows systematic deviation from linearity (Fig. 1B). However, a Hill plot of the data is highly linear (Fig. 1C). From both direct fitting of the cooperative saturation kinetics model and the Hill plot, it is possible to extract a cooperativity constant, a parameter that measures the extent of cooperativity of the system. Both methods of fitting are in good agreement, 1.97 ± 0.15 and 1.91 ± 0.08, respectively. This magnitude of cooperativity is at least comparable with most known cooperative enzymes and suggests that maximal histone acetylation activity of the SAGA complex is facilitated through the binding of multiple substrates.
Fig. 1.
Acetylation of nucleosomal arrays by the SAGA complex is cooperative with respect to array concentration. (A) Initial velocity of nucleosomal array acetylation per enzyme as a function of nucleosome concentration, where [SAGA]0 = 1.0 nM and [acetyl-CoA]0 = 4.0 μM. For each nucleosomal array concentration the initial velocity is the average of 3 trials, with the associated SD indicated by the error bars. These data points were fit to both a Michaelis–Menten model (dashed line) and a cooperative saturation kinetics model (solid line). The cooperative saturation fit gives a cooperativity constant of 1.97 ± 0.15. (B) Linear fit of the data from A by using the Lineweaver–Burk formulation of the Michaelis–Menten model. R = 0.983. (C) Linear fit of the data from A by using the Hill plot formulation of the cooperative saturation kinetics model. Data were fit where the Hill plot demonstrates linearity, from 10 to 90% saturation (34). Cooperativity constant is 1.91 ± 0.08. R = 0.997.
Cooperativity Results from Interactions Within the Nucleosome.
What is it about the nucleosomal substrate that creates such cooperativity of acetylation? Overall, the acetylation reaction consists of 2 substrates, nucleosomes and acetyl-CoA. Binding of nucleosome substrate by the SAGA complex could change subsequent kinetic activity by either changing enzyme activity toward additional nucleosome substrates or toward acetyl-CoA. The simplest model of the latter would be that binding of the nucleosome substrate could organize the acetyl-CoA binding site, increasing its affinity for the cofactor at higher nucleosome concentration. Previous kinetics studies with the catalytic core of Gcn5 and H3 peptide substrates argue against this possibility because substrate binding is ordered with acetyl-CoA binding preceding H3 tail binding (20). However, the more complex nature of both the nucleosome substrate and the SAGA enzyme complex in our reactions could change the overall reaction mechanism. To explore this possibility, initial-velocity kinetics assays were performed to determine whether the binding of acetyl-CoA was changed at different concentrations of nucleosomal array substrates (Fig. 2A). At both a low (15 nM) and high (150 nM) concentration of nucleosomal substrate, the Km for acetyl-CoA is nearly identical, being 4.04 μM and 4.16 μM, respectively. This result suggests that cooperativity of nucleosome acetylation does not come from changes in binding of acetyl-CoA, but instead from changes in activity toward nucleosomes.
Fig. 2.
Mononucleosomes, but not histone H3 tails alone, demonstrate cooperative acetylation. (A) Initial velocity of nucleosomal array acetylation per enzyme as a function of acetyl-CoA concentration. Assays were performed with a total nucleosome concentration of either 15 nM (solid line) or 150 nM (dashed line). Data were fit to the Michaelis–Menten model. Km values were determined to be 4.04 μM and 4.16 μM, respectively. (B) Initial velocity of mononucleosome acetylation per enzyme as a function of nucleosome concentration. Data were fit to a cooperative saturation model to give a cooperativity constant of 1.76 ± 0.17. (C) Amino acid sequences of the histone H3 peptide and proteins used in this work. (Upper) Sequence of wild-type, recombinant H3 histone. (Lower) Highlighted regions of this sequence that differ among the H3 histones and peptide analyzed. Deviations from the wild-type sequence are indicated in red. Side-chain acetylated lysine is abbreviated as X. (D) Initial velocity of histone H3 tail peptide acetylation per enzyme as a function of peptide concentration as determined from the average of 2 independent trials. SAGA concentration is 10 nM, and acetyl-CoA concentration is 10 μM. Data were fit to a cooperative saturation model to give a cooperativity constant of 0.97.
