Abstract
Mass spectrometry (MS) has been revolutionized by electrospray ionization (ESI), which is sufficiently “gentle” to introduce nonvolatile biomolecules such as proteins and nucleic acids (RNA or DNA) into the gas phase without breaking covalent bonds. Although in some cases noncovalent bonding can be maintained sufficiently for ESI/MS characterization of the solution structure of large protein complexes and native enzyme/substrate binding, the new gaseous environment can ultimately cause dramatic structural alterations. The temporal (picoseconds to minutes) evolution of native protein structure during and after transfer into the gas phase, as proposed here based on a variety of studies, can involve side-chain collapse, unfolding, and refolding into new, non-native structures. Control of individual experimental factors allows optimization for specific research objectives.
Keywords: electrospray ionization, gaseous proteins, mass spectrometry, protein conformations, proteomics
Until the late 1980s, it was believed by many researchers in the field of mass spectrometry (MS) that the transfer of large biomolecules into the gas phase, without breaking covalent bonds, was just impossible. Then biomolecular MS was revolutionized by the introduction of the “soft” desorption/ionization methods matrix-assisted laser desorption (MALDI) (1) and electrospray ionization (ESI) (2) that produced intact biomolecular ions. Even more surprising was early evidence that ESI could preserve noncovalent bonds, producing masses corresponding to undissociated protein complexes (3–5). These examples for retention during ESI of noncovalent bonding initiated great enthusiasm about the potential of mass spectrometry for studying biologically relevant systems, but at the same time raised the question of whether or not ESI is generally capable of transferring biomolecules and their noncovalently bound complexes into the gas phase while preserving their solution structure. Ever since, the effect of gaseous environment on noncovalent bonding and higher-order biomolecular structure has been hotly debated. Should the unique structure of a native protein survive the removal of its aqueous environment, with the loss of hydrophobic bonding offset by the strengthening of its electrostatic interactions? In his high-impact 1997 review article, Joseph Loo (6) says about the use of ESI for the study of noncovalent complexes that “there are three camps of opinion: believers, nonbelievers, and undecided. I am a cautious believer.” Today, in 2008, substantial evidence for retention of native structure has been presented for ESI of systems such as large protein (7–9) and nucleic acid (10, 11) complexes. However, globular proteins such as cytochrome c and ubiquitin (12) appear to undergo a temporal evolution of structures after ESI: side-chain collapse, unfolding, and refolding into multiple gaseous structures in 1012 to 102 s. Solution structure and experimental factors can affect each of these steps, indicating ESI optimization possibilities for specific control of the resulting gaseous conformers for a variety of protein studies.
ESI MS has been used to characterize complex protein machinery such as GroEL (≈800 kDa) (13) or to determine relative binding energies of protein/ligand complexes (6, 14). Here, it is imperative that any modifications to the solution structure occurring in the gas phase do not appreciably affect the measurement, e.g., the binding energies in the gas phase must be proportional to those in solution. On the other hand, characterizing the structural transition from solution to gas phase and the stable gaseous conformers is crucial for a fundamental understanding of how chemical environment affects biomolecular structure and stability and for developing efficient strategies for manipulation of gaseous ion structure in sophisticated MS experiments. The combined data from a wide variety of experiments and computational results now allow us to draw a clearer picture of how desolvation affects biomolecular structure and stability and lead us to believe that temporal evolution of structure during and after the phase transition is the key factor for understanding apparently conflicting MS data. Now possibly Loo's question in 1997 can be rephrased: “For how long, under what conditions, and to what extent, can solution structure be retained without solvent?”
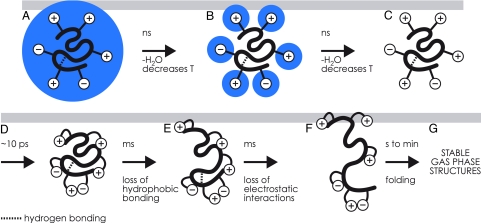
The evidence to be discussed for a stepwise structural evolution is summarized in Fig. 1. It covers the removal of water molecules from the native protein conformation and the period of picoseconds to many minutes in which the stable gaseous structures are formed.
Fig. 1.
Stepwise evolution after ESI of the structure of a globular protein (e.g., cytochrome c, ubiquitin). (A) Native protein covered with a monolayer of H2O, followed by nanosecond H2O loss and concomitant cooling. (B) Native protein with exterior ionic functionalities still hydrated undergoes ns H2O loss and cooling. (C) The dry protein undergoes ≈10-ps collapse of its exterior ionic functionalities. (D–G) The exterior-collapsed “near-native” protein undergoes thermal re-equilibration (D), millisecond loss of hydrophobic bonding (E), and millisecond loss of electrostatic interactions (F); the transiently unfolded ions form new noncovalent bonds in seconds, folding to more stable gaseous ion structures (G); these stabilize to energy minima conformers in minutes.
Final Water Loss from the Protein and Its Charged Side Chains
In the transition from solution to gas phase during ESI MS, biomolecules such as proteins experience vastly different physicochemical environments that can potentially affect their structure. The ESI process starts with the production from solution of electrically charged droplets that undergo a series of successive evaporation and droplet fission events at atmospheric pressure and ambient temperature. Accumulating evidence suggests that ESI of native, globular proteins proceeds via the charge residue model, in which solvent evaporation from very small (nanometer-sized) droplets containing only one protein (or protein complex) eventually results in the formation of “dry” ions (15).
Because of small droplet size, the final desolvation step has yet escaped examination by experiment, but was recently studied with molecular dynamics (MD) simulations for native cytochrome c surrounded by a monolayer of water molecules (16). This study finds no appreciable changes in the overall protein structure during water evaporation on a nanosecond time scale. Moreover, the last water molecules to evaporate aggregate around charged sites that point away from the protein surface in the native structure (Fig. 1 A and B), thereby stabilizing the solution structure until the very last stages of desolvation. In the absence of energy exchange with the environment, solvent evaporation was also found to result in a significant reduction of protein ion temperature and an associated decrease in water evaporation rate. For a cytochrome c ion initially surrounded by 182 water molecules at 110°C, after 200 ps of evaporation it contains 102 H2Os at 80°C, and after 400 ps it contains 81 H2Os at 65°C (16).
