Abstract
The Gram-negative bacterium Vibrio cholerae is the causative agent of a severe diarrheal disease that afflicts three to five million persons annually, causing up to 200,000 deaths. Nearly all V. cholerae strains produce a large multifunctional-autoprocessing RTX toxin (MARTXVc), which contributes significantly to the pathogenesis of cholera in model systems. The actin cross-linking domain (ACD) of MARTXVc directly catalyzes a covalent cross-linking of monomeric G-actin into oligomeric chains and causes cell rounding, but the nature of the cross-linked bond and the mechanism of the actin cytoskeleton disruption remained elusive. To elucidate the mechanism of ACD action and effect on actin, we identified the covalent cross-link bond between actin protomers using limited proteolysis, X-ray crystallography, and mass spectrometry. We report here that ACD catalyzes the formation of an intermolecular iso-peptide bond between residues E270 and K50 located in the hydrophobic and the DNaseI-binding loops of actin, respectively. Mutagenesis studies confirm that no other residues on actin can be cross-linked by ACD both in vitro and in vivo. This cross-linking locks actin protomers into an orientation different from that of F-actin, resulting in strong inhibition of actin polymerization. This report describes a microbial toxin mechanism acting via iso-peptide bond cross-linking between host proteins and is, to the best of our knowledge, the only known example of a peptide linkage between nonterminal glutamate and lysine side chains.
Keywords: cross-linking, X-ray crystallography, mass spectrometry
As a vital component of every eukaryotic cell, the actin cytoskeleton is a frequent target for microbial toxins. By impairing actin-orchestrated functions via production of specific toxins, pathogenic bacteria manage to escape immune cell surveillance and not only overcome cell barriers but also hijack the actin cytoskeleton for efficient invasion and dissemination. These toxins modulate activities of host proteins either via direct binding to or covalent modifications of their targets. Most of the toxins acting on the actin cytoskeleton shift the equilibrium between polymerized F- and monomeric G-actin by targeting actin adaptor and regulatory proteins such as the Arp2/3 complex and RhoGTPases (1). A lower number of protein toxins affect actin directly rather than through other components of the cytoskeleton. Two types of covalent modifications of actin by two distinct groups of bacterial toxins have been described so far. The first group consists of a family of interrelated toxins with ADP-ribosyltransferase activity (such as C2 toxin of Clostridium botulinum, Iota toxin of C. perfringens, and SpvB of Salmonella enterica), which block actin polymerization by adding an ADP-ribose moiety to Arg-177 of G-actin, thereby resulting in sterical clashes between actin protomers (2, 3).
The second group is represented by MARTXVc [multifunctional-autoprocessing repeats-in-toxins (RTX) toxin of V. cholerae], a toxin produced by nearly all environmental and clinical isolates of V. cholerae, including the current pandemic El Tor O1 and O139 strains (4). This toxin has been associated with V. cholerae virulence in both pulmonary and intestinal infection models and has been demonstrated to contribute to cholera pathogenesis by preventing clearance of the bacterium from the intestine at the earliest stages of infection (5–7). We have shown recently that the actin cross-linking domain (ACD) of MARTXVc directly catalyzes a covalent cross-linking of monomeric G-actin in a Mg2+-ATP dependent manner, causing disassembly of the stress fibers and cell rounding (8–11), but the nature of the cross-linked bond and the mechanism of the actin cytoskeleton disruption remained elusive.
In this study, we found that ACD covalently cross-links actin monomers into arrays of oligomers by forming an iso-peptide bond between highly conserved K50 and E270 residues of adjacent actin molecules. Mutagenesis studies on yeast actin and mammalian cytoplasmic β-actin unequivocally demonstrated that these are the only actin residues cross-linked by ACD both in vitro and in vivo. Actin polymerization assays and electron microscopy imaging showed that the ACD-catalyzed cross-linking inhibits efficiently actin polymerization. At the cellular level this inhibition results in the disassembly of stress fibers and cell rounding.
Results and Discussion
Mapping of the Cross-Linked Sites on Actin by Limited Proteolysis.
To reveal the mechanism of actin cross-linking by ACD, we limited the extent of the cross-linking to yield mainly dimers (10). We then used limited proteolysis of purified ACD cross-linked actin dimers and uncross-linked monomers to identify the actin peptides that are covalently linked. Under well-defined conditions, subtilisin, trypsin, and GluC cleave actin after amino acids 47, 62/68, and 226, respectively (12–14). By analyzing the migration pattern of proteolytic fragments of the cross-linked dimers by SDS/PAGE, we mapped the cross-linking sites to peptides 48–68 on one protomer and 227–375 on another protomer (Fig. 1 A and B).
Fig. 1.