What in the nucleosomal array is required to create cooperativity of acetylation? Nucleosomal arrays are very complicated substrates because they present many potential binding interactions, including 2 copies each of the histone tails, the histone globular domain, and the nucleosomal DNA. Moreover, the oligomeric nature of nucleosomal arrays potentially allows binding interactions to occur between nucleosomes in both a SAGA-dependent and -independent manner. To simplify the system, initial rate kinetic experiments were performed on mononucleosomes that constitute a single repeat unit of the studied nucleosomal arrays (Fig. 2B). Analysis of initial velocities reveals that acetylation of the mononucleosomes is also highly cooperative (cooperativity constant of 1.76 ± 0.17), similar to the complete nucleosomal arrays (Table 1). Thus, cooperative acetylation does not require contiguous strands of nucleosomes. Similarly, formation of higher-order chromatin structure, which cannot be adopted by mononucleosomes, is not necessary for substrate cooperativity.
Table 1.
Summary of kinetic constants determined in this work
| Substrate | [Half-saturation], nM | kcat,app, min−1 | Cooperativity coefficient |
|---|---|---|---|
| Wild-type 208-12 arrays | 33.6 ± 5.0 | 1.42 ± 0.15 | 1.97 ± 0.15 |
| Single H3 tail arrays | 79.5 ± 9.2 | 1.54 ± 0.12 | 0.86 ± 0.13 |
| His6-tagged H3 arrays | 36.2 ± 2.9 | 1.48 ± 0.04 | 1.86 ± 0.07 |
| Wild-type 196-1 monos | 17.3 ± 4.9 | 1.02 ± 0.11 | 1.76 ± 0.17 |
| H3 tail-swap monos | 85.7 ± 5.7 | 0.84 ± 0.06 | 1.02 ± 0.07 |
| Tetra-Ala H3/WT H3 monos | 38.9 ± 6.9 | 1.07 ± 0.07 | 0.95 ± 0.03 |
| Tetra-Ac H3/WT H3 monos | 11.5 ± 2.4 | 1.12 ± 0.08 | 0.99 ± 0.10 |
| H3 tail peptide | 130,000 | 0.52 | 0.97 |
Kinetic constants were determined from the average of three independent trials, except with the H3 tail peptide, which was the average of two trials. Half-saturation concentration was calculated from K1/n. The apparent turnover rate constant, kcat,app, was determined from Vmax/[E]0 and reflects an initial concentration of acetyl-CoA of 4.0 μM. For the H3 tail peptide, in which [acetyl-CoA]0 is 10.0 μM, the apparent turnover rate constant has been normalized to be directly comparable with the other assay.
Because mononucleosomes are sufficient to induce cooperativity of SAGA-mediated acetylation, we investigated whether an even simpler substrate could function equivalently. Initial-velocity kinetics assays were performed on a histone H3 peptide (Fig. 2 C and D). Unlike the mononucleosome substrate, the H3 tail peptide was noncooperative (cooperativity constant of 0.97), fitting a saturation curve resembling the hyperbolic Michaelis–Menten equation. This finding suggests that cooperative acetylation requires more than the H3 tail in isolation. Because the nucleosome presents a pair of H3 tails in the context of a nucleosome, we wondered whether a nucleosome with a single H3 tail would suffice to restore the cooperative behavior.
Asymmetric Histone Octamer Generation.
To generate nucleosomes with a single H3 tail, we developed a general strategy for making asymmetric octamers (Fig. 3A). In this strategy, histone octamers are assembled from constituent histones similar to standard protocols (21). However, for the histone that is ultimately to be present in 2 different forms, in our case a full-length H3 histone and an amino-terminal truncated H3 histone (Fig. 2C), they are introduced as a mixture, with an untagged form present in vast excess of a tagged form. For our single H3 tail octamer preparation, statistically 3 forms of octamers are expected: a majority of octamers with 2 truncated H3 tails and no tags; a small population of octamers with a single full-length, tagged H3 histone and a single truncated H3; and a very small population of octamers with 2 full-length, tagged histones. Octamers containing no tag can then be removed by using affinity purification, to give a final mix of octamers where the majority consists of asymmetric octamers with a single, tagged H3 tail. In this final mixture, some symmetric octamer with 2 full-length tails will be present. However, varying the ratio of tagged and untagged histone initially can control the amount of this octamer. For example, by using a 9:1 ratio of untagged to tagged histone, a little more than 1 in 20 octamers that have been affinity-purified contain 2 tails, a level we deemed acceptable for our experiments. For the affinity purification, we opted to use a His6 tag/nickel resin strategy because others had shown that octamers could be isolated under these conditions (22).
Fig. 3.