In reality, however, the partially hydrated protein ions from ESI are in constant energy exchange with their environment via inelastic collisions with gas molecules and surfaces and via blackbody infrared radiation. Because the energy exchange process may be too slow for full desolvation of the protein (or protein complex) ions before entering the mass spectrometer, most ESI mass spectrometers allow for additional heating in the region interfacing atmospheric pressure and the first vacuum stage (for example, a heated capillary inlet or a counterflow of heated gas). Kebarle has estimated a time scale of hundreds of microseconds for the formation of nanometer-sized droplets from initial ESI droplets of 3 μm diameter, with droplet temperatures ≈10°C lower than that of the ambient gas as a result of evaporative cooling (17), although the resulting ions may have somewhat higher temperatures (18). Solvent evaporation from and fission of larger droplets (20–50 μm in diameter) studied in “ping-pong” experiments was incomplete even after 250 ms (19). However, only parent, not offspring, droplet behavior has so far been monitored in experiments.
Collapse of Charged Side Chains (Picoseconds)
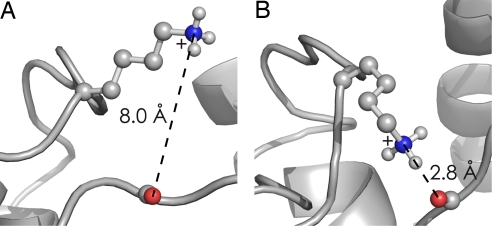
According to new MD calculations on native cytochrome c ions, full desolvation (Fig. 1C) is followed almost instantaneously, on a time scale of a few picoseconds, by specific structural changes on the exterior of the native protein (20). These generally involve charged side chains (Fig. 1D) such as protonated lysine or arginine and deprotonated glutamic or aspartic acid residues. The simulations show that charged side chains form salt links between each other, or ionic hydrogen bonds with backbone heteroatoms (e.g., Fig. 2; N+H3 of Lys-79 to amide oxygen of Tyr-48). The number of electrostatic interactions of the 21 positively and 12 negatively charged side chains in equine cytochrome c increased from 11 in solution to ≈35 after 10 ps, with some of the charged side chains participating in more than one electrostatic interaction. The multiple interactions result in a cross-link-like network of electrostatic interactions on the protein surface, thereby stabilizing the native fold. This surface interaction, along with the ion cooling resulting from solvent evaporation, produces transiently stabilized protein ions; the backbone fold was found to stay essentially the same as that in solution during the 4-ns duration of calculations (20). A common feature of proteins denatured by addition of organic solvent such as methanol is a high α-helix content of their solution structure; collapse of the charged side chains should strengthen the H-bonding of α-helices in the gas phase by interaction of charge with the helix dipole (21–23) (see below).
Fig. 2.
In ESI of native cytochrome c, juxtaposition of the ε-N+H3 of Lys-79 versus the amide oxygen of Tyr-48. (A) Local native structure just after complete H2O loss. (B) Ten picoseconds later, the near-instantaneous collapse forming ionic hydrogen bonds between these adjacent (in structure, but not sequence) sites. [Reproduced with permission from ref. 20 (Copyright 2008, Wiley).]
Loss of Hydrophobic Interactions and Subsequent Dissociation of Electrostatic Bonds (Milliseconds)
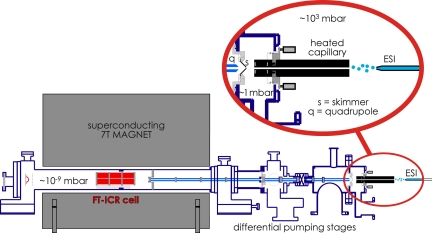
Consistent with the MD results, experimental evidence also indicates that this basic structure (Fig. 1D) can remain essentially unchanged for up to milliseconds. Ion transfer into the MS vacuum yields not only cooling by solvent evaporation and adiabatic expansion but also heating by collisions and surface contact (18, 24). In our ESI system (Fig. 3), an inlet capillary heats the ion beam as well as drops the pressure from atmospheric to ≈1 mbar. The ion residence time in a similar capillary was estimated by Mirza and Chait (25) as ≈103 s. Stepwise unfolding here of cytochrome c is demonstrated by the effect of capillary temperature on fragment ion abundance in native electron capture dissociation (NECD) (26, 27).
Fig. 3.
Electrospray of a protein solution (from right) at atmospheric pressure with ≈1-kV acceleration of ESI droplets into a heated capillary. Exiting ions are accelerated (≈50 V) into a lower pressure (≈1 mbar) region to undergo variable low energy (<1 eV) collisions, pass through a skimmer into a lower pressure region for possible higher energy (>1 eV) collisions, and are conducted into and trapped inside the measurement cell (≈10−9 mbar) of the Fourier-transform mass spectrometer.
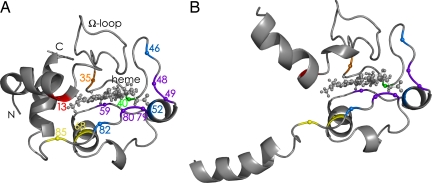
NECD spectra of cytochrome c show fragment ions from protein backbone cleavage next to residues that are in contact with the heme in the native structure, whereas no fragment ions are found for nonheme proteins under the exact same experimental conditions. This unusual phenomenon was observed with ESI of relatively concentrated (>40 μM), aqueous cytochrome c solutions at nondenaturing pH (26), from which it was concluded that noncovalently bound dimers of native protein are required for NECD. Analysis of the fragment ion charge states suggested that proton transfer occurs between the two monomers (26–28), thereupon causing electron transfer via the heme group and backbone cleavage in a mechanism similar to that of electron capture dissociation (ECD) (29). “Asymmetric charge partitioning” was also observed with collisionally activated dissociation (CAD) of cytochrome c homodimers (30). Apparently, one of the monomers in the dimer unfolds faster than the other, causing proton transfer from the still compact monomer (Fig. 1D) to the partially unfolded monomer (Fig. 1E). The lack of NECD cleavage products from the terminal helices and the Ω-loop (Fig. 4) identified the regions of cytochrome c that unfold first (Fig. 4) to start the charge asymmetry process.
