Abstract
The Dax-1 protein is an enigmatic nuclear receptor that lacks an expected DNA binding domain, yet functions as a potent corepressor of nuclear receptors. Here we report the structure of Dax-1 bound to one of its targets, liver receptor homolog 1 (LRH-1). Unexpectedly, Dax-1 binds to LRH-1 using a new module, a repressor helix built from a family conserved sequence motif, PCFXXLP. Mutations in this repressor helix that are linked with human endocrine disorders dissociate the complex and attenuate Dax-1 function. The structure of the Dax-1:LRH-1 complex provides the molecular mechanism for the function of Dax-1 as a potent transcriptional repressor.
Keywords: Dax-1, LRH-1, nuclear receptor, regulation, structure
The orphan nuclear receptor DAX-1/Dax-1 (dosage-sensitive sex-reversal adrenal hypoplasia congenital critical region on the X chromosome gene 1) (1) is well known for its role in human pathophysiology. Duplication of the DAX-1 gene causes phenotypic sex reversal in XY individuals (2), and mutations in DAX-1 are responsible for adrenal hypoplasia congenita, an inherited disorder of adrenal gland development (3). During embryogenesis, Dax-1 functions to direct cell differentiation in testes and adrenal tissues (1). In adult physiology, Dax-1 acts as a global repressor of many nuclear receptors, including SF-1, Nur77, ERRγ, ER, AR, PR, and LRH-1 (4–13). Dax-1 also is indispensable to maintaining the pluripotent state of embryonic stem cells (14, 15).
There is a little information on either the structure or regulatory mechanisms of Dax-1. Dax-1 belongs to a unique family of nuclear receptors (NR0B1) that lack the essential DNA binding domain. Instead, the human Dax-1 N terminus consists of three sequence repeats that include the LXXL/ML motif (“LXXL/ML boxes” 1–3) (16). This unique N-terminal extension is thought to play a role in subcellular distribution and nuclear localization of Dax-1 (17). No homologues for the N-terminal region of Dax-1 are known, but its C-terminal domain is a clear homologue of the nuclear receptor ligand-binding domain (LBD) (1). To date, no hormone for Dax-1 has been identified, and the mechanism of its function as corepressor remains under debate (4, 5, 7–9, 12, 18–20). The elucidation of Dax-1 mechanisms has been frustrated by a lack of high-resolution structural information. Here we report the first structure of Dax-1 bound to its physiological target, nuclear receptor liver receptor homolog 1 (LRH-1; NR5A2).
LRH-1 was first discovered in the liver and intestine, where it regulates genes controlling bile acid synthesis and cholesterol homeostasis (21–24). Recently, LRH-1 was found in human steroidogenic tissues and was shown to activate transcription of genes encoding steroidogenic enzymes (25). In particular, regulation of the CYP19A gene encoding aromatase, which converts androgens to estrogens, gives LRH-1 a pivotal role in estrogen signaling (25–28). Similar to Dax-1, LRH-1 is indispensable to maintaining the pluripotent state of embryonic stem cells (29).
Unlike other nuclear receptors that function as homodimers or heterodimers, LRH-1 binds DNA with high affinity as a monomer (30, 31). In contrast to hormone-controlled nuclear receptors, physiological ligands for LRH-1 have not yet been identified, consistent with the fact that NR5A receptors activate reporter gene transcription in the absence of exogenously added ligands (32). Structural studies of LRH-1 (33–37) have revealed its LBD in the active conformation and suggested phosphatidylinositols as potential candidate hormones for this receptor (34); however, whether these ligands stabilize the LBD or function as regulating hormones remains to be determined. The hinge region preceding the LRH-1 LBD provides additional sites for receptor regulation through posttranslational modification (38).
Recent studies have found that the two targets of our work, LRH-1 and Dax-1, are coexpressed in the ovary, where they regulate production of steroid hormones (39–42). These findings show that Dax-1 is a key physiological regulator of LRH-1 transcriptional activity and LRH-1-mediated steroidogenesis. The present work provides the first structural and functional analysis of a regulatory Dax-1:LRH-1 assembly and suggests a mechanism for Dax-1 function as a potent transcriptional repressor.