Generation of nucleosomes with asymmetric histone H3 composition. (A) Scheme for generation of histone octamers containing a single H3 tail. H3 histones are depicted as blue-gray wedges, whereas histones H2A, H2B, and H4 are a lighter orange. H3 histone with HHHHHH denotes full-length H3 histone with a His6 amino-terminal tag. H3 histone with a short tail denotes amino-terminal truncated H3 histone. The first arrow indicates octamer assembly, whereas the second indicates affinity purification of the tagged octamers. (B) Histone composition of octamers prepared as described in A before and after affinity purification. Histones were resolved on an 18% SDS/polyacrylamide gel and stained with Coomassie blue. (C) Characterization of nucleosomal arrays prepared from single H3 tail octamers. 208-12 5S rDNA nucleosomal arrays prepared at varying molar ratios of single tail octamer to octamer positioning site were digested with EcoRI and then separated on a native 4% polyacrylamide gel and stained with ethidium bromide. Nuc and Naked indicate 5S mononucleosomes and free DNA, respectively.
Employing this asymmetric octamer strategy resulted in the desired single H3 tail octamers (Fig. 3B). Before nickel affinity purification, assembly of histone octamers using a 9:1 ratio of tailless H3 to His6-tagged full-length H3 resulted in a population with very little tagged histone. However, after nickel affinity purification, the level of tagged, full-length H3 was equal to that of the truncated histone, as would be expected for octamers containing a single full-length H3 tail. These asymmetric single H3 tail octamers formed nucleosomal arrays, saturating the 208-12 5S rDNA template in a manner similar to recombinant wild-type octamers (Fig. 3C).
Substrate Cooperativity Requires Dual H3 Tail Interactions.
Initial-velocity kinetic analysis of single H3 tail nucleosomal arrays was performed (Fig. 4A), and this substrate was found to be noncooperative in SAGA-mediated acetylation (cooperativity constant of 0.86 ± 0.13). This loss of cooperativity was caused by the truncation of the H3 tail and not by the presence of the His6 tag because arrays containing 2 full-length H3 histones with the His6 tag exhibit full cooperativity (Fig. 4B). These results indicate that both H3 tails within a nucleosome must be present to generate cooperative acetylation and that the presence of multiple full-length H3 tails in single copy across different nucleosomes is insufficient to generate cooperativity.
Fig. 4.
Truncation of a single-histone H3 tail or repositioning of both H3 tails results in loss of cooperativity of substrate acetylation. (A) Initial velocity of nucleosomal arrays per enzyme containing octamers with a single H3 tail as a function of nucleosome concentration. Three independent trials with differing nucleosome concentration were performed. Shown is a representative trial. Each trial was fit to a cooperative saturation model to give an average cooperativity constant of 0.86 ± 0.13. (B) Initial velocity of nucleosomal arrays per enzyme containing 2 copies of the amino-terminal His6-tagged H3 histones as a function of nucleosome concentration. Data were fit to a cooperative saturation model to give a cooperativity constant of 1.86 ± 0.07. (C) Relative orientation of the H3 and H4 histone tails based on mononucleosome structure 1AOI.11 DNA is depicted in white, H3 and H3′ histones in blue, and H4 and H4′ histones in green. The points at which the H3 and H4 tails exit the nucleosome are labeled in yellow. The histone tails, histones H2A and histone H2B, are omitted for clarity. (Lower) The point at which the H4 tail exits the nucleosome occurs behind the DNA and is not labeled. (D) Initial velocity per enzyme of histone H3 tail-swap mononucleosome as a function of nucleosome concentration. These mononucleosomes replace the H3 histones with tailless H3 histone, and histone H4 with the H3gH4 fusion histone. Data were fit to a cooperative saturation model to give a cooperativity constant of 1.02 ± 0.07.
The simplest explanation for why both histone H3 tails are necessary for cooperative acetylation is that the SAGA complex must bind both tails during the course of the reaction. Presumably, this tail binding is made possible by having binding domains in the enzyme complex located in a specific spatial orientation. To test this idea explicitly, we investigated the effect of moving the histone H3 tails to a new position.
In the nucleosome each pair of histone tails extends past the nucleosomal DNA at different locations (23). The H3 tails exit the nucleosome near the DNA entry/exit points and between gyres of the DNA (Fig. 4C). To a large extent, this puts the exit point of the H3 tails in the same plane defined by the trajectory of the nucleosomal DNA. In contrast, the histone H4 tails exit the nucleosome on opposite faces of the plane defined by the nucleosomal DNA. Thus, moving the H3 tail to the location of the H4 tail would present a different binding surface to the SAGA complex and potentially disrupt the cooperativity of acetylation. To prepare such a tail-swap nucleosome, recombinant histone containing the amino-terminal tail of histone H3, fused to the globular domain of histone H4, H3gH4 (Fig. 2C), was expressed and purified (plasmid was a gift from J. Hansen, Colorado State University, Fort Collins, CO). Octamers and mononucleosomes were assembled from H3gH4, H2A, H2B, tailless H3, and 5S rDNA. Tailless H3 was used to ensure that the major target of acetylation was the H3 tail of H3gH4. Initial-velocity kinetics were measured for the tail-swap mononucleosome (Fig. 4D). Fitting of the data to a cooperative saturation model resulted in a cooperativity constant of 1.02 ± 0.07, i.e., noncooperative binding. Thus, cooperativity of nucleosome acetylation not only requires 2 tails, but also requires that they be located in their normal spatial context.