Fig. 4.
Cytochrome c structures. (A) Native state, based on NMR data (31). (B) Initial gas-phase unfolding, based on NECD data (27). The order of regional stability in the gas phase, based on the reverse of the order of unfolding determined in NECD experiments, is Y48/T49 = W59 = K79/M80 > F46 = N52 = F82 > T40 > L68 = I85 > L35 > K13.
These regions are generally stabilized by hydrophobic bonding between each other and the heme and are the most stable “fold-ons” in the native solution structure (32, 33). In the gas phase, however, these become the least stable (Fig. 1E). Now the order of disappearance of specific NECD fragment ions with increasing capillary temperature directly indicates the order of further unfolding (Fig. 1F). This further unfolding involves separation of electrostatic protein/heme interactions, which have been strengthened by removal of water that could otherwise compete for (ionic) hydrogen bonding. By loss of hydrophobic bonding and strengthening of electrostatic interactions, removal of water has reversed the order of regional protein stability, making the newly desolvated gaseous ions (Fig. 1 C and D) thermodynamically unstable. This reversal causes disintegration of the native fold, making possible the formation of new intramolecular interactions that are necessary for the formation of stable gas-phase structures.
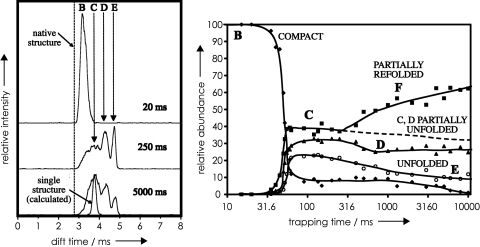
Elegant ion mobility spectrometry (IMS) studies by Clemmer and coworkers (34) also show unfolding of cytochrome c ions in the milliseconds time domain (Fig. 5). The IMS drift time provides collision cross-section values for the corresponding ions. In Fig. 5 Left, the conformer assigned as B of cytochrome c 9+ ions has a drift time nearly as short as that calculated for the native structure. B represents the major conformer present when the ions are stored for up to 30 ms, corresponding to the limited structural evolution of Fig. 1 A to D. Longer storage allows rearrangement into multiple conformers with longer drift times such as unfolded conformer E, consistent with the unfolding of Fig. 1 E and F. The substantially longer times required for conformer changes in these IMS experiments versus those observed with NECD could result from lower IMS ion temperatures and/or collisional activation in the NECD experiment.
Fig. 5.
Ion mobility drift times of 9+ cytochrome c ions stored after ESI at 27°C and 1.3 mbar for various trapping times. Drift times are the smallest for the most compact ions and are directly related to ion collision cross sections. (Left) Drift times characterizing compact conformer B, partially unfolded conformers C and D, and unfolded conformer E. (Right) Relative conformer abundance as a function of trapping time. [Reproduced with permission from ref. 34 (Copyright 2005, Elsevier).]
New Noncovalent Bond Formation (Seconds to Minutes)
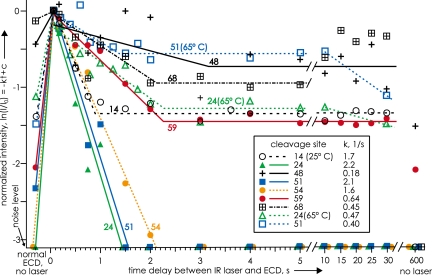
In Fig. 5, however, after ion storage for fractions of a second, the proportion of the most open conformers decreases in favor of more compact forms (Fig. 1 F and G). “Drying” the native conformer has caused hydrophobic and electrostatic bonds to dissociate until the structure is sufficiently open (Fig. 1F) to allow refolding into more stable structures. To study the kinetics of such gaseous ion refolding, 7+ ubiquitin ions were stored and thermally equilibrated in the cell of a Fourier-transform mass spectrometer (21), presumably to form an approximation of the “stable gas-phase structures” of Fig. 1G. These ions were unfolded with a 0.25-s pulse of a CO2 IR laser, and their ECD spectra measured every 0.2 s (Fig. 6). Conformer denaturation, shown by increasing abundance of all ECD fragments, occurred in <0.2 s, after which first-order refolding in the regions around residues 24, 51, and 54 occurred in 1 s. Although refolding in other regions occurred more slowly, and by separate kinetic steps (including the regions around residues 24 and 51 at 65°C), most returned to near initial values in 10 min. However, the regions around residues 51 and 48 underwent refolding into conformers allowing less and no ECD product formation, respectively. Although laser activation did not immediately unfold the residue 68 region, it then folded, unfolded, and dramatically refolded to prevent all ECD fragment ion separation.
Fig. 6.
Relative log intensity values of ECD products from cleavage next to indicated residues of ubiquitin 7+ ions versus time after unfolding by an IR laser pulse (from ref. 21). Apparent first-order rate constants for initial refolding that prevents ECD product separation at the indicated residues are shown in the box. Note that overall refolding can be both single-step and two-step, even folding and unfolding, not necessarily returning to the same degree of folding as before unfolding with the IR laser, and with a temperature increase changing the folding process from one-step to two-step. [Reproduced with permission from ref. 21 (Copyright 2002, American Chemical Society).]
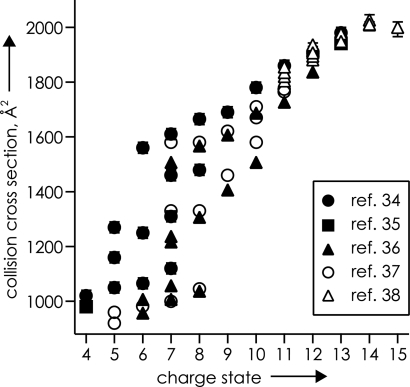
This variety of new conformers was formed by activating and cooling 7+ ubiquitin ions that already had substantial opportunity for equilibration. Thus many conformers must be in local energy minima on the very bottom of the 32 kJ/mol energy well for the overall unfolding of 7+ ubiquitin ions (21). IMS cross-section measurements (34–40) also indicate that a variety of ubiquitin ion conformers are formed by ESI not only for 7+ ions, but also for most charge states from 4+ to 15+ (Fig. 7).