Results
Preparation and Characterization of the (Dax-1)2:LRH-1 Heterotrimer.
To evaluate whether Dax-1 can bind to LRH-1 in vitro, we performed the standard GST pull-down assay using bacterially expressed and purified GST-LRH-1 LBD fusion protein and in vitro transcribed and translated 35S-labeled full-length Dax-1. The results of this experiment show that Dax-1 interacted with LRH-1 LBD in the absence of any added hormones or coregulatory proteins. Moreover, under the same conditions, the observed Dax-1-LRH-1 binding exceeded the analogous interactions with nuclear receptor SF-1, another functional target of Dax-1 (supporting information (SI) Fig. S1).
Because multiple regions of Dax-1 have been reported to bind nuclear receptors (4, 5, 7–9, 12), we assessed the binding of five different fragments of Dax-1 to LRH-1 LBD: its N-terminal region (aa 1–208), the LBD (aa 205–472), the LBD with preceding LXXL/ML repeats (aa 138–472 and 70–472), and full-length Dax-1 (aa 1–472). Of these fragments, only the putative Dax-1 LBD produced a stable complex with LRH-1 (Fig. S2). Further biochemical analyses of the purified Dax-1:LRH-1 complex showed that the assembly is a heterotrimer with a Dax-1:LRH-1 ratio of 2:1 (Fig. S3 A and B). Consistent with these data, analytical ultracentrifugation revealed the presence of a single protein species with a molecular mass of ≈90 kDa, which agrees with the calculated molecular mass of the (Dax-1)2:LRH-1 heterotrimer.
We characterized the binding affinity of Dax-1 LBD for LRH-1. Direct binding experiments using surface plasmon resonance showed that these proteins interact with high affinity (Kd = 0.9 +/- 0.1 μM; Fig. S3C), comparable to the reported affinities of other nuclear receptors for their functional regulators (34, 43–45). Notably, the binding affinity of Dax-1 LBD for LRH-1 exceeds the affinity of Dax-1 peptides containing the N-terminal LXXL/ML motifs for LRH-1 (34) by more than 30-fold. This comparative analysis suggests that the observed interactions between Dax-1 and LRH-1 LBDs are functionally relevant.
The purified Dax:LRH-1 complex was crystallized, and its three-dimensional structure was solved using the molecular replacement method. Consistent with the biochemical data, the asymmetric unit of the crystalline complex revealed one (Dax-1)2:LRH-1 heterotrimer (Fig. 1). The structure of the complex was refined to 3.0 Å with Rfree/R values of 26.1/23.4 (Fig. S4) and included amino acid residues 318–559 for LRH-1 LBD and 250–315, 353–472 and 250–312, 353–472 for the first and second molecules of Dax-1, respectively. The N-terminal region (aa 205–249) of both Dax-1 molecules was not visible on the electron density maps of the complex and not included in the structure. Details of the structure determination and refinement are summarized in Table S1.
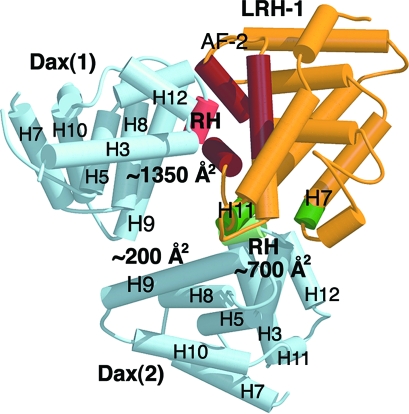
Fig. 1.
The three-dimensional structure of the Dax-1:LRH-1 complex. LRH-1 is shown in yellow; the two Dax-1 molecules are shown in blue. The structural elements involved in binding of the first Dax-1 to LRH-1 are shown in rose (RH site) and dark red (AF-2 region). The structural elements at the second Dax-1-LRH-1 interface are shown in light and dark green and are indicated.
Dax-1 Dimerization and Its Determinants.