Mapping the cross-linked sites on actin. (A) Actin monomers (m) and ACD cross-linked dimers (d) were subjected to limited proteolysis by GluC, trypsin, and subtilisin. The expected molecular mass of proteolytic peptides and the soybean trypsin inhibitor (in blue) are indicated. (B) Summary of the results from (A) with an indication of cleavage sites, the location of the covalent cross-link bond (yellow bar), and the masses of proteolytic peptides. Red and blue rectangles represent two protomers in the ACD cross-linked actin dimer. (C) Ribbon representation of two crystal structures of the ACD cross-linked actin dimer (AD) in complex with GS1 and with both GS1 and DNaseI (GS1 and DNaseI are not shown) and the recent Holmes model of actin filament (20). Both dimers are oriented in a way that displays better the proximity of the cross-liked peptides. Subdomains 1 to 4 of each protomer are colored blue, yellow, red, and green, respectively. Dotted black line indicates disordered residues 40–49. The cross-linked peptides 48–61 and 257–284, mapped by limited proteolysis and mass spectrometry of the biotin-labeled cross-linked peptide, are shown in black. Taking peptide 48–61 as the only constraint for the localization of one of the cross-linked residues, K50 and E270 were identified as the only two residues within 15-Å radius from each other in both crystal structures as detailed in text. (D) Tryptic peptides of cross-linked actin dimer were analyzed by ESI-FTMS. The 1095.9 (M + 5H)5+ peak in MS spectra, corresponding to the cross-linked peptide, was fragmented in the FT-ICR cell. The most intense of the 104 ions that could be matched to the sequence are annotated in blue (listed in Tables S1 and S2). The masses from resulting high resolution MS/MS spectrum were assigned to peptide fragments by using MS2Assign software. The inset shows the base line separation of the quintuply charged remainder of the precursor. (E) Schematic representation of the cross-linked peptides with the number of ions found for each bond. Of 47 bonds, product ions defining 43 were found. Importantly, 10 ions can be found related to a fragmentation next to the amino acids involved in the cross-linking. This identifies unambiguously the site of cross-linking.
Crystallographic Determination of Residues Involved in ACD Cross-Linking.
Next, we produced crystals and determined the atomic structure of two different complexes of ACD cross-linked actin dimers (AD): one with gelsolin segment 1 (GS1) alone (Fig. 1C; AD-GS1) and another with GS1 together with DNaseI (Fig. 1C; AD-GS1-DNaseI). Unfortunately, the resolution of the diffraction pattern (3.2 Å and 3.9 Å, respectively) was insufficient to identify the cross-link on the basis of a connecting electron density between two protomers. However, it was possible to identify amino acid residues involved in the cross-link by examining the actin residues involved in forming crystal lattice contacts. Using the constraints that one of the cross-linked residues belongs to the peptide 48–68 and assuming that the cross-linked residues should fall within 15 Å of each other, we found that K50 and E270 are the only pair of residues that satisfy these requirements in both crystals. The K50-E270 distance is 8.2 Å and 11.7 Å in AD-GS1 and in AD-GS1-DNaseI complexes, respectively.
Mass Spectrometric Identification of ACD Cross-Linked Residues.
To facilitate mass spectrometry identification of the cross-linked actin residues, we modified actin so that the cross-linked polypeptides could be isolated easily after limited proteolysis. To this end, we introduced Cys at position 59 of yeast actin (which is cross-linked by ACD as well as mammalians actins), and substituted the reactive C374 with Ala. This made it possible to chemically attach a biotin affinity tag to residue 59, which lies within the 48–68 peptide identified earlier. Using a combination of affinity chromatography, MALDI-TOF MS, and high resolution/high mass accuracy ESI-Fourier transform mass spectrometry (ESI-FTMS), and a mixture of ACD cross-linked oligomers as a target, we detected a biotinylated tryptic peptide with a neutral mass 5,852.75627 Da (Mr) that was unique to cross-linked actin oligomers compared with a parallel sample of uncross-linked actin. Only one theoretically predicted cross-linked product matches this mass to within 1 Da, allowing for a loss of water (18.01056 Da) during cross-linking and addition of a biotin molecule with a hydrolyzed maleimide ring (543.23628). In agreement with limited proteolysis and crystallography data, this product encompasses peptide 40–61 linked to peptide 257–284 (0.0058 Da or 1 ppm error; both peptides are shown in black on Fig. 1C).
Similar to cross-linking of yeast actin, a peak specific for the cross-linked rabbit skeletal dimeric actin was detected at 5,475.5 Da. Because the peptide mixture was relatively complex, and the cross-linked peptide ion signal was of low intensity, the peptides were fractionated by strong cation-exchange chromatography. Fractions containing the cross-linked peptide were pooled and analyzed by ESI-FTMS for high resolution MS/MS and MS/MS/MS (MS3) measurements (Fig. 1D). A total of 104 ions matched peptide fragments with high accuracy [see supporting information (SI) Tables S1 and S2] and 43 of 47 peptide bonds were cleaved, achieving high sequence coverage (Fig. 1E). More importantly, 10 product ions determine unambiguously that the peptides are cross-linked at K50-E270. The assignment of the spectrum verifies that E270 loses H2O during cross-linking, but also establishes that the N-terminal C257 was modified by the addition of one carbon (from the mass accuracy of the b-ion series spanning this amino acid, a Cys-to-Asp substitution can be excluded). To determine whether the modification was caused by ACD or a sample processing artifact, digested monomeric skeletal actin was fractionated and analyzed by ESI-quadruple-TOF MS/MS. The MS/MS data showed that the modification of C257 was present also in the ACD-untreated sample, suggesting that it was introduced during sample preparation (data not shown). Addition of a carbon atom to an N-terminal Cys can be explained by conversion of Cys to thioproline during trypsin digestion. The assignment of the cross-link to K50 and E270 is consistent with the results from limited proteolysis and crystallography.