Fig. 7.
Unique collision cross-section values of ubiquitin ions of various charge states, indicating multiple gaseous conformers, from the references indicated. [Reproduced with permission from ref. 12 (Copyright 2006, Wiley).]
Thus with sufficient time and activation, singular native protein conformations passing into the gas phase can undergo side-chain collapse, unfolding, and folding to a variety of 3D structures substantially different from the structure tailored by evolution for a specific purpose in solution. The extent and timing of the events in Fig. 1 are controlled by a variety of experimental factors and the nature of the biomolecules being electrosprayed. Optimization of these factors depends on the objectives of the specific ESI application, as discussed further below.
Structural Features of Multiprotonated Gaseous Biomolecules
Basic knowledge of the effect of solvent removal on the secondary and tertiary structure of proteins, DNA, RNA, and other biomolecules should contribute to the understanding of their biological functions. A variety of methods have provided important data for the characterization of gaseous conformations of proteins (12, 41): H/D exchange labels exposed reactive sites (42–44), IMS (see above) directly indicates their collisional cross section (34–40, 45, 46); ECD (see above) of backbone bonds does not destroy, and thus helps characterize, noncovalent bonding (21, 29, 47); and gas-phase infrared photodissociation (IRPD) spectroscopy measuring O–H/N–H stretching vibrations directly characterizes H-bonding (22, 48). The initial side-chain collapse (Fig. 1 C and D) illustrates the common high tendency found for ionic H-bonding of the protonated residues Lys and Arg to amide oxygen or nitrogen of the gaseous protein backbone. This finding offers an explanation for an apparent dichotomy of early results of two experimental approaches. Increasing the charge of an ion increases its collision cross-section (Fig. 7), with ≈1,000 and ≈2,000 Å2 values (35–39) for the 6+ and 13+ ions, respectively, of ubiquitin (nominally, the 13+ ion is fully protonated, with seven Lys, four Arg, one His, and N-terminal NH2 as protonation sites) (12). In contrast, the extent of gaseous H/D exchange dramatically decreases with increasing charge, with ≈70 D atoms exchanged for 7+ and ≈16 for 13+ ions (22, 43). These amino/imino groups of the side chains undergo ready H/D exchange when neutral, but adding a proton that binds them to the backbone apparently protects most of their H atoms (even four of five guanidinium Hs on Arg) from exchange. The ≈2,000 Å2 cross-section values found for the 13+ ions correspond to that calculated for an α-helical structure (35–39); ECD of the 13+ ubiquitin ions is highly specific for cleavage at the second, third, and fourth residues toward the N terminus from a basic residue, corresponding to collapse of its side-chain N+–H to the nearest amide oxygens in an α-helix (21, 23).
More detailed evidence of this interaction comes from 3,050–3,800 cm1 (O–H/N–H stretching region) IRPD spectra of several electrosprayed protein ions (e.g., mellitin, ubiquitin, cytochrome c, and ribonuclease) that show absorption only in the 3,300–3,400-cm1 region (ref. 22 and unpublished work) that mainly represents amide A bands. The N+-H bonding to the α-helix appears to increase its charge density sufficiently so that other side-chain OH/NH groups are also strongly H-bonded, red-shifting their absorbencies by >300 cm1 to <3,050 cm1. In fact, these H-bonding tendencies appear to be relatively insensitive to total charge, with only minor qualitative differences in the IRPD spectra of 7+ to 13+ ubiquitin ions.
From IMS (34–40, 45) and ECD (21–23) evidence, H+ removal from an α-helical peptide or protein ion allows bending at the charge removal site. From the 13+ ubiquitin ions that apparently have a linear α-helical structure, the 11+ ions have several bent isomers, the 9+ are folded over at one end (stabilized by the opposite dipoles of the overlapping helices), and the 7+, 6+ ions have both ends folded over into a three-helix bundle (22) whose compact ion cross-section closely resembles that calculated for the native structure (37–40). Despite the reduction of total charge, the 7+, 6+ conformer (or mixture of similar conformers) has apparently maintained a sufficiently high charge density to retain the IRPD spectral characteristics of extensive hydrogen bonding. Removal of water molecules that solvate the charge sites in solution not only allows side-chain collapse to the central structure, but by removing charge and thus lowering electrostatic repulsion, the anhydrous secondary structure such as a helix can itself collapse further to a variety of more compact conformers (Fig. 7) in local energy minima (Fig. 1 F and G) that bear little resemblance to the native structure of ubiquitin.
Other techniques for further characterization of such gaseous conformations are obviously needed. Some reviewers (49, 50) have suggested trying direct assays of native protein functions, such as measuring fluorescence of a green fluorescent protein, binding of CO to hemoglobin, or gas-phase enzymology. The transition from solution to gas-phase behavior could even be probed with protein/complex/(H2O)n ions of different n values.
Gaseous Protein Complexes
Electrospray at very low temperatures, into a very cold mass spectrometer, could in principle result in preservation of solution structure on at least a limited time scale. However, freezing the ESI droplets prevents desolvation, which is an endoergic reaction and requires energy input to proceed (24). Joseph Loo (6) pointed out that “there is a very fine line between sufficient desolvation of the gas-phase complex and dissociation of the complex.” Nevertheless, transfer of a native biomolecular structure to a gaseous environment may not immediately result in global conformational changes if ion activation by collisions with background gas and surfaces is outweighed by ion cooling from solvent evaporation and adiabatic expansion (24). As discussed above, a native fold can actually be stabilized, although transiently, by formation of new electrostatic interactions on the protein surface (Fig. 1D). For protein complexes, some of the new electrostatic interactions (Fig. 1D) could form between complex partners and thus stabilize the complex in the gas phase. Depending on experimental conditions and nature of the complex, a time window of ≈103 s can be available for complex detection in the mass spectrometer. Then ESI MS can give information on complex stoichiometry and, after complex dissociation (MS/MS), reveal the identity of binding partners (6–11).