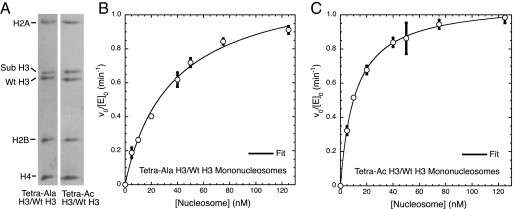
Because substrate cooperativity is lost with elimination of a single tail, we sought to define the factors necessary for cooperativity at the amino acid level. Further, although experiments with both single-tail and tail-swapped nucleosomes show loss of substrate cooperativity, both substrates employ H3 histone truncated at the amino terminus. The absence of these residues can affect the properties of nucleosomes because mononucleosomes lacking the H3 tail demonstrate enhanced octamer mobility, whereas full-length H3 histones and H3 histones with smaller tail truncations do not (24). To address both of these issues, we desired nucleosomes containing octamers with 1 copy each of a full-length wild-type H3 tail and a full-length H3 tail that cannot be acetylated. Tetra-Ala H3 histone, containing a His6 amino-terminal tag and K9A, K14A, K18A, K23A point mutations to prevent SAGA-mediated acetylation (Fig. 2C), was generated in high yield by native chemical ligation [supporting information (SI) Fig. S1] (25). Asymmetric octamers containing equal amounts of wild-type H3 and tetra-Ala H3 were produced in a manner similar to the single-tail octamers (Fig. 5A). SAGA-dependent initial velocities were determined with mononucleosomes containing the tetra-Ala H3/WT H3 octamers (Fig. 5B). Fitting of these data indicates that cooperativity of SAGA acetylation has been lost (cooperativity constant 0.95 ± 0.03). This both reinforces the single-tail and tail-swapped nucleosome results and suggests that the lysine residues of both tails must be present to achieve substrate cooperativity.
Fig. 5.
Loss of free lysines on a single H3 histone tail result in loss of cooperativity of substrate acetylation. (A) Characterization of tetra-Ala H3/WT H3 and tetra-Ac H3/WT octamers. Histones were resolved on a TAU gel and stained with Coomassie blue. (B) Initial velocity per enzyme of mononucleosomes containing tetra-Ala H3/WT H3 octamers as a function of nucleosome concentration. Data were fit to a cooperative saturation model to give a cooperativity constant of 0.95 ± 0.03. (C) Initial velocity per enzyme of mononucleosomes containing tetra-Ac H3/WT H3 octamers as a function of nucleosome concentration. Data were fit to a cooperative saturation model to give a cooperativity constant of 0.99 ± 0.10.
To determine whether this requirement is the result of direct recognition of the lysine side-chain ε-amines or because of recognition of the ultimate acetylated product, asymmetric mononucleosomes preacetylated on a single tail at lysine-9, -14, -18, and -23 were prepared by using the same strategy as above (Fig. 5A and Fig. S1). The loss of substrate cooperativity exhibited by these preacetylated nucleosomes (cooperativity constant 0.99 ± 0.10; Fig. 5C) indicates that acetylation of the target sites of the SAGA complex in 1 tail of a nucleosome complex disrupts cooperative acetylation and suggests that cooperativity is mediated by the recognition of free lysine side chains in the H3 histone tail.
Discussion
In enzyme kinetics, substrate cooperativity can occur when the initial activity of an enzyme toward a substrate, be it binding or turnover, activates the system toward further reaction with substrate. This work shows that, in the case of SAGA-mediated histone acetylation, this activation requires that within a nucleosome both H3 tails exist in their proper orientation and that the target sites of SAGA acetylation are present and unacetylated. To understand this behavior mechanistically, several models were investigated.