Yeast and Mammalian Actins with Mutations at K50 and/or E270 Are Resistant to Cross-Linking by ACD.
Although mass spectrometry and X-ray crystallography converge on identifying K50 and E270 as the actin residues cross-linked in the ACD-catalyzed reaction, we could not exclude that other residues are also cross-linked. Therefore, to test this possibility, we produced yeast actins with K50C, E270Q, and E270D mutations along with a mutation C374A. In an in vitro cross-linking reaction in the presence of Mg2+-ATP, ACD failed to cross-link K50C/C374A and E270D/C374A (Fig. 2A), as well as E270Q/C374A (data not shown) mutant actins. In parallel experiments, WT yeast actin (Fig. 2A), as well as C374A, Q59C/C374A, I43C/C374A, M47C/C374A, and S265C/C374A mutant actins (Fig. S1), were cross-linked efficiently. Importantly, ACD cross-linked a mixed population of K50C/C374A and E270D/C374A to yield dimers (Fig. 2A), confirming that (i) the proteins are properly folded and are capable of cross-linking, but lack the residues essential for higher order oligomer formation; and (ii) all actin protomers in the higher order oligomers are cross-linked via the same E270-K50 iso-peptide bond.
Fig. 2.
Point mutations at K50 and E270 residues abolish cross-linking of actin. (A) Purified S. cerevisiae wild-type actin, K50C/C374A, E270D/C374A mutant actins (10 μM), and the mixture of two mutants (5 μM each) were cross-linked at 1:250 mole ratio of ACD to actin in the presence of 1 mM MgCl2 and 15 μM Kabiramide C (to prevent polymerization) for the time spans indicated at the bottom of the SDS/PAGE gel. Numbers on the left side of the gel indicate the number of actin molecules corresponding to a given protein band. (B) HeLa cells were transiently transfected to express wild-type and mutant Myc-tagged β-actin as indicated at top. All cells were treated with rtxA+ V. cholerae KFV119 for 90 min before cells were collected for western blotting using α-myc antibody.
To demonstrate that the MARTXVc holotoxin cross-links the same residues in vivo, we transiently transfected HeLa cells with a plasmid encoding Myc-tagged human β-actin. Upon addition of rtxA+ V. cholerae, Myc-actin was cross-linked as efficiently as native actin (data not shown). By contrast, K50A and E270A, as well as K50R, E270D, and E270Q, Myc-actin mutants were poorly incorporated into higher order oligomers, most likely because of the premature termination of the MARTXVc-catalyzed oligomerization of native wild-type actin. Yet, mutants with changes at other residues in the hydrophobic and DNaseI loops, E276A and Q41N, respectively, were cross-linked as efficiently as wild-type Myc-actin. A double Myc-actin mutant with both K50A and E270A mutations was not incorporated into native actin oligomers (Fig. 2B). Together, our results demonstrate unambiguously that ACD of MARTXVc catalyzes a unique reaction: the formation of an intermolecular iso-peptide bond between γ-carboxyl group of glutamic acid 270 and ε-amino group of lysine 50 on actin both in vitro and in vivo.
To the best of our knowledge, ACD-catalyzed cross-linking is the only example of iso-peptide bond formation between a carboxyl group of a nonterminal residue (glutamate) of one polypeptide chain and a primary amine group of another nonterminal residue (lysine) on another polypeptide chain. In all other known cases, either a carboxyl group of a C-terminal residue is involved (as in ubiquitination), or an amide group of an internal glutamine residue is involved in a transamidation reaction (as in fibrin cross-linking with factor XIIIa). Although cytoplasmic β-actin has 26 glutamate and 19 lysine residues, some of which are in a proximity of E270 and K50, only the latter two serve as substrates for ACD. This is in contrast to FrpC, a Neisseria meningitidis RTX toxin, which after a cleavage-linked activation, promiscuously attaches its C-terminal aspartate to a choice of lysines in an intramolecular cross-linking reaction (15).
ACD-Induced Cross-Linking Precludes Actin Polymerization into Filaments.