Protein/Ligand Relative Binding Energies
However, “solution binding affinities may or may not be accurate predictors of their stability in vacuo” (50). Also in his 1997 review, Loo (6) raised the important question: “Can a gas-phase measurement reveal solution-phase binding characteristics?” At the time, encouraging and discouraging examples from gas-phase experiments using CAD and blackbody infrared radiative dissociation (BIRD) had been published. Among the discouraging examples were complexes with weak electrostatic binding in solution that become unusually strong in the gas phase, up to a point where covalent bond breaking is favored over complex dissociation. For other systems, it was found that ligand hydrophobicity, although greatly affecting solution binding, did not affect protein complex stability in the gas phase. Loo (6) concluded that “although some features of the solution structure may be preserved by the gas-phase ions, the stability of the gas-phase complex ion may not be reflected by the solution-phase binding constant.” In their 2002 review on the use of MS for quantitative determination of noncovalent binding interactions, Zenobi and coworkers (14) state that “with a few exceptions, no agreement exists between solution-phase and gas-phase binding energies.” Then in 2003, the Klassen group (51) reported evidence from BIRD experiments that protein–carbohydrate complexes from nonspecific association during ESI can actually be more stable in the gas phase than the specific complexes that were transferred intact from solution. It is now generally agreed that the strengthening of electrostatic interactions and the loss of hydrophobic effect in the gaseous environment of a mass spectrometer result in vastly altered strengths of interactions compared with those in solution (41), consistent with the above NECD data showing an essentially reversed order of stability in the gas phase. Moreover, as discussed above, the new electrostatic interactions formed almost immediately after desolvation (Fig. 1D) can result in additional links between protein and ligand and further transfigure binding energies.
Prefolding Dissociation of Large Proteins
As described elsewhere in this issue of PNAS (52), most MS protein analysis (“proteomics”) is done by first digesting (e.g., with trypsin) the protein mixture, subjecting this more complex peptide mixture to MS, and using MS/MS of the separated peptide molecular ions to obtain extensive sequence information. In this “bottom-up” approach, these protein fragment sequences are matched against the possible proteins predicted by their precursor DNA; matching peptide sequences can give up to 95% sequence coverage and 99% identification reliability. The “top-down” approach has far higher identification reliability and capabilities for locating sequence errors and posttranslational modifications (PTMs), although it requires instrumentation such as Fourier-transform MS of far higher resolving power and expense than instruments used previously for bottom-up proteomics (52–54). In this, individual molecular ions of a protein in a mixture are separated by MS-I and dissociated (MS-II) to give fragment ions indicative of that protein's sequence and PTM positions. ECD (29) is especially helpful for MS-II, yielding fragment ions containing either the N or C terminus that often provide complete sequence coverage and detailed PTM positioning. A serious problem is that discussed above with Fig. 1 F and G; the extensive folding taking place in ≥1 s in the Fourier-transform MS ion cell makes protein ions highly stable. Proteins larger than ≈50 kDa, although electrosprayed under denaturing conditions, yield few or no fragment ions by CAD, IRMPD, or other ion dissociation methods. The prefolding dissociation (PFD) approach instead activates the ions <1 s after ESI while they are still in open, more labile conformations (Fig. 1F), which provides useful fragmentation information from proteins as large as 229 kDa (55). Activation to delay the folding and dissociate bonds is accomplished by combinations of capillary heating and ion acceleration in both the ≈1- and 103-mbar regions before and after the skimmer (Fig. 3); the latter collision energies generally effect dissociations of noncovalent and covalent bonds, respectively.
Combinations of these three activation steps in 21 PFD mass spectra gave product ions from 287 different interresidue cleavages of formylglycinimide ribonucleotide amidotransferase (PurL), 1,314 residues, covering 238 and 248 residues of the N and C termini, respectively (37% overall coverage). Although no cleavages were in the 828-residue center of the protein, the fragment ions found provided detailed sequence information (cleavage at 59% of interresidue bonds) in the termini. For example, the predicted N-terminal Met was shown to be absent, correcting the predicted molecular weight value to 143,504 versus the 143,500 ± 23 experimental value. For the human complement C4 glycoprotein (1,714 residues) of three subproteins (longest, 767 residues) joined by S–S bonds, the PFD spectra identified all 27 Cys residues as having –SH or S–S bonds. ESI of mycocerosic acid synthase, 2,153 residues, gave molecular weight = 228,934 ± 60 versus 228,936 predicted after correcting for a missing N-terminal Met. This sequence information was shown in five PFD spectra indicating 62 cleavages up to 134 and 182 residues from the N and C termini, respectively (15% coverage).
Note that the smaller 144-kDa linear protein underwent greater (37%) denaturation in the steps of Fig. 1 D–F under the vigorous CAD conditions (e.g., capillary temperatures up to 345°C) (55).
For these solution-denatured large ions, the further collapse of the ions in Fig. 1D has been suggested to form a “ball of spaghetti” structure (55), for which vigorous activation either extends its free ends or keeps them extended, consistent with the dominant PFD fragmentation in the protein termini. We are now attempting to affect ECD on the exterior of the ball of spaghetti to create new ends; with activation, these could unravel and fragment to provide sequence/PTM characterization of the interior of the protein.
Electrospray Additives
A serendipitous discovery of the PFD investigation was that specific additives to the electrospray protein solution could increase the number of backbone cleavages by 50% (55). The most effective additives are the ammonium salts of saturated dicarboxylic acids such as N+H4−O2CCH2CH2CO2−N+H4 and derivatives. Related unsaturated and aromatic compounds are less effective, whereas similar sulfonates, phosphates, polyhydroxy compounds, and other small molecules are far less effective or inactive. A possible explanation is that in solution the additive forms multiple noncovalent bonds to one or more basic residues, displacing water, so that the solvent monolayer of the structure in Fig. 1B structure will instead consist mainly of the far more strongly bonded additive molecules of far lower volatility, some even of net negative charge. For a denatured protein with negligible tertiary structure, this should not only prevent immediate side-chain collapse (Fig. 1 C and D), but also delay the participation of these charged side chains in electrostatic folding to the very stable gas-phase structures (Fig. 1 F and G) of these large proteins. However, the volatility of the additives is sufficient that none remain in the PFD mass spectra (see above) to confuse interpretation.
