Abstract
This paper shows that for microbial communities, “fences make good neighbors.” Communities of soil microorganisms perform critical functions: controlling climate, enhancing crop production, and remediation of environmental contamination. Microbial communities in the oral cavity and the gut are of high biomedical interest. Understanding and harnessing the function of these communities is difficult: artificial microbial communities in the laboratory become unstable because of “winner-takes-all” competition among species. We constructed a community of three different species of wild-type soil bacteria with syntrophic interactions using a microfluidic device to control spatial structure and chemical communication. We found that defined microscale spatial structure is both necessary and sufficient for the stable coexistence of interacting bacterial species in the synthetic community. A mathematical model describes how spatial structure can balance the competition and positive interactions within the community, even when the rates of production and consumption of nutrients by species are mismatched, by exploiting nonlinearities of these processes. These findings provide experimental and modeling evidence for a class of communities that require microscale spatial structure for stability, and these results predict that controlling spatial structure may enable harnessing the function of natural and synthetic multispecies communities in the laboratory.
Keywords: microbial, microscale, model, stability, microfluidic
Microbial communities perform a wide range of functions, such as nitrogen processing in the soil, decomposition of organic matter in the carbon cycle, and remediation of environmental contamination. These communities and their interactions with the human host are of high biomedical significance. The stability and function of these communities require balancing competition and positive interactions among multiple species (1–5). Although the interactions within several symbiotic communities are well characterized, in general it is not understood how this balance is achieved within most communities of microbes. Under homogeneous laboratory conditions, most attempts to co-culture multiple microbial species do not result in stable communities, in part because of lopsided competition for nutrients among the species. In nature, microbial communities inhabit matrices with intricate spatial structure (6, 7). Many species of soil bacteria coexist as microcolonies separated by a few hundred micrometers (8, 9). This spatial structure has been hypothesized to be important in microbial ecology (10–13). However, on this small scale spatial structure is difficult to control in natural environments. Furthermore, microscale spatial structure has not been controlled and varied experimentally to understand its effect on the stability of bacterial communities.
To test experimentally the role of microscale spatial structure in bacterial communities, we constructed a synthetic community of three species of wild-type bacteria and used a microfluidic device based on previously described devices (14–17) to control spatial structure and chemical communication within this community. Using three soil bacteria, Azotobacter vinelandii (Av), Bacillus licheniformis (Bl), and Paenibacillus curdlanolyticus (Pc), we designed this community to survive under nutrient-limited conditions by reciprocal syntrophy. We refer to the interactions within the community as “reciprocal syntrophy” because each species performs a unique function required for the survival of the entire community (Fig. 1A). Only Av supplies nitrogen sources by fixing gaseous nitrogen into amino acids with a molybdenum-coupled nitrogenase under aerobic conditions (18, 19), only Bl reduces antibiotic pressure by degrading penicillin G with β-lactamases (20), and only Pc provides a carbon energy source, such as glucose, by using cellulases to cleave carboxymethyl-cellulose (21). This community is purely synthetic—we have no evidence that these species interact in nature.
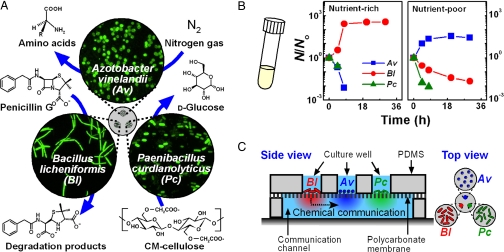
Fig. 1.
A synthetic community of three bacterial species requires spatial structure to maintain stable coexistence. (A) A schematic drawing of the wild-type soil bacteria and their functions used to create a synthetic community with syntrophic interactions. (B) Graphs show the survival ratio of each species (N/No) as a function of time when cultured in well-mixed conditions in a test tube in nutrient-rich TSB/1771 (Left) and nutrient-poor CP (Right) media, indicating instability of the community under spatially unstructured conditions. (C) A schematic drawing of the microfluidic device used to co-culture the three species stably by imposing spatial structure with three culture wells and a communication channel.