Initially we considered cases where binding of a single unmodified or acetylated H3 tail facilitates acetylation of the second tail in the same nucleosomes. However, neither explicit rate equation solutions nor kinetic simulations based on our experimental results generated robust cooperativity (Figs. S2–S4). Instead, they conformed to Michaelis–Menten kinetics, where initial H3 tail binding could increase the overall apparent affinity for substrate. One reason that such a situation does not lead to cooperativity is because, although binding of the first H3 tail is directly related to the overall substrate concentration, increased binding of the second H3 tail is caused by an increase of effective concentration of the substrate and is not improved by increasing the actual substrate concentration. It is important to note that even though this model does not generate cooperative behavior, it could still be an important aspect of the overall kinetic mechanism.
An alternative kinetic model is that intramolecular binding of a nucleosome via both H3 tails facilitates intermolecular binding of a second nucleosome. In our experiments, this second nucleosome would be either another mononucleosome or a nucleosome located in a different nucleosomal array. Explicit rate equation solutions and kinetic simulations confirm that such models do readily generate substrate cooperativity consistent with our experimental results (Figs. S5 and S6). In our current working model (Fig. S5D), the binding of both unacetylated H3 tails within a nucleosome induces an allosteric change in the SAGA complex, facilitating intermolecular binding of another nucleosome and maximal acetylation activity. In this model, intramolecular binding of a second nucleosome within the same array could also occur. However, because this pathway is noncooperative it cannot occur exclusively, otherwise no positive cooperativity would be observed.
In the SAGA complex numerous protein domains are potential candidates for mediating binding to nucleosomes. Domains known to bind unmodified H3 tails include the active site of Gcn5 and the SANT domain of Ada2 (9). Furthermore, although binding of H3 tails is essential for substrate cooperativity, interactions with the whole-histone protein or other aspects of nucleosome structure, such as nucleosomal DNA binding by the SWIRM domain (26), could be required. Binding of multiple nucleosomes is consistent with either a monomeric or multimeric SAGA complex (Fig. S5 A and B). Although isolated SAGA complex is monomeric (17), the complex might oligomerize in its biological context. Additionally, binding of the second nucleosome may only require interactions with a portion of it.
In vitro, nanomolar concentrations of nucleosomes are necessary to reveal kinetic cooperativity, whereas in vivo, average nucleosome concentrations are substantially higher. Within the nucleus, such modulation of presaturation binding of SAGA to nucleosomes may still be significant because nucleosome affinity could be significantly reduced by factors such as chromatin compaction and chromatin-associated proteins and because the effective concentration of nucleosomes could be reduced if cellular nucleosome are not freely mobile. More importantly, our kinetic analysis reveals a mechanistic aspect of SAGA activity that, although only observable at nanomolar concentrations, would occur over all concentrations. Specifically, it shows that the SAGA complex binds, and potentially acetylates, multiple noncontiguous nucleosomes. In our experiments, these binding interactions occurred intermolecularly. In the cell nuclei, this could occur either between different chromosomes or between nucleosomes that are separated by large distances in sequence, but that are organized to be close in 3-dimensional space. Such spatial colocalization of noncontiguous genes has been proposed as an important mechanism of coregulation of gene expression. A number of potential examples exist, including “transcriptional factories” of multiple copies of RNA polymerase II and associated transcriptional machinery (16) and localization of coregulated genes to nuclear pore complexes (27).
One of the best-characterized examples of spatial coregulation comes from chromatin that interacts with proteins associated with nuclear architecture. Kohwi-Shigematsu and coworkers (28) have shown that during thymocyte differentiation, the DNA-binding protein SATB1 is both necessary for organizing multiple cytokine genes into loop structures, where these gene promoters become colocalized to the hub of these loops, and is required for efficient coexpression of these genes (28). Further, SATB1 is essential for robust acetylation of coregulated promoters because loss of SATB1 reduces histone H3 K9 and K14 acetylation, presumably due in part to loss of chromosomal loop structure (29). These observations are consistent with the idea that SAGA cooperativity could promote coregulation of colocalized genes. Recruiting a coactivator complex like SAGA to the promoter of 1 of the coregulated genes would not only increase the effective enzyme concentration to promote acetylation of nucleosomes in the other spatially proximal coregulated promoters, but the dual H3–tail interaction at nucleosomes in the original promoter could effectively couple its acetylation to its functional interaction with other coregulated promoters. This high degree of cooperativity would not only help to couple the activation of coregulated genes, but might also help to protect these promoters from global, and potentially noncooperative, histone deacetylation.