Such additives can also be useful for MS studies in which it is desirable to retain the solution conformation in the gas phase. In drug discovery, a common technique for lead compound identification is by ESI characterization of noncovalent ligand–macromolecular target interactions. (6–11, 14) Here, it is necessary during electrospray to preserve these noncovalent interactions, which could be disrupted in the unfolding steps of Fig. 1 D–F. As reported by Klassen and coworkers (56), the solution-stable complex of serine protease trypsin and its competitive binding inhibitor benzamidine does not survive ESI/MS introduction (56). However, ESI solution additives such as imidazole can preserve the complex, possibly because of enhanced cooling from imidazole evaporation that delays the dissociation. In a recent innovative approach to a similar problem, Robinson and colleagues (57, 58) show that detergent micelles can protect membrane protein complexes from dissociating during ESI. The micelles could prevent or retard all of the steps in Fig. 1 C–G that disintegrate the native fold.
Conclusions
Can ESI transfer biomolecules and their noncovalently bound complexes into the gas phase while preserving their solution structure? For several cases, the answer now to Loo's cogent 1997 question (6) is a “yes,” at least to the degree necessary to answer important questions. But the opposite question, “can the solution structure be lost?” may also deserve a yes. In fact, a variety of research approaches now appear to provide a far broader understanding of the changes in protein ion conformation that can be effected by its transfer from solution to the gas phase, as summarized in Fig. 1. The picoseconds side-chain collapse (Fig. 1 C and D) on the protein surface can transiently stabilize the native fold of large, compact protein complexes. Critical conformer stabilization during ESI can also be provided by noncovalent binding of intermolecular additives (56–58). For a native structure with substantial stabilization by hydrophobic bonding in solution, ESI denaturing can form an unstable transient structure (Fig. 1 D–F) that then refolds and equilibrates (e.g., stronger electrostatic bonds) to a more stable gaseous conformer (Fig. 1 F and G). The native solution structure can bear little resemblance to that of this refolded counterpart, with the study of such structures representing an exciting extension of the field of gaseous ion chemistry. For applications needing a protein conformation less stable than either the solution form or the equilibrated gaseous conformers, the most open conformers appear to be formed during entrance into the mass spectrometer, tens of milliseconds after ESI (Fig. 1F), making possible top-down dissociation methods (PFD) effective for >200-kDa proteins (55).
Of basic importance, this stepwise visualization of the effect of solvent loss on protein conformation should not only provide further insight into the role of the aqueous environment on the stability and function of conformation, but also serve as a temporal framework for further experimental studies of these effects using new methodologies.
Acknowledgments.
This work was supported by Austrian Fonds zur Förderung der wissenschaftlichen Forschung Grants V59 and Y372 to K.B. and the U.S. National Institute of General Medical Sciences, National Institutes of Health, Grant GM 16609 to F.W.M.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.Karas M, Bachmann D, Bahr U, Hillenkamp F. Matrix-assisted ultraviolet laser desorption of nonvolatile compounds. Int J Mass Spectrom. 1987;78:53–68. [Google Scholar]
- 2.Fenn JB, Mann M, Meng CK, Wong SF, Whitehouse CM. Electrospray ionization for mass spectrometry of large biomolecules. Science. 1989;246:64–71. doi: 10.1126/science.2675315. [DOI] [PubMed] [Google Scholar]
- 3.Ganem B, Li Y-T, Henion JD. Detection of noncovalent receptor–ligand complexes by mass spectrometry. J Am Chem Soc. 1991;113:6294–6296. [Google Scholar]
- 4.Ganem B, Li Y-T, Henion JD. Observation of noncovalent enzyme–substrate and enzyme–product complexes by ion spray mass spectrometry. J Am Chem Soc. 1991;113:7818–7819. [Google Scholar]
- 5.Katta V, Chait BT. Observation of the heme–globin complex in native myoglobin by electrospray ionization mass spectrometry. J Am Chem Soc. 1991;113:8534–8535. [Google Scholar]
- 6.Loo JA. Studying noncovalent protein complexes by electrospray ionization mass spectrometry. Mass Spectrom Rev. 1997;16:1–23. doi: 10.1002/(SICI)1098-2787(1997)16:1<1::AID-MAS1>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 7.Heck AJR, van den Heuvel RHH. Investigation of intact protein complexes by mass spectrometry. Mass Spectrom Rev. 2004;23:368–389. doi: 10.1002/mas.10081. [DOI] [PubMed] [Google Scholar]
- 8.van den Heuvel RHH, Heck AJR. Native protein mass spectrometry: From intact oligomers to functional machineries. Curr Opin Chem Biol. 2004;8:519–526. doi: 10.1016/j.cbpa.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 9.Benesch JLP, Robinson CVR. Mass spectrometry of macromolecular assemblies: Preservation and dissociation. Curr Opin Struct Biol. 2006;16:245–251. doi: 10.1016/j.sbi.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 10.Hofstadler SA, Griffey RH. Analysis of noncovalent complexes of DNA and RNA by mass spectrometry. Chem Rev. 2001;101:377–390. doi: 10.1021/cr990105o. [DOI] [PubMed] [Google Scholar]