Results and Discussion
First, we attempted to co-culture all three species of the community under well-mixed conditions in a test tube in a nutrient-rich or nutrient-poor medium (Fig. 1B). Here, the nutrient-rich medium was a mixture of trypticase soy broth (TSB) and 1771 media [see supporting information (SI) Text: Cultivation of Microorganisms and Culture Media], and the nutrient-poor medium was a cellulose/penicillin medium (CP), which contained the β-lactam antibiotic penicillin G as the antibiotic pressure, carboxymethyl cellulose as the only carbon source, and N2 from the atmosphere as the nitrogen source. We confirmed that Av, Bl, and Pc cannot maintain viability over time when cultured individually in a nutrient-limited medium (Fig. S1A). In this co-culture, we found that the community was unstable regardless of nutrient availability (Fig. 1B). In the nutrient-rich medium, the population size of Bl increased rapidly, and the population sizes of Av and Pc decreased rapidly below the limit of detection. In the CP medium, the population size of Av increased, and the population sizes of Bl and Pc decreased. Control experiments in monoculture demonstrated that Bl grew faster at high concentrations of nutrient-rich medium; in contrast, Av grew faster at very low concentrations of nutrient rich medium, suggesting that Av had the higher affinity for a growth-limiting substrate (Fig. S1B). In addition, we confirmed that neither the presence of heat-killed Bl nor the degradation products of penicillin G had a strong effect on the viability of Av or Pc cells (Fig. S1 C and D). Spent medium from one monoculture did not show toxicity to another monoculture. These results indicate that, although the community has the potential for reciprocal syntrophic interactions, this potential is not realized under well-mixed culture conditions.
It is known that space influences interactions between groups of bacteria (22–26), and that some bacterial communities spontaneously develop spatial structures (27–31). To test whether the synthetic community can be stabilized by imposing a specific spatial structure, we used a microfluidic (14–17, 24, 32) device that localized each bacterial species into an individual culture well separated from a microfluidic communication channel by a nano-porous membrane (Fig. 1C). This device spatially localized each species so that bacteria were unable to migrate from one well to another (Fig. S2 A and B), while allowing chemical communication among the species (Fig. S2 D and E). Control experiments indicated that the bacteria remained confined in a culture well and that chemicals were exchanged via diffusion through the communication channel (Figs. S2 and S3). The device supported growth of all three species when Av, Bl, and Pc were separated into individual culture wells of the same device and were cultured in a nutrient-rich medium (Fig. S4).
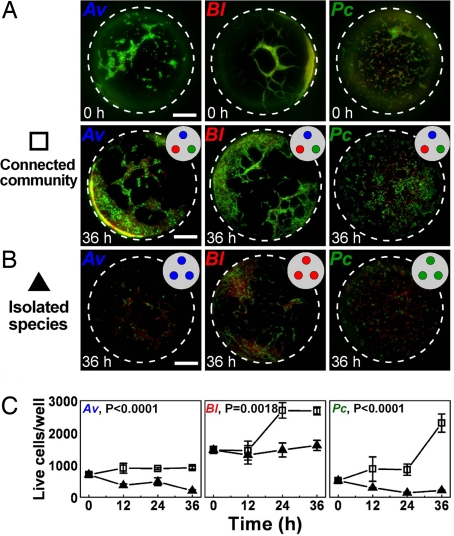
To test whether spatial structure stabilizes syntrophic interactions within this synthetic community in CP media, cells of Av, Bl, and Pc were cultured as either a connected community of all three species (Fig. 2A) or as isolated species (Fig. 2B) in the microfluidic device for 36 h. When all three species were cultured in connected individual wells of the microfluidic device, the community was stable, and each species increased in population size over time (Fig. 2 A and C). In contrast, when each species was cultured alone in the microfluidic device, the isolated species were unstable, and the population size of each species decreased or remained at initial levels (Fig. 2 C and D). Similar results were obtained when only two members of the community were cultured in the microfluidic device (Fig. S5): live-cell numbers in cultures of two-member communities were significantly less than those in cultures of all three community members. Therefore, spatial structure stabilized the community and facilitated syntrophic interactions between community members. It would be interesting to investigate the influence of spatial structure on stability of communities over long time scales. This investigation would require building an open system (24), supplying nutrients and removing waste, and also monitoring genotypic changes in the bacterial population.
Fig. 2.