The ACD-cross-linked residues E270 and K50 are located in the hydrophobic loop and the DNaseI-binding loop of actin, respectively. Both loops are distant from the actin binding sites of major G-actin binding proteins, such as thymosin β4 and profilin. This correlates with our recent findings that both proteins do not inhibit actin cross-linking by ACD (10). Both loops are mobile, unstructured, and protrude out of the body of the actin molecule, making them good substrates for enzymes. This does not explain, however, the disrupting effect of cross-linking on the actin cytoskeleton because introduction of cross-links generally stabilizes, rather than destabilizes, polymerized actin (16, 17). In general, the net effect of cross-linking on polymerization depends on whether constraints introduced by a covalent bond allow protomers to adopt F-actin-like orientation. In three different models of F-actin, including the most recent one fitted without the assumption of perfect helical symmetry (18–20), the distances between Cα atoms of K50 and E270 range between 17 to 25 Å (Table S3). When these residues are cross-linked, our data indicate that the actin protomers can adapt orientation similar to, but yet different from, the lateral dimer orientation expected in F-actin (Fig. 1C). This altered orientation of actin monomers is likely to prevent undisturbed polymerization of cross-linked oligomers in the absence of filament stabilizing drugs or proteins.
To test this prediction, we monitored actin polymerization in the presence of ACD. Pyrene fluorescence assay (data not shown), light scattering, sedimentation, and electron microscopy (Fig. 3 A–C) proved independently that ACD-catalyzed cross-linking competes with and blocks actin polymerization. A small fraction of cross-linked dimers, but not higher order oligomers, copelleted with the polymerized, uncross-linked actin (Fig. 3B), indicating that conformational distortions accumulate upon oligomer growth and prevent incorporation of cross-linked oligomers into polymerized actin, that is, their participation in filament elongation. To confirm that the elongation step is impaired for the ACD cross-linked actin oligomers, we used chemically cross-linked oligomers as nuclei seeds to initiate the polymerization. It is well documented that actin oligomers, chemically cross-linked in longitudinal and lateral F-actin-like orientations (by azidonitrophenyl putrescine and N,N′-p-phenylene dimaleimide, respectively), can serve as nuclei for filament assembly and, thereby, accelerate the bulk polymerization of actin [Fig. S2; (21, 22)]. Yet, these nuclei failed to rescue the polymerization of the ACD-cross-linked oligomers, indicating that the latter species do not participate in the elongation process.
Fig. 3.
Inhibition of actin polymerization by the ACD induced cross-linking. (A–C) Polymerization and cross-linking of 10 μM rabbit skeletal actin in the presence or absence of ACD were initiated simultaneously by adding 1.0 mM MgCl2 and 50 mM KCl and monitored by light scattering (A), sedimentation assay (B), and electron microscopy (C). Compared with polymerization of actin in the absence of ACD, the polymerization was strongly inhibited at 1:1000 mole ratio of ACD to actin and it was blocked completely at 1:100 mole ratio to actin. ACD cross-linked actin oligomers form aggregates (C), which do not pellet during ultracentrifugation (B). In all cases, s and p designate supernatant and pellet fractions, respectively. (D–F) The increase in light scattering upon addition of 15 μM phalloidin (red trace) or 10 μM cofilin (blue trace) indicates that the polymerization of ACD cross-linked actin oligomers (at 1:100 mole ratio of ACD to actin) can be rescued by these agents. To estimate the extent of polymerization and the appearance of the filaments, samples from (D) were analyzed by a sedimentation assay (E) and electron microscopy (F).
To summarize, polymerization and cross-linking reactions compete with each other for actin: Cross-linked actin does not polymerize (Figs. 3 A–C and 4), while polymerized actin cannot be cross-linked efficiently by ACD (10).
Fig. 4.
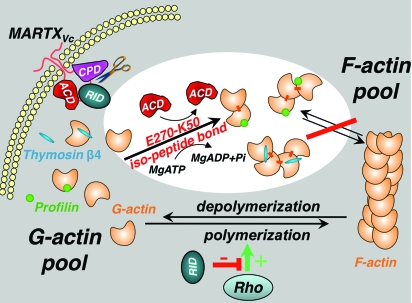
Mechanism of the actin cytoskeleton disruption by MARTXVc. Upon transport through the cytoplasmic membrane of the host cell, the cysteine protease domain (CPD) of MARTXVc cleaves and releases into the cytoplasm functional domains, the Rho-inactivation domain (RID) and the actin cross-linking domain (ACD). RID shifts equilibrium from F- to G-actin by affecting Rho signaling via an unknown mechanism. ACD uses the enriched G-actin pool, maintained by thymosin β4 and profilin, as a substrate for covalent cross-linking dependent on the hydrolysis of ATP. In the resulting oligomers of actin, residue K50 of each actin protomer is connected via an iso-peptide bond with E270 of an adjacent protomer in a conformation incompatible with polymerization. This results in irreversible disruption of the actin cytoskeleton. The white background highlights the mechanism of action elucidated for ACD in the present study.
Phalloidin and Cofilin Independently Rescue the Polymerization of Cross-Linked Oligomers.