- 11.Hofstadler SA, Sannes-Lowery KA. Applications of ESI-MS in drug discovery: Interrogation of noncovalent complexes. Nat Rev Drug Discovery. 2006;5:585–595. doi: 10.1038/nrd2083. [DOI] [PubMed] [Google Scholar]
- 12.Breuker K. In: Principles of Mass Spectrometry Applied to Biomolecules. Laskin J, Lifshitz C, editors. Hoboken, NJ: Wiley; 2006. pp. 177–212. [Google Scholar]
- 13.van Duijn E, et al. Monitoring macromolecular complexes involved in the chaperonin-assisted protein folding cycle by mass spectrometry. Nat Methods. 2005;23:372–376. doi: 10.1038/nmeth753. [DOI] [PubMed] [Google Scholar]
- 14.Daniel JM, Friess SD, Rajagopalan S, Wendt S, Zenobi R. Quantitative determination of noncovalent binding interactions using soft ionization mass spectrometry. Int J Mass Spectrom. 2002;216:1–27. [Google Scholar]
- 15.Kebarle P, Peschke M. On the mechanisms by which the charged droplets produced by electrospray lead to gas-phase ions. Anal Chim Acta. 2000;406:11–35. [Google Scholar]
- 16.Steinberg MZ, Breuker K, Elber R, Gerber RB. The dynamics of water evaporation from partially solvated cytochrome c in the gas phase. Phys Chem Chem Phys. 2007;9:4690–4697. doi: 10.1039/b705905a. [DOI] [PubMed] [Google Scholar]
- 17.Kebarle P, Tang L. From ions in solution to ions in the gas phase. Anal Chem. 1993;65:972A–986A. [Google Scholar]
- 18.Drahos L, Heeren RMA, Collette C, De Pauw E, Vékey K. Thermal energy distribution observed in electrospray ionization. J Mass Spectrom. 1999;34:1373–1379. doi: 10.1002/(SICI)1096-9888(199912)34:12<1373::AID-JMS907>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 19.Smith JN, Flagan RC, Beauchamp JL. Droplet evaporation and discharge dynamics in electrospray ionization. J Phys Chem A. 2002;106:9957–9967. [Google Scholar]
- 20.Steinberg MZ, Elber R, McLafferty FW, Gerber RB, Breuker K. Early structural evolution of native cytochrome c after solvent removal. ChemBioChem. 2008;9:2417–2423. doi: 10.1002/cbic.200800167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Breuker K, Oh H-B, Horn DM, Cerda BA, McLafferty FW. Detailed unfolding and folding of gaseous ubiquitin ions characterized by electron capture dissociation. J Am Chem Soc. 2002;124:6407–6420. doi: 10.1021/ja012267j. [DOI] [PubMed] [Google Scholar]
- 22.Oh H-B, et al. Secondary and tertiary structures of gaseous protein ions characterized by electron capture dissociation mass spectrometry and photofragment spectroscopy. Proc Natl Acad Sci USA. 2002;99:15863–15868. doi: 10.1073/pnas.212643599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Breuker K, Oh H-B, Lin C, Carpenter BK, McLafferty FW. Nonergodic and conformational control of the electron capture dissociation of protein cations. Proc Natl Acad Sci USA. 2004;101:14011–14016. doi: 10.1073/pnas.0406095101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gabelica V, De Pauw E. Internal energy and fragmentation of ions produced in electrospray sources. Mass Spectrom Rev. 2005;24:566–587. doi: 10.1002/mas.20027. [DOI] [PubMed] [Google Scholar]
- 25.Mirza UA, Chait BT. Do proteins denature during droplet evolution in electrospray ionization? Int J Mass Spectrom Ion Processes. 1997;162:173–181. [Google Scholar]
- 26.Breuker K, McLafferty FW. Native electron capture dissociation for the structural characterization of noncovalent interactions in native cytochrome c. Angew Chem Int Ed. 2003;42:4900–4904. doi: 10.1002/anie.200351705. [DOI] [PubMed] [Google Scholar]
- 27.Breuker K, McLafferty FW. The thermal unfolding of native cytochrome c in the transition from solution to gas phase probed by native electron capture dissociation. Angew Chem Int Ed. 2005;44:4911–4914. doi: 10.1002/anie.200500668. [DOI] [PubMed] [Google Scholar]
- 28.Breuker K. Segmental charge distributions of cytochrome c on transfer into the gas phase. Int J Mass Spectrom. 2006;253:249–255. [Google Scholar]
- 29.Zubarev RA, Kelleher NL, McLafferty FW. Electron capture dissociation of multiply charged protein cations. A nonergodic process. J Am Chem Soc. 1998;120:3265–3266. [Google Scholar]
- 30.Jurchen JC, Williams ER. Origin of asymmetric charge partitioning in the dissociation of gas-phase protein homodimers. J Am Chem Soc. 2003;125:2817–2826. doi: 10.1021/ja0211508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Banci L, et al. Solution structure of oxidized horse heart cytochrome c. Biochemistry. 1997;36:9867–9877. doi: 10.1021/bi970724w. [DOI] [PubMed] [Google Scholar]
- 32.Krishna MMG, Lin Y, Rumbley JN, Englander SW. Cooperative omega loops in cytochrome c: Role in folding and function. J Mol Biol. 2003;331:29–36. doi: 10.1016/s0022-2836(03)00697-1. [DOI] [PubMed] [Google Scholar]
- 33.Maity H, Maity M, Englander SW. How cytochrome c folds, and why: Submolecular foldon units and their stepwise sequential stabilization. J Mol Biol. 2004;343:223–233. doi: 10.1016/j.jmb.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 34.Badman ER, Myung S, Clemmer DE. Evidence for unfolding and refolding of gas-phase cytochrome c ions in a Paul trap. J Am Soc Mass Spectrom. 2005;16:1493–1497. doi: 10.1016/j.jasms.2005.04.013. [DOI] [PubMed] [Google Scholar]
- 35.Valentine SJ, Counterman AE, Clemmer DE. Conformer-dependent proton transfer reactions of ubiquitin ions. J Am Soc Mass Spectrom. 1997;8:954–961. [Google Scholar]