Stability of the community in the nutrient-poor CP medium requires communication among the three species. (A) Fluorescence images of all three species in the microfluidic device at t = 0 (Top) and at t = 36 h (Bottom). Each species was cultured in an individual culture well of the microfluidic device. (B) Fluorescence images of an isolated species in the microfluidic device at t = 36 h. The same species occupied all three culture wells. Images at t = 0 were similar to those for the three species community at t = 0 (A, Top) and are not shown. Bacteria were stained with a fluorescent dye to indicate live (green) and dead (red) cells. Scale bars represent 50 μm. (C) Graphs comparing the number of live bacteria over time in devices containing all three species, each in an individual well (open squares) and in devices containing a single species in all three wells (closed triangles). Error bars represent standard error with n = 3, except for Av, 0 h (n = 4) and Bl, community, 24 h; Pc, community, 12 h; Pc, community, 36 h; and Pc, isolated species, 36 h (n = 2). P values were calculated by using two-way ANOVA.
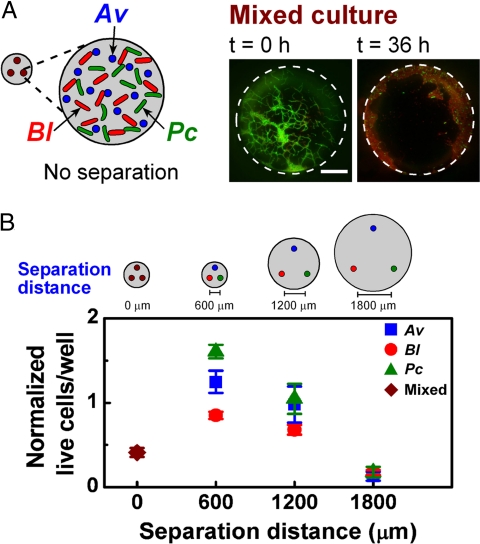
To test the influence of changes in spatial structure on this stability, we varied the distance between the individual, constant-size culture wells of the microfluidic device and proportionally changed the diameter of the communication channel between the wells (Fig. 3). When all wells were inoculated with a mixture of all three species, effectively reducing the separation distance between species to a few micrometers, the community experienced a significant, overall population decline in 36 h (Fig. 3A). We could not always reliably differentiate Pc from Av, but we did not find any cells that resembled Pc in the mixture after 36 h. A similar decline was observed when each species was inoculated individually into a culture well separated from the other wells by 1800 μm (Fig. 3B). Interestingly, the community coexisted stably only at intermediate separation distances on the order of a few hundreds of micrometers (Fig. 3B). These results suggest that a specific spatial structure is required for the stability of the community.
Fig. 3.
Synthetic community coexists only at intermediate separations. (A) A schematic drawing (Left) of a mixed culture of all three species in each well of the microfluidic device and representative fluorescent images (Right) of a culture well containing all three species over time. Bacteria were stained to indicate live (green) and dead (red) cells. Scale bar represents 50 μm. (B) Graph comparing the normalized number of live cells of each species in devices with culture wells separated by four different distances. In the 0-μm separation distance, total numbers of all three species were counted. Error bars represent standard errors.
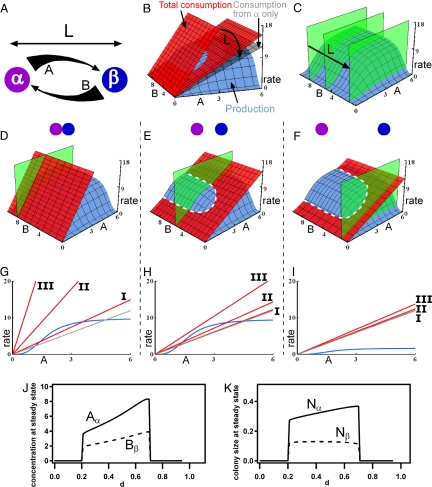
Next, to interpret better the effect of spatial structure, we developed a simple mathematical model describing the role of spatial separation in modulating production, consumption, and diffusion of molecules that regulate the functions of neighboring colonies within a community. To illustrate the model, we use the exchange of essential nutrients between the colonies as an example (e.g., when a colony of species α produces nutrient A, and a colony of species β produces nutrient B). Colonies are separated by distance L (m). The full model, which takes into account both nutrient fluxes and colony growth (Fig. S6 and S7), provides the same overall conclusions as the simpler model below that focuses on nutrient fluxes.