The cooperativity of SAGA-mediated nucleosome acetylation demonstrates the functional utilization of the same pair of histone tails. It seems likely that other enzyme complexes that interact with nucleosomes might exploit such homotypic cross-tail interactions within the nucleosome. For example, the catalytic core of the SAGA complex, Gcn5, Ada2, and Ada3, is shared with a number of other histone acetyltransferase complexes in yeast as well as in higher eukaryotes (2). Additionally, many other chromatin-remodeling complexes contain multiple potential histone tail interaction domains, such as the SANT (30), chromo (31), bromo (32), and PHD finger domains (33). Further studies using tools such as asymmetric nucleosomes should help to address to what extent other systems use cross-talk between identical pairs of histone tails and how such cross-talk ultimately influences biological function.
Materials and Methods
DNA Template and Full-Length Recombinant Histone Preparation.
These reagents were prepared by using standard techniques. Details are provided in the SI Text.
Tetra-Ala and Tetra-Ac H3 Preparation.
Side-chain-protected, resin-bound peptides, HHHHHHARTKQTARASTGGAAPRAQLATAA and HHHHHHARTKQTARK(Ac)STGG K(Ac)APR K(Ac)QLAT K(Ac)A, were synthesized by Fmoc-based solid-phase peptide synthesis on Cl-Trityl support (Baylor Protein Chemistry Core Laboratory). Generation of fully deprotected thioester peptide was performed as described with modifications to the thioesterification step (25). Specifically, the peptide carboxyl terminus (5 mM) was activated with N,N′-dicyclohexylcarbodiimide (100 mM) and then reacted with benzyl mercaptan (100 mM) at 25 °C for 3 h, all in DMSO.
Peptide ligation was performed as described in ref. 25. To remove unligated core histone, the crude reaction mixture was dissolved into 70% acetonitrile, diluted 5-fold into buffer B [100 mM NaH2PO4, 10 mM Tris, 8 M urea, 5 mM DTT (pH 8.0)], incubated with Ni–nitriloacetate (NTA) resin (Qiagen) for 45 min at room temperature. The resin was then washed twice with buffer C (buffer B at pH 6.3) and eluted with buffer D (buffer B at pH 4.5). The eluted fractions were then neutralized by adding 1 M Tris (pH 8.0). Ligated histone H3 was quantified by comparison with a known quantity of Xenopus recombinant His6-tagged H3 histone on an 18% SDS/polyacrylamide gel, stained by Coomassie blue. The identity of the ligated histones was confirmed by MALDI-TOF mass spectrometry.
Histone Octamer Preparation.
Octamers were prepared largely according to standard protocols (21). For asymmetric octamers, 0.02 μmol of His6-tagged H3 was combined with 0.18 μmol of untagged H3, 0.20 μmol of H4, and 0.22 μmol of H2A and H2B. Gel filtration-purified octamers mix was incubated with Ni-NTA resin at 2 M salt for 1 h at 4 °C. The resin was then washed twice with wash buffer [150 mM NaH2PO4, 2 M NaCl, 20 mM imidazole (pH 8.0)] and eluted with elution buffer (wash buffer with 250 mM imidazole). The concentration of octamers was determined by comparison with a known quantity of WT octamers on an 18% SDS/polyacrylamide gel, stained by Coomassie blue.
Nucleosomal Arrays and Mononucleosomes Reconstitutions.
Mononucleosomes and nucleosomal arrays were assembled with DNA template and purified histone octamers according to standard protocol with little modifications (18, 21). The level of saturation of the purified arrays was verified by EcoRI digestion, followed by a 4% native polyacrylamide gel analysis with ethidium bromide staining. Mononucleosomes were analyzed by running and staining the 4% native polyacrylamide gel directly. Mononucleosome and array concentrations were determined by DNA absorbance.
SAGA Complex Purification.
The yeast strain (FY2031) encoding TAP-tagged Spt7 was a generous gift from Fred Winston, Harvard Medical School, Boston, MA. SAGA complex was isolated by tandem affinity purification (17). SAGA composition was confirmed by silver staining. SAGA concentration was determined by comparative Western blotting against known amounts of recombinant Gcn5 by using anti-Gcn5 antibody (sc-9078; Santa Cruz Biotechnology).
Kinetic Assays.