The fungal toxin phalloidin and actin-binding protein cofilin can rescue via different mechanisms the polymerization of actins whose self-assembly was impaired by mutations, limited proteolysis, or chemical modifications (23–26). Therefore, we used these two actin-interacting partners to test the rescue of filament assembly for ACD cross-linked actin. Indeed, addition of phalloidin or cofilin partially rescued polymerization of ACD cross-linked actin oligomers as indicated by: (i) increase of light scattering, (ii) presence of actin oligomers in the pellet fractions after ultracentrifugation, and (iii) appearance of short filaments in electron microscopy images (Fig. 3 D–F). Clearly, the observed rescue was incomplete, because the filaments assembled with the ACD cross-linked actin oligomers were short (140 ± 78 nm; n = 200 and 58 ± 27 nm; n = 100 for phalloidin and cofilin stabilized filaments, respectively) and only approximately one-half of the total actin was polymerized in both cases after one hour incubation (Fig. 3E). This rescue could occur because the hydrophobic loop and the DNaseI-binding loop, to which E270 and K50 belong, have sufficient structural flexibility to be cross-linked but yet allow the formation of appropriate lateral contacts. Cofilin has been shown to rescue polymerization of impaired actins (26), perhaps by both the rearrangement of contacts between actin protomers [for recent review see (27)] and a direct “stapling” of protomers by cofilin (26, 28). Also, the ability of cofilin to increase significantly the structural flexibility of the DNaseI-binding loop in F-actin (29) may contribute to the stabilization of actin filaments assembled from ACD cross-linked oligomers. Based on our data, one can speculate that cofilin might have a protective role against ACD-induced actin cytoskeleton disassembly. However, the net in vivo effect of cofilin is hard to predict because of its complex relationships with the cross-linking. Previously, we have shown that cofilin inhibits the cross-linking of G-actin by ACD, but at the same time, it facilitates the cross-linking of F-actin, probably by increasing the rate of treadmilling. In turn, treadmilling provides a pool of G-actin that may be used as a substrate for cross-linking. Therefore, it will be interesting to explore in future which of the cofilin effects are prevalent in living cells.
Despite the unique nature of the toxicity, the ability to cross-link actin appears to be common among V. cholerae isolates and related species. Sequences homologous to ACD of MARTXVc have been identified in several other toxins (11, 30). Two ACDs are also domains within MARTX toxins produced by the aquatic pathogens Aeromonas hydrophilia (31) and V. vulnificus serogroup E isolates (32). One is found within Type 6 secretion effector Vgr-G1 produced by environmental V. cholerae isolates (11, 33). In all cases, evidence is emerging that toxins with actin cross-linking activities are associated with evasion of phagocytosis (7, 32, 33). Three of these ACDs have been shown to cross-link actin in tissue culture cells (11, 33) (and our unpublished observations) and therefore the mechanism of actin cross-linking by MARTXVc presented here might be applicable to all of these toxins. Interestingly, both E270 and K50 residues, which are cross-linked by ACD, are highly conserved throughout the evolution, suggesting that most actins in living organisms are potentially vulnerable to bacterial toxins with MARTXVc ACD-like activities.
In conclusion, ACD-catalyzed cross-linking is an example of a microbial toxin that functions to destabilize assembly of a cellular actin filament structure by introduction of an iso-peptide bond between structural elements of two actin molecules (summarized in Fig. 4). All bacterial toxin enzymes characterized so far belong to one of the following enzymatic groups: proteases, adenylate cyclases, ADP-ribosyltransferases, glucosylating enzymes, deoxyribonucleases, metalloproteases, RNA N-glycosidases, transglutaminases, phospholipases, and deamidases. MARTXVc extends this list. Because MARTXVc consumes ATP as an energy source to produce an iso-peptide bond between protein molecules, according to the IUBMB classification, it has the typical features of ligases, the least numerous of the six major classes of enzymes. The ACD-catalyzed ligation reaction results in actin oligomers in an orientation that, because of spatial constraints, fail to polymerize into filaments unless supported by filament stabilizing factors. At the cellular level, these events result in the disassembly of stress fibers and cell rounding. Investigations are now on the way to determine the details of the actin-ACD interactions, the catalytic mechanism of actin cross-linking by ACD, as well as the precise sequence of events leading to the actin cytoskeleton disruption at the cellular level.
Materials and Methods
Protein Preparation.
Rabbit skeletal actin was purified as described (34). G-actin was maintained in buffer A [5 mM Tris (pH8.0), 0.2 mM ATP, and 0.2 mM CaCl2 buffer with 5 mM β-ME]. Yeast actins were purified as described previously (16), except that 30% sucrose was added to the DNaseI column elution buffer for protein stability. DNaseI was purchased from Bio-World. ACD cross-linked actin dimer was prepared as described previously (8) and used for X-ray crystallography, limited proteolysis, and mass spectrometry experiments. Azidonitrophenyl putrescine and N,N′-p-phenylene dimaleimide cross-linked actin species were produced by standard procedures (22, 35). Gelsolin segment 1 (GS1), and yeast and human cofilin were expressed in E. coli and purified as described earlier (36, 37). Recombinant fusion protein of ACD with the Bacillus anthracis lethal factor (LFNACD) was purified as previously described (8). LFN does not interfere with actin-cross-linking activity of ACD (8).