Nonlinearity must be present for spatial effects to be observed (34), and we used a nonlinear production (24, 35–37) function approximated as the product of two Hill functions (Fig. 4, blue plane):
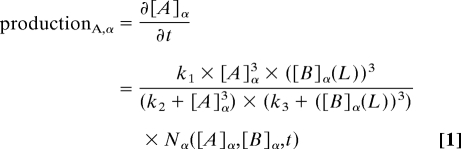 |
where [X] χ (M) is the concentration of the nutrient X at colony χ, Nχ is the number cells in colony χ, and ki are constants (Table S1). Eq. 1 is an example of a nonlinear equation with a two-component threshold for activation that saturates at high concentrations. For our analysis the exact form of the equation is not critical. The same conclusions are obtained from any other set of equations that represent the same general shapes of the curves. In addition to the nonlinearity of the production curve, many other nonlinearities may give rise to spatial effects (38). For simplicity, consumption of nutrients is taken to be linear where the rate of consumption of nutrient A by colony α is defined as
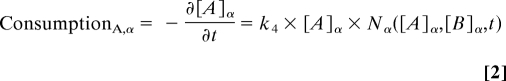 |
and the rate of consumption of nutrient A by colony β is defined as
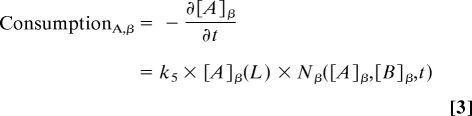 |
The total consumption of A is the sum of Eqs. 2 and 3 (Fig. 4, red plane). The shapes of linear consumption and nonlinear production curves of glucose by Pc were confirmed experimentally (Fig. S8). We simplify production of B by species β to a constant source of nutrient B that diffuses from β to α (in Fig. 4, the green plane represents [B]α). The concentrations [A]β and [B]α are functions of L so [A]β ≈ [A]α and [B]α ≈ [B]β when α and β are close together, and [A]β ≈ [B]α ≈ 0 when they are far apart. These concentration values mimic the profile of a diffusive gradient connecting colonies α and β (Fig. 4A).
Fig. 4.
Mathematical model of a two-species syntrophic community. In all panels, the green plane represents the concentration of nutrient B at species α, [B]α; the blue surfaces and curves represent the production rate of nutrient A; the red plane and lines represent the total rate of consumption of A; and the gray planes and lines represent the rate of consumption of A by species α only. (A) A schematic diagram showing that colony α produces nutrient A and colony β produces nutrient B, establishing gradients over the distance between the colonies L (m). The thickness of the arrows represents the continuous change of concentrations of A and B. (B) 3D rate plot of changes of consumption of A as a function of L. (C) 3D plot of changes in [B]α as a function of L. (D–I) 3D rate plots and 2D sections of the state of the community when colonies of species α and β are separated by small (D and G), intermediate (E and H), and large (F and I) L. The steady-state concentration of nutrient A occurs where the consumption and production curves intersect, shown by the dashed white line in E and F, at the given concentration of nutrient B. (D–F) 3D rate plots from a Class II community predict a non-zero steady state only at intermediate L, indicating that the synthetic community experimentally tested here is Class II. (G–I) 2D sections of representative production and consumption curves for Class I, II, and III communities. (J and K) Steady-state concentrations of A and B and colony sizes of α and β as a function of distance parameter d; see SI Text for details.
The system is stable at a distance L only when the combined consumption rate of a nutrient is matched by the production rate. This criterion is met only when the three surfaces for [B]α, production of A and consumption of A in Fig. 4 cross at a non-zero point. As L is increased, the two effects compete: the consumption of nutrient A by species β decreases (Fig. 4B), and [B]α decreases (Fig. 4C). The spatial dependence observed in Fig. 3 is recapitulated in this model. For small L, [B]α is high, but mutual consumption exceeds production, and the criterion for stability is not satisfied (Fig. 4D). For larger L, both [B]α and the consumption rate decrease (Fig. 4E), and a stable steady state occurs. The nonlinearity (saturability) of the production curve is critical in this model: species α is insensitive to a decrease in [B]α but is sensitive to a decrease in the consumption of nutrient A. No stable steady state is found at very large L (Fig. 4F): the consumption rate of nutrient A by species β becomes insignificant, the total consumption rate approaches the rate of consumption of nutrient A by species α only (Fig. 4, gray plane), and [B]α becomes so low that it becomes limiting and no stable steady state is possible.