SAGA histone acetyltransferase activity was measured by using a radioactive P81 filter-binding assay (19). Assay conditions were chosen to ensure that measured rates of acetylation were initial rates and that substrate was in excess of enzyme while minimizing data deviation and reagent consumption. Specifically, nucleosomal arrays or mononucleosomes were incubated with acetyl-CoA (1.33 μM [3H]acetyl-CoA and 2.66 μM cold acetyl-CoA, final concentration) in the histone acetyltransferase buffer [final concentration of 50 mM Tris (pH 7.5), 5% (vol/vol) glycerol, 0.125 mM EDTA, 50 mM KCl, 1 mM DTT, 1 mM PMSF, 10 mM sodium butyrate) at 30 °C for 5 min. Reactions were initiated with SAGA (1 nM final). Time points were taken from zero to 1.5 or 3.0 min and quenched by spotting the samples (8 μL) onto P81 phosphocellulose filters. Filters were then washed 3 times in 300 mL of washing solution (4 mM Na2CO3, 46 mM NaHCO3) and then 10 min in 200 mL of acetone. The amount of [3H]acetate incorporated into the substrates was determined by scintillation counting. To convert cpm to moles, a 60% counting efficiency, a 71% postwash nucleosome retention, and a 10 Ci/mmol specific activity were used. Initial rates for different concentrations of substrates were determined by linear fit. Kinetic constants describing initial rates as a function of substrate concentration were obtained either by fitting the data to the Michaelis–Menten equation,
or to a cooperative saturation kinetics model,
Supplementary Material
Acknowledgments.
We thank F. Winston (Harvard Medical School) for the tagged SAGA strain; J. C. Hansen (Colorado State University, Fort Collins, CO) for the H3gH4 plasmid; and G. K. Amarasinghe, H. J. Fromm, M. S. Hargrove, J. J. Hayes, J. K. Hines, R. B. Honzato, and K. M. Johansen for helpful discussion. This work was supported by National Institutes of Health Grant GM79663 (to M.A.S.-K.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.L.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0804530105/DCSupplemental.
References
- 1.Baker SP, Grant PA. The SAGA continues: Expanding the cellular role of a transcriptional coactivator complex. Oncogene. 2007;26:5329–5340. doi: 10.1038/sj.onc.1210603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee KK, Workman JL. Histone acetyltransferase complexes: One size doesn't fit all. Nat Rev Mol Cell Biol. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- 3.Huisinga KL, Pugh BF. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae. Mol Cell. 2004;13:573–585. doi: 10.1016/s1097-2765(04)00087-5. [DOI] [PubMed] [Google Scholar]
- 4.Robert F, et al. Global position and recruitment of HATs and HDACs in the yeast genome. Mol Cell. 2004;16:199–209. doi: 10.1016/j.molcel.2004.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Govind CK, Zhang F, Qiu H, Hofmeyer K, Hinnebusch AG. Gcn5 promotes acetylation, eviction, and methylation of nucleosomes in transcribed coding regions. Mol Cell. 2007;25:31–42. doi: 10.1016/j.molcel.2006.11.020. [DOI] [PubMed] [Google Scholar]
- 6.Grant PA, et al. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: Characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 7.Grant PA, et al. Expanded lysine acetylation specificity of Gcn5 in native complexes. J Biol Chem. 1999;274:5895–5900. doi: 10.1074/jbc.274.9.5895. [DOI] [PubMed] [Google Scholar]
- 8.Rojas JR, et al. Structure of Tetrahymena GCN5 bound to coenzyme A and a histone H3 peptide. Nature. 1999;401:93–98. doi: 10.1038/43487. [DOI] [PubMed] [Google Scholar]
- 9.Boyer LA, et al. Essential role for the SANT domain in the functioning of multiple chromatin remodeling enzymes. Mol Cell. 2002;10:935–942. doi: 10.1016/s1097-2765(02)00634-2. [DOI] [PubMed] [Google Scholar]
- 10.Hassan AH, et al. Selective recognition of acetylated histones by bromodomains in transcriptional coactivators. Biochem J. 2007;402:125–133. doi: 10.1042/BJ20060907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luger K, Mader AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 Å resolution. Nature. 1997;389:251–260. doi: 10.1038/38444. [DOI] [PubMed] [Google Scholar]
- 12.Yuan GC, et al. Genome-scale identification of nucleosome positions in S. cerevisiae. Science. 2005;309:626–630. doi: 10.1126/science.1112178. [DOI] [PubMed] [Google Scholar]