Actin Cross-Linking with ACD.
ACD is not active in the absence of Mg2+ (8). Therefore, the in vitro cross-linking was initiated by adding 1 mM MgCl2 and 50 mM KCl to a mixture of ACD with either S. cerevisiae actin or skeletal rabbit actin. In vivo cross-linking of actin in V. cholerae-treated, transiently transfected cells was based on previously described procedure (8), as detailed in the SI Methods.
Proteolytic Digestions.
Limited proteolysis of actin is detailed in the SI Methods. Briefly, actin (5 μM in buffer A) was digested with subtilisin and trypsin at an enzyme to protein mass ratio of 1:2,000 and 1:50, respectively, at 25°C. Cleavage of 18 μM actin with GluC was performed at an enzyme/protein mass ratio of 1:50 in buffer A supplemented with 1% SDS as described previously (14).
Light Scattering and Fluorescence Measurements.
Light scattering and fluorescence measurements were performed in a PTI spectrofluorometer (Photon Technology Industries Co). When pyrene-actin (5% of total actin) fluorescence and light scattering were monitored simultaneously, the excitation wavelength was set at 364 nm, with the emission wavelengths at two different channels set at 406 and 364 nm. Light scattering alone was measured with the excitation and emission wavelengths set at 325 nm. Sedimentation and electron microscopy experiments are described in detail in the SI Methods.
Preparation of Samples for Mass Spectrometry.
For mass spectrometry samples of cross-linked and uncross-linked control rabbit actin were digested with trypsin at a protease:protein ratio of 1:20 in 50 mM NH4HCO3 and 30% DMSO (38). The cross-linked peptides were enriched on strong cation exchange microcolumns packed with PolySULFOETHYL Aspartamide (PolyLC, Inc.) stationary phase in P10 pipette tips (39). Eluted peptides, resuspended in 5% formic acid, were loaded onto C18 (20-μm porous R2, Applied Biosystems) microcolumns and eluted directly into electrospray capillaries. The cross-linked peptides were eluted into a GlassTip nanoESI emitter (New Objective) and analyzed with an LTQ-FT Ultra mass spectrometer (Thermo Fischer Scientific). The MS and the MS/MS spectra were measured in the ICR cell with a resolution of 100,000, and peptides were fragmented by using IRMPD within the ICR cell. Three of the product ions were further fragmented (MS3) in the LTQ linear ion trap by CID and detected by the ion multiplier detector. Preparation of biotinylated ACD cross-linked peptides in yeast actin mutant Q59C/C374A are described in the SI Methods and Table S4.
Crystallization.
Crystals of the purified actin dimer (AD) in complex with GS1 alone or GS1 and with DNaseI were grown by a hanging-drop vapor-diffusion method. The reservoir solutions contained 6% PEG 5000 MME, 7% tacsimate, pH 7.0 (Hampton Research), and 0.1 M Hepes, pH 7.0, for actin-GS1 complex, and 1.1 M ammonium sulfate, 0.1 M Bis-Tris, pH 6.5, and 0.06 M sodium chloride for the actin-DNaseI-GS1 complex. The coordinates of the final models and the merged structure factors have been deposited with the Protein Data Bank. The corresponding PDB codes are 3CJB and 3CJC for AD-GS1 and AD-DNaseI-GS1 complexes, respectively. Data collection and structure determination were performed by standard procedures detailed in the SI Methods and Table S5.
Supplementary Material
Acknowledgments.
We thank the staff at the Advanced Light Source beamline 8.2.2 for assistance, especially Corie Ralston for data collection on one of the complexes, and Dr. Gerard Marriott for the gift of Kabiramide C. This work was supported by National Institutes of Health Grants GM077190 (to E.R.), AI051490 (to K.J.S.), and RR020004 (to J.A.L.); National Science Foundation Grant MCB0316269 (to E.R.); an Investigator in the Pathogenesis of Infectious Disease Award from the Burroughs Wellcome Fund (to K.J.S.); Biological and Environmental Research program of the Department of Energy Office of Science (T.O.Y.); the Howard Hughes Medical Institute (M.R.S.); and National Institutes of Health/National Center for Research Resources High-End Instrumentation Program Grant S10 RR023045 (to J.A.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0808082105/DCSupplemental.