This model predicts three classes of microbial communities (Fig. 4 G, H, and I) with obligate interactions. A Class I community is stable in a well-mixed environment (Fig. 4G), as well as at intermediate separations (Fig. 4H), because production rates are sufficiently high to accommodate consumption by all species. However, a Class I community is not stable beyond a maximum separation where metabolic coupling is lost (Fig. 4I). A Class II community is not stable when colonies are well mixed or too close, because consumption exceeds production (Fig. 4 D and G). As the species become spatially separated, interspecific competition is reduced, and the community becomes stable (Fig. 4 E and H) until the maximum separation is reached, and then metabolic coupling is lost (Fig. 4 F and I). Class II communities require spatial structure for stability; the synthetic community described in this paper is Class II. Simulations of the full model that includes colony growth (see SI Text: Full Mathematical Model Including Colony Growth) demonstrated that for a Class II community non-zero steady state values of nutrients and colony sizes are observed only for intermediate separations (Fig. 4 J and K). A Class III community is not stable at any separation distance, because the distance at which the colonies are metabolically decoupled is smaller than the distance at which cross-consumption is sufficiently reduced (Fig. 4 G and I).
This work experimentally shows that examining microscale spatial structure and transport in natural environments may be essential to understand how communities of microbes interact (39) and perform community-level functions in natural ecosystems (40) and how species diversity of microbial communities is maintained on the microscale. Several Class I communities have been characterized and cultured (41), but the number of communities in nature vastly exceeds the number of communities cultured in the laboratory. Metabolic requirements for a Class II community are less restrictive. It remains to be established whether many natural communities are Class II; if so, they would not be culturable by traditional methods but could be cultured by methods that control spatial structure, such as the microfluidic devices (14, 24, 32, 33). It is possible that many unculturable species of microbes may require growth factors that could be provided by cultivation within a Class II community. Class II communities also can be constructed synthetically, as was done here, by choosing strains that do not necessarily coexist in nature but that possess desired complementary functions. Although here a synthetic community of three different species is stabilized by the spatial structure of an equilateral triangle a few hundred micrometers in size, other communities may be stable at different distances and in different geometric arrangements. Mathematical modeling suggests that communities can be stabilized by a range of spatial structures, indicating that this mechanism may be applicable in a variety of natural and laboratory settings. Using spatial structure, rather than matching metabolic and growth rates, also would expand the range of systems amendable to synthetic biology approaches (22, 23). Control of spatial structures in the laboratory may be used to understand better naturally occurring communities that have environmental and biomedical relevance, to harness the functions of natural microbial communities, and to create synthetic communities with new functions.
Materials and Methods
See SI Text for materials, more detailed procedures, and additional data.
Fabrication of Devices.
The microfluidic devices were fabricated by using multilayer soft lithography in polydimethylsiloxane (PDMS) (14–16). The culture wells and the communication channel were separated by a polycarbonate membrane with 0.2-μm pores and were bonded together by using PDMS prepolymer (16). The device used here consisted of two layers—a well layer and a channel layer. The well layer was designed with three culture wells, 200 μm in diameter and 150 μm high, separated by 600 μm. The channel layer was designed with a circular channel 1100 μm in diameter and 150 μm high. For the experiments shown in Fig. 3B, the well layer was designed with wells 200 μm in diameter and 150 μm high separated by 1200 μm or 1800 μm, and the channel layer was designed with a circular channel 2200 μm or 3300 μm in diameter and 150 μm high. Each layer was prepared by pouring 5 ml of PDMS (Sylgard, Dow Corning) prepolymer (10:1, silicone elastomer to the curing agent, Sylgard) onto the patterned wafer. A flat, silanized PDMS support was placed over the prepolymer, and air bubbles were removed. Next a 5-kg weight was placed on the PDMS support, and the setup was cured at 60 °C for 6 h. The well layer with the PDMS support was stamped on a glass slide (75 × 50 × 1 mm, Fisher Scientific), spin-coated with a layer of PDMS prepolymer ∼ 10 μm thick (1:2 elastomer to curing agent; 3,500 rpm for 30 sec in a Laurell Model WS-400A-GNPP/LITE rotor), attached to a nanoporous polycarbonate (PC) membrane (Isopore, 0.2-μm GTBP, Millipore), and incubated in a dry oven (110 °C) for 1 min. After the PDMS support was removed, the channel layer was stamped on a glass slide, spin-coated with PDMS prepolymer, and the channel layer was aligned with the culture wells of the well layer below the PC membrane. The assembled device (Fig. 1C) was cured at 60 °C for 3 h. For sterility, all microfluidic devices were soaked in a 20% ethanol solution under UV exposure overnight.