- 36.Hoaglund CS, Valentine SJ, Sporleder CR, Reilly JP, Clemmer DE. Three-dimensional ion mobility/TOFMS analysis of electrosprayed biomolecules. Anal Chem. 1998;70:2236–2242. doi: 10.1021/ac980059c. [DOI] [PubMed] [Google Scholar]
- 37.Li J, Taraszka JA, Counterman AE, Clemmer DE. Influence of solvent composition and capillary temperature on the conformations of electrosprayed ions: Unfolding of compact ubiquitin conformers from pseudonative and denatured solutions. Int J Mass Spectrom. 1999;185/186/187:37–47. [Google Scholar]
- 38.Purves RW, Barnett DA, Ells B, Guevremont R. Investigation of bovine ubiquitin conformers separated by high-field asymmetric waveform ion mobility spectrometry: Cross-section measurements using energy-loss experiments with a triple quadrupole mass spectrometer. J Am Soc Mass Spectrom. 2000;11:738–745. doi: 10.1016/S1044-0305(00)00136-7. [DOI] [PubMed] [Google Scholar]
- 39.Purves RW, Barnett DA, Ells B, Guevremont R. Elongated conformers of charge states +11 to +15 of bovine ubiquitin studied using ESI-FAIMS-MS. J Am Soc Mass Spectrom. 2001;12:894–901. doi: 10.1016/S1044-0305(01)00272-0. [DOI] [PubMed] [Google Scholar]
- 40.Koeniger SL, Merenbloom SI, Clemmer DE. Evidence for many resolvable structures within conformation types of electrosprayed ubiquitin ions. J Phys Chem B. 2006;110:7017–7021. doi: 10.1021/jp056165h. [DOI] [PubMed] [Google Scholar]
- 41.Breuker K. The study of protein–ligand interactions by mass spectrometry: A personal view. Int J Mass Spectrom. 2004;239:33–41. [Google Scholar]
- 42.McLafferty FW, Guan Z, Haupts U, Wood TD, Kelleher NL. Gaseous conformational structures of cytochrome c. J Am Chem Soc. 1998;120:4732–4740. [Google Scholar]
- 43.Freitas MA, Hendrickson CL, Emmett MR, Marshall AG. Gas-phase bovine ubiquitin cation conformations resolved by gas-phase hydrogen/deuterium exchange rate and extent. Int J Mass Spectrom. 1999;185/186/187:565–575. [Google Scholar]
- 44.Robinson EW, Williams ER. Multidimensional separations of ubiquitin conformers in the gas phase: Relating ion cross-sections to H/D exchange measurements. J Am Soc Mass Spectrom. 2005;16:1427–1437. doi: 10.1016/j.jasms.2005.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kohtani M, Jarrold MF. Water molecule adsorption on short alanine peptides: How short is the shortest gas-phase alanine-based helix? J Am Chem Soc. 2004;126:8454–8458. doi: 10.1021/ja049298+. [DOI] [PubMed] [Google Scholar]
- 46.Hoaglund-Hyzer CS, Counterman AE, Clemmer DE. Anhydrous protein ions. Chem Rev. 1999;99:3037–3079. doi: 10.1021/cr980139g. [DOI] [PubMed] [Google Scholar]
- 47.Horn DM, Breuker K, Frank AJ, McLafferty FW. Kinetic intermediates in the folding of gaseous protein ions characterized by electron capture dissociation mass spectrometry. J Am Chem Soc. 2001;123:9792–9799. doi: 10.1021/ja003143u. [DOI] [PubMed] [Google Scholar]
- 48.Oh H-B, et al. Infrared photodissociation spectroscopy of electrosprayed ions in a Fourier-transform mass spectrometer. J Am Chem Soc. 2005;127:4076–4083. doi: 10.1021/ja040136n. [DOI] [PubMed] [Google Scholar]
- 49.He F, Ramirez J, Lebrilla CB. Evidence for enzymatic activity in the absence of solvent in gas-phase complexes of lysozyme and oligosaccharides. Int J Mass Spectrom. 1999;193:103–114. [Google Scholar]
- 50.Yin S, Xie Y, Loo JA. Mass spectrometry of protein–ligand complexes: Enhanced gas-phase stability of ribonuclease-nucleotide complexes. J Am Soc Mass Spectrom. 2008;19:1199–1208. doi: 10.1016/j.jasms.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang W, Kitova EN, Klassen JS. Bioactive recognition sites may not be energetically preferred in protein–carbohydrate complexes in the gas phase. J Am Chem Soc. 2003;125:13630–13631. doi: 10.1021/ja036998q. [DOI] [PubMed] [Google Scholar]
- 52.Mann M, Kelleher NL. Precision proteomics: The case for high resolution and high mass accuracy. Proc Natl Acad Soc USA. 2008;105:18132–18138. doi: 10.1073/pnas.0800788105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McLafferty FW, et al. Top-down mass spectrometry, a powerful complement to the high capabilities of proteolysis proteomics. FEBS J. 2007;274:6256–6268. doi: 10.1111/j.1742-4658.2007.06147.x. [DOI] [PubMed] [Google Scholar]
- 54.Breuker K, Jin M, Han X, Jiang H, McLafferty FW. Top-down identification and characterization of biomolecules by mass spectrometry. J Am Soc Mass Spectrom. 2008;19:1045–1053. doi: 10.1016/j.jasms.2008.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Han X, Jin M, Breuker K, McLafferty FW. Extending top-down mass spectrometry to proteins with masses >200 kDa. Science. 2006;314:109–112. doi: 10.1126/science.1128868. [DOI] [PubMed] [Google Scholar]
- 56.Sun J, Kitova EN, Klassen JS. Method for stabilizing protein–ligand complexes in nanoelectrospray ionization mass spectrometry. Anal Chem. 2007;79:416–425. doi: 10.1021/ac061109d. [DOI] [PubMed] [Google Scholar]
- 57.Barrera NP, Di Bartolo N, Booth PJ, Robinson CV. Micelles protect membrane complexes from solution to vacuum. Science. 2008;321:243–246. doi: 10.1126/science.1159292. [DOI] [PubMed] [Google Scholar]
- 58.Zhou Z, et al. Mass spectrometry reveals modularity and a complete subunit interaction map of the eukaryotic translation factor eIF3. Proc Natl Acad Sci USA. 2008;105:18139–18144. doi: 10.1073/pnas.0801313105. [DOI] [PMC free article] [PubMed] [Google Scholar]