- 13.Bednar J, et al. Nucleosomes, linker DNA, and linker histone form a unique structural motif that directs the higher-order folding and compaction of chromatin. Proc Natl Acad Sci USA. 1998;95:14173–14178. doi: 10.1073/pnas.95.24.14173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schalch T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–141. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 15.Gordon F, Luger K, Hansen JC. The core histone N-terminal tail domains function independently and additively during salt-dependent oligomerization of nucleosomal arrays. J Biol Chem. 2005;280:33701–33706. doi: 10.1074/jbc.M507048200. [DOI] [PubMed] [Google Scholar]
- 16.Fraser P. Transcriptional control thrown for a loop. Curr Opin Genet Dev. 2006;16:490–495. doi: 10.1016/j.gde.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 17.Wu PY, Ruhlmann C, Winston F, Schultz P. Molecular architecture of the S. cerevisiae SAGA complex. Mol Cell. 2004;15:199–208. doi: 10.1016/j.molcel.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 18.Carruthers LM, Tse C, Walker KP, 3rd, Hansen JC. Assembly of defined nucleosomal and chromatin arrays from pure components. Methods Enzymol. 1999;304:19–35. doi: 10.1016/s0076-6879(99)04004-5. [DOI] [PubMed] [Google Scholar]
- 19.Eberharter A, John S, Grant PA, Utley RT, Workman JL. Identification and analysis of yeast nucleosomal histone acetyltransferase complexes. Methods. 1998;15:315–321. doi: 10.1006/meth.1998.0635. [DOI] [PubMed] [Google Scholar]
- 20.Tanner KG, Langer MR, Kim Y, Denu JM. Kinetic mechanism of the histone acetyltransferase GCN5 from yeast. J Biol Chem. 2000;275:22048–22055. doi: 10.1074/jbc.M002893200. [DOI] [PubMed] [Google Scholar]
- 21.Luger K, Rechsteiner TJ, Richmond TJ. Preparation of nucleosome core particle from recombinant histones. Methods Enzymol. 1999;304:3–19. doi: 10.1016/s0076-6879(99)04003-3. [DOI] [PubMed] [Google Scholar]
- 22.Viens A, et al. Analysis of human histone H2AZ deposition in vivo argues against its direct role in epigenetic templating mechanisms. Mol Cell Biol. 2006;26:5325–5335. doi: 10.1128/MCB.00584-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Luger K, Richmond TJ. The histone tails of the nucleosome. Curr Opin Gen Dev. 1998;8:140–146. doi: 10.1016/s0959-437x(98)80134-2. [DOI] [PubMed] [Google Scholar]
- 24.Ferreira H, Somers J, Webster R, Flaus A, Owen-Hughes T. Histone tails and the H3 αN helix regulate nucleosome mobility and stability. Mol Cell Biol. 2007;27:4037–4048. doi: 10.1128/MCB.02229-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shogren-Knaak MA, Peterson CL. Creating designer histones by native chemical ligation. Methods Enzymol. 2004;375:62–76. doi: 10.1016/s0076-6879(03)75004-6. [DOI] [PubMed] [Google Scholar]
- 26.Qian C, et al. Structure and chromosomal DNA binding of the SWIRM domain. Nat Struct Mol Biol. 2005;12:1078–1085. doi: 10.1038/nsmb1022. [DOI] [PubMed] [Google Scholar]
- 27.Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nat Rev Genet. 2007;8:507–517. doi: 10.1038/nrg2122. [DOI] [PubMed] [Google Scholar]
- 28.Cai S, Lee CC, Kohwi-Shigematsu T. SATB1 packages densely looped, transcriptionally active chromatin for coordinated expression of cytokine genes. Nat Genet. 2006;38:1278–1288. doi: 10.1038/ng1913. [DOI] [PubMed] [Google Scholar]
- 29.Cai S, Han HJ, Kohwi-Shigematsu T. Tissue-specific nuclear architecture and gene expression regulated by SATB1. Nat Genet. 2003;34:42–51. doi: 10.1038/ng1146. [DOI] [PubMed] [Google Scholar]
- 30.Boyer LA, Latek RR, Peterson CL. The SANT domain: A unique histone tail-binding module? Nat Rev Mol Cell Biol. 2004;5:158–163. doi: 10.1038/nrm1314. [DOI] [PubMed] [Google Scholar]
- 31.Brehm A, Tufteland KR, Aasland R, Becker PB. The many colours of chromodomains. Bioessays. 2004;26:133–140. doi: 10.1002/bies.10392. [DOI] [PubMed] [Google Scholar]
- 32.Zeng L, Zhou MM. Bromodomain: An acetyllysine-binding domain. FEBS Lett. 2002;513:124–128. doi: 10.1016/s0014-5793(01)03309-9. [DOI] [PubMed] [Google Scholar]
- 33.Bienz M. The PHD finger, a nuclear protein-interaction domain. Trends Biochem Sci. 2006;31:35–40. doi: 10.1016/j.tibs.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 34.Fersht A. Structure and Mechanism in Protein Sci: A Guide to Enzyme Catalysis and Protein Folding. New York: Freeman; 1998. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.