Cultivation of Microorganisms and Culture Media.
Bacterial strains of Av (ATCC 12837), Bl (ATCC 25972), and Pc (ATCC 51899) at exponential phase were inoculated individually into individual culture wells in the microfluidic device at a density of ∼ 500-1000 live cells/well. The number of live cells loaded into each well varied by ± 10%. The inoculated device was placed over a droplet of appropriate medium on a siliconized glass cover slide, and the medium filled the communication channel below the wells. The device was inverted and incubated at 30 °C. The low-nutrient antibiotic medium (CP medium) contained carboxymethyl cellulose (1 g/L) as a sole carbon source, no nitrogen source, and penicillin G (100 mg/L). The nutrient-rich medium was a mixture of TSB and 1771 media in a 4:1 (vol/vol) ratio. The number of viable cells in macroscale cultures was estimated by agar plate counting of colony-forming units; in contrast, the number of live cells in a microfluidic device was counted manually after the live/dead staining with solutions of SYTO9 (Molecular Probes), which stained live cells green, and propidium iodide, which stained dead cells red.
Data Acquisition and Analysis of Microscopic Images.
Images of bacteria stained with live/dead dye were acquired by using an epi-fluorescence microscope (Leica) with either GFP (L5) or Texas red (TX2) filter sets. After fluorescent images taken by both filters were processed with appropriate background scales, the GFP and Texas red images were overlaid using MetaMorph image software (Molecular Devices). Intensity profiles of all fluorescent images in the main text are provided in Fig. S9.
Mathematical Modeling.
Mathematical modeling was performed using Mathematica software (Mathematica 6.0, Wolfram Research Inc.). The equations for each curve in Fig. 4 are found in the main text with rate constants: k1 = 10, k2 = 5, k3 = 5, k4 = 2, and k5 = 2. The constants k1, k4, and k5 have general units of (time × colony size)1, whereas k2 and k3 have units of (concentration)3. Two-dimensional slices were taken at [B]α = 8 for small colony spacing, [B]α = 5 for intermediate colony spacing, and [B]α = 1 for far colony spacing.
Statistical Analysis.
Statistical analysis was performed by using two-way ANOVA with standard weighted-means analysis, where independent variables were time and community composition. P-values indicate the combined comparison of both variables. All error bars indicate standard errors.
Supplementary Material
Acknowledgments.
We thank Elizabeth W. Boyd and Jessica M. Price for assistance with writing and editing this paper. This work was supported by the National Institutes of Health Director's Pioneer Award 1DP1OD003584 to R.F.I., the Leo P. Kadanoff and Stuart A. Rice Fellowship from the Chicago Materials Research Science and Engineering Center (Chicago MRSEC) to H.J.K., and the Yen Fellowship to J.W.C. Some of this work was performed at the Chicago MRSEC microfluidic facility (funded by the National Science Foundation).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 18075.
This article contains supporting information online at www.pnas.org/cgi/content/full/0807935105/DCSupplemental.
References
- 1.Dechesne A, Or D, Smets BF. Limited diffusive fluxes of substrate facilitate coexistence of two competing bacterial strains. FEMS Microbiol Ecol. 2008;64:1–8. doi: 10.1111/j.1574-6941.2008.00446.x. [DOI] [PubMed] [Google Scholar]
- 2.Kaeberlein T, Lewis K, Epstein SS. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science. 2002;296:1127–1129. doi: 10.1126/science.1070633. [DOI] [PubMed] [Google Scholar]
- 3.Ohno M, et al. Establishing the independent culture of a strictly symbiotic bacterium Symbiobacterium thermophilum from its supporting Bacillus strain. Biosci Biotechnol Biochem. 1999;63:1083–1090. doi: 10.1271/bbb.63.1083. [DOI] [PubMed] [Google Scholar]