References
- 1.Barbieri JT, Riese MJ, Aktories K. Bacterial toxins that modify the actin cytoskeleton. Annu Rev Cell Dev Biol. 2002;18:315–344. doi: 10.1146/annurev.cellbio.18.012502.134748. [DOI] [PubMed] [Google Scholar]
- 2.Wegner A, Aktories K. Adp-ribosylated actin caps the barbed ends of actin-filaments. J Biol Chem. 1988;263:13739–13742. [PubMed] [Google Scholar]
- 3.Margarit SM, Davidson W, Frego L, Stebbins CE. A steric antagonism of actin polymerization by a Salmonella virulence protein. Structure. 2006;14:1219–1229. doi: 10.1016/j.str.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 4.Cordero CL, Sozhamannan S, Satchell KJF. RTX toxin actin cross-linking activity in clinical and environmental isolates of Vibrio cholerae. J Clin Microbiol. 2007;45:2289–2292. doi: 10.1128/JCM.00349-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fullner KJ, et al. The contribution of accessory toxins of Vibrio cholerae O1 El Tor to the proinflammatory response in a murine pulmonary cholera model. J Exp Med. 2002;195:1455–1462. doi: 10.1084/jem.20020318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olivier V, Haines GK, Satchell KJF. Hemolysin and the multifunctional autoprocessing RTX toxin are virulence factors during intestinal infection of mice with Vibrio cholerae El tor O1 strains. Infect Immun. 2007;75:5035–5042. doi: 10.1128/IAI.00506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olivier V, Salzman NH, Satchell KJF. Prolonged colonization of mice by Vibrio cholerae El tor O1 depends on accessory toxins. Infect Immun. 2007;75:5043–5051. doi: 10.1128/IAI.00508-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cordero CL, Kudryashov DS, Reisler E, Satchell KJF. The actin cross-linking domain of the Vibrio cholerae RTX toxin directly catalyzes the covalent cross-linking of actin. J Biol Chem. 2006;281:32366–32374. doi: 10.1074/jbc.M605275200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fullner KJ, Mekalanos JJ. In vivo covalent cross-linking of cellular actin by the Vibrio cholerae RTX toxin. EMBO J. 2000;19:5315–5323. doi: 10.1093/emboj/19.20.5315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kudryashov DS, Cordero CL, Reisler E, Satchell KJF. Characterization of the enzymatic activity of the actin cross-linking domain from the Vibrio cholerae MARTX(Vc) toxin. J Biol Chem. 2008;283:445–452. doi: 10.1074/jbc.M703910200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sheahan KL, Cordero CL, Satchell KJF. Identification of a domain within the multifunctional Vibrio cholerae RTX toxin that covalently cross-links actin. Proc Natl Acad Sci USA. 2004;101:9798–9803. doi: 10.1073/pnas.0401104101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwyter D, Phillips M, Reisler E. Subtilisin-cleaved actin - Polymerization and interaction with myosin subfragment-1. Biochemistry. 1989;28:5889–5895. doi: 10.1021/bi00440a027. [DOI] [PubMed] [Google Scholar]
- 13.Jacobson GR, Rosenbusch JP. Atp binding to a protease-resistant core of actin. Proc Natl Acad Sci USA. 1976;73:2742–2746. doi: 10.1073/pnas.73.8.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roustan C, et al. Conformational-changes induced by Mg-2+ on actin monomers - An immunological attempt to localize the affected region. Febs Lett. 1985;181:119–123. doi: 10.1016/0014-5793(85)81125-x. [DOI] [PubMed] [Google Scholar]
- 15.Osicka R, et al. A novel “clip-and-link” activity of repeat in toxin (RTX) proteins from gram-negative pathogens - Covalent protein cross-linking by an Asp-Lys isopeptide bond upon calcium-dependent processing at an Asp-Pro bond. J Biol Chem. 2004;279:24944–24956. doi: 10.1074/jbc.M314013200. [DOI] [PubMed] [Google Scholar]
- 16.Kim E, Reisler E. Intermolecular dynamics and function in actin filaments. Biophys Chem. 2000;86:191–201. doi: 10.1016/s0301-4622(00)00143-5. [DOI] [PubMed] [Google Scholar]
- 17.Knight P, Offer G. P-Nn'-Phenylenebismaleimide, a specific cross-linking agent for F-actin. Biochem J. 1978;175:1023–1032. doi: 10.1042/bj1751023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cong Y, et al. Crystallographic conformers of actin in a biologically active bundle of filaments. J Mol Biol. 2008;375:331–336. doi: 10.1016/j.jmb.2007.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Holmes KC, Popp D, Gebhard W, Kabsch W. Atomic model of the actin filament. Nature. 1990;347:44–49. doi: 10.1038/347044a0. [DOI] [PubMed] [Google Scholar]
- 20.Holmes KC, Angert I, Kull FJ, Jahn W, Schroder RR. Electron cryo-microscopy shows how strong binding of myosin to actin releases nucleotide. Nature. 2003;425:423–427. doi: 10.1038/nature02005. [DOI] [PubMed] [Google Scholar]