- 4.Ferrari BC, Binnerup SJ, Gillings M. Microcolony cultivation on a soil substrate membrane system selects for previously uncultured soil bacteria. Appl Environ Microbiol. 2005;71:8714–8720. doi: 10.1128/AEM.71.12.8714-8720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fernandez AS, et al. Flexible community structure correlates with stable community function in methanogenic bioreactor communities perturbed by glucose. Appl Environ Microbiol. 2000;66:4058–4067. doi: 10.1128/aem.66.9.4058-4067.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dechesne A, et al. A novel method for characterizing the microscale 3D spatial distribution of bacteria in soil. Soil Biology & Biochemistry. 2003;35:1537–1546. [Google Scholar]
- 7.Young IM, Crawford JW. Interactions and self-organization in the soil-microbe complex. Science. 2004;304:1634–1637. doi: 10.1126/science.1097394. [DOI] [PubMed] [Google Scholar]
- 8.Grundmann GL, et al. Spatial modeling of nitrifier microhabitats in soil. Soil Science Society of America Journal. 2001;65:1709–1716. [Google Scholar]
- 9.Nunan N, Wu KJ, Young IM, Crawford JW, Ritz K. Spatial distribution of bacterial communities and their relationships with the micro-architecture of soil. FEMS Microbiol Ecol. 2003;44:203–215. doi: 10.1016/S0168-6496(03)00027-8. [DOI] [PubMed] [Google Scholar]
- 10.Leibold MA, et al. The metacommunity concept: A framework for multi-scale community ecology. Ecol Lett. 2004;7:601–613. [Google Scholar]
- 11.Treves DS, Xia B, Zhou J, Tiedje JM. A two-species test of the hypothesis that spatial isolation influences microbial diversity in soil. Microb Ecol. 2003;45:20–28. doi: 10.1007/s00248-002-1044-x. [DOI] [PubMed] [Google Scholar]
- 12.Tilman D, Kareiva P. Spatial Ecology: The Role of Space and Population Dynamics and Interspecific Interactions. Princeton Univ Press; 1997. [Google Scholar]
- 13.Tilman D. Resource Competition and Community Structure. Princeton Univ Press; 1982. [PubMed] [Google Scholar]
- 14.Abhyankar VV, Beebe DJ. Spatiotemporal micropatterning of cells on arbitrary substrates. Anal Chem. 2007;79:4066–4073. doi: 10.1021/ac062371p. [DOI] [PubMed] [Google Scholar]
- 15.Anderson JR, et al. Fabrication of topologically complex three-dimensional microfluidic systems in PDMS by rapid prototyping. Anal Chem. 2000;72:3158–3164. doi: 10.1021/ac9912294. [DOI] [PubMed] [Google Scholar]
- 16.Chueh BH, et al. Leakage-free bonding of porous membranes into layered microfluidic array systems. Anal Chem. 2007;79:3504–3508. doi: 10.1021/ac062118p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ismagilov RF, Ng JMK, Kenis PJA, Whitesides GM. Microfluidic arrays of fluid-fluid diffusional contacts as detection elements and combinatorial tools. Anal Chem. 2001;73:5207–5213. doi: 10.1021/ac010502a. [DOI] [PubMed] [Google Scholar]
- 18.Gonzalezlopez J, Martineztoledo MV, Rodelas B, Pozo C, Salmeron V. Production of amino-acids by free-living heterotrophic nitrogen-fixing bacteria. Amino Acids. 1995;8:15–21. doi: 10.1007/BF00806540. [DOI] [PubMed] [Google Scholar]
- 19.Post E, Kleiner D, Oelze J. Whole cell respiration and nitrogenase activities in Azotobacter-Vinelandii growing in oxygen controlled continuous culture. Arch Microbiol. 1983;134:68–72. doi: 10.1007/BF00429410. [DOI] [PubMed] [Google Scholar]
- 20.Sargent MG, Ghosh BK, Lampen JO. Characteristics of penicillinase release by washed cells of Bacillus licheniformis. J Bacteriol. 1968;96:1231–1239. doi: 10.1128/jb.96.4.1231-1239.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pason P, Kyu KL, Ratanakhanokchai K. Paenibacillus curdlanolyticus strain B-6 xylanolytic-cellulolytic enzyme system that degrades insoluble polysaccharides. Appl Environ Microbiol. 2006;72:2483–2490. doi: 10.1128/AEM.72.4.2483-2490.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brenner K, Karig DK, Weiss R, Arnold FH. Engineered bidirectional communication mediates a consensus in a microbial biofilm consortium. Proc Natl Acad Sci USA. 2007;104:17300–17304. doi: 10.1073/pnas.0704256104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Basu S, Gerchman Y, Collins C, Arnold F, Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–1134. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- 24.Keymer JE, Galajda P, Muldoon C, Park S, Austin RH. Bacterial metapopulations in nanofabricated landscapes. Proc Natl Acad Sci USA. 2006;103:17290–17295. doi: 10.1073/pnas.0607971103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Codeco CT, Grover JP. Competition along a spatial gradient of resource supply: A microbial experimental model. The American Naturalist. 2001;157:300–315. doi: 10.1086/319195. [DOI] [PubMed] [Google Scholar]