- 21.Kim E, Phillips M, Hegyi G, Muhlrad A, Reisler E. Intrastrand cross-linked actin between Gln-41 and Cys-374. II. Properties of cross-linked oligomers. Biochemistry. 1998;37:17793–17800. doi: 10.1021/bi9812874. [DOI] [PubMed] [Google Scholar]
- 22.Millonig R, Salvo H, Aebi U. Probing actin polymerization by intermolecular cross-linking. J Cell Biol. 1988;106:785–796. doi: 10.1083/jcb.106.3.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khaitlina SY, Moraczewska J, Strzeleckagolaszewska H. The actin actin interactions involving the N-terminus of the Dnase-I-binding loop are crucial for stabilization of the actin filament. Eur J Biochem. 1993;218:911–920. doi: 10.1111/j.1432-1033.1993.tb18447.x. [DOI] [PubMed] [Google Scholar]
- 24.Kuang B, Rubenstein PA. Beryllium fluoride and phalloidin restore polymerizability of a mutant yeast actin (V266G,L267G) with severely decreased hydrophobicity in a subdomain 3/4 loop. J Biol Chem. 1997;272:1237–1247. doi: 10.1074/jbc.272.2.1237. [DOI] [PubMed] [Google Scholar]
- 25.Kudryashov DS, Phillips M, Reisler E. Formation and destabilization of actin filaments with tetramethylrhodamine-modified actin. Biophys J. 2004;87:1136–1145. doi: 10.1529/biophysj.104.042242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kudryashov DS, et al. Cofilin cross-bridges adjacent actin protomers and replaces part of the longitudinal F-actin interface. J Mol Biol. 2006;358:785–797. doi: 10.1016/j.jmb.2006.02.029. [DOI] [PubMed] [Google Scholar]
- 27.Ono S. Mechanism of depolymerization and severing of actin filaments and its significance in cytoskeletal dynamics. Int Rev Cytol. 2007;258:1–82. doi: 10.1016/S0074-7696(07)58001-0. [DOI] [PubMed] [Google Scholar]
- 28.Ono S, et al. The C-terminal tail of UNC-60B (actin depolymerizing factor/cofilin) is critical for maintaining its stable association with F-actin and is implicated in the second actin-binding site. J Biol Chem. 2001;276:5952–5958. doi: 10.1074/jbc.M007563200. [DOI] [PubMed] [Google Scholar]
- 29.Muhlrad A, et al. Cofilin induced conformational changes in F-actin expose subdomain 2 to proteolysis. J Mol Biol. 2004;342:1559–1567. doi: 10.1016/j.jmb.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 30.Seshadri R, et al. Genome sequence of Aeromonas hydrophila ATCC 7966(T): Jack of all trades. J Bacteriol. 2006;188:8272–8282. doi: 10.1128/JB.00621-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Satchell KJF. MARTX, multifunctional autoprocessing repeats-in-toxin toxins. Infect Immun. 2007;75:5079–5084. doi: 10.1128/IAI.00525-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee CT, et al. A common virulence plasmid in biotype 2 Vibrio vulnificus and its dissemination aided by a conjugal plasmid. J Bacteriol. 2008;190:1638–1648. doi: 10.1128/JB.01484-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pukatzki S, Ma AT, Revel AT, Sturtevant D, Mekalanos JJ. Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc Natl Acad Sci USA. 2007;104:15508–15513. doi: 10.1073/pnas.0706532104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Spudich JA, Watt S. The regulation of rabbit skeletal muscle contraction. I. Biochemical studies of the interaction of the tropomyosin-troponin complex with actin and the proteolytic fragments of myosin. J Biol Chem. 1971;246:4866–4871. [PubMed] [Google Scholar]
- 35.Hegyi G, et al. Intrastrand cross-linked actin between Gln-41 and Cys-374. I. Mapping of sites cross-linked in F-actin by N-(4-azido-2-nitrophenyl) putrescine. Biochemistry. 1998;37:17784–17792. doi: 10.1021/bi981285j. [DOI] [PubMed] [Google Scholar]
- 36.Bobkov AA, et al. Structural effects of cofilin on longitudinal contacts in F-actin. J Mol Biol. 2002;323:739–750. doi: 10.1016/s0022-2836(02)01008-2. [DOI] [PubMed] [Google Scholar]
- 37.Goldsmith SC, Guan JQ, Almo S, Chance M. Synchrotron protein footprinting: A technique to investigate protein–protein interactions. J Biomol Struct Dyn. 2001;19:405–418. doi: 10.1080/07391102.2001.10506750. [DOI] [PubMed] [Google Scholar]
- 38.Ytterberg AJ, Peltier JB, Van Wijk KJ. Protein profiling of plastoglobules in chloroplasts and chromoplasts. A surprising site for differential accumulation of metabolic enzymes. Plant Physiol. 2006;140:984–997. doi: 10.1104/pp.105.076083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gobom J, Nordhoff E, Mirgorodskaya E, Ekman R, Roepstorff P. Sample purification and preparation technique based on nano-scale reversed-phase columns for the sensitive analysis of complex peptide mixtures by matrix-assisted laser desorption/ionization mass spectrometry. J Mass Spectrom. 1999;34:105–116. doi: 10.1002/(SICI)1096-9888(199902)34:2<105::AID-JMS768>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.