- 26.Allison SD. Cheaters, diffusion and nutrients constrain decomposition by microbial enzymes in spatially structured environments. Ecol Lett. 2005;8:626–635. [Google Scholar]
- 27.Hansen SK, Rainey PB, Haagensen JAJ, Molin S. Evolution of species interactions in a biofilm community. Nature. 2007;445:533–536. doi: 10.1038/nature05514. [DOI] [PubMed] [Google Scholar]
- 28.Hassell MP, Comins HN, May RM. Species coexistence and self-organizing spatial dynamics. Nature. 1994;370:290–292. [Google Scholar]
- 29.Kerr B, Riley MA, Feldman MW, Bohannan BJM. Local dispersal promotes biodiversity in a real-life game of rock-paper-scissors. Nature. 2002;418:171–174. doi: 10.1038/nature00823. [DOI] [PubMed] [Google Scholar]
- 30.Nielsen AT, Tolker-Nielsen T, Barken KB, Molin S. Role of commensal relationships on the spatial structure of a surface-attached microbial consortium. Environmental Microbiology. 2000;2:59–68. doi: 10.1046/j.1462-2920.2000.00084.x. [DOI] [PubMed] [Google Scholar]
- 31.Rainey PB, Travisano M. Adaptive radiation in a heterogeneous environment. Nature. 1998;394:69–72. doi: 10.1038/27900. [DOI] [PubMed] [Google Scholar]
- 32.Ingham CJ, et al. The micro-Petri dish, a million-well growth chip for the culture and high-throughput screening of microorganisms. Proc Natl Acad Sci USA. 2007;104:18217–18222. doi: 10.1073/pnas.0701693104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weibel DB, DiLuzio WR, Whitesides GM. Microfabrication meets microbiology. Nature Reviews. Microbiology. 2007;5:209–218. doi: 10.1038/nrmicro1616. [DOI] [PubMed] [Google Scholar]
- 34.Pompano RR, Li H-W, Ismagilov RF. Rate of mixing controls rate and outcome of autocatalytic processes: Theory and microfluidic experiments with chemical reactions and blood coagulation. Biophys J. 2008;95:1531–1543. doi: 10.1529/biophysj.108.129486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bodini S, Nunziangeli L, Santori F. Influence of amino acids on low-density Escherichia coli responses to nutrient downshifts. J Bacteriol. 2007;189:3099–3105. doi: 10.1128/JB.01753-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bader FG. Analysis of double-substrate limited growth. Biotechnology and Bioengineering. 1978;20:183–202. doi: 10.1002/bit.260200203. [DOI] [PubMed] [Google Scholar]
- 37.Zinn M, Witholt B, Egli T. Dual nutrient limited growth: Models, experimental observations, and applications. J Biotechnol. 2004;113:263–279. doi: 10.1016/j.jbiotec.2004.03.030. [DOI] [PubMed] [Google Scholar]
- 38.Tyson JJ, Chen KC, Novak B. Sniffers, buzzers, toggles and blinkers: Dynamics of regulatory and signaling pathways in the cell. Curr Opin Cell Biol. 2003;15:221–231. doi: 10.1016/s0955-0674(03)00017-6. [DOI] [PubMed] [Google Scholar]
- 39.Price-Whelan A, Dietrich LEP, Newman DK. Rethinking ‘secondary’ metabolism: Physiological roles for phenazine antibiotics. Nature Chemical Biology. 2006;2:71–78. doi: 10.1038/nchembio764. [DOI] [PubMed] [Google Scholar]
- 40.Cosgrove L, McGeechan PL, Robson GD, Handley PS. Fungal communities associated with degradation of polyester polyurethane in soil. Appl Environ Microbiol. 2007;73:5817–5824. doi: 10.1128/AEM.01083-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kato S, Haruta S, Cui ZJ, Ishii M, Igarashi Y. Stable coexistence of five bacterial strains as a cellulose-degrading community. Appl Environ Microbiol. 2005;71:7099–7106. doi: 10.1128/AEM.71.11.7099-7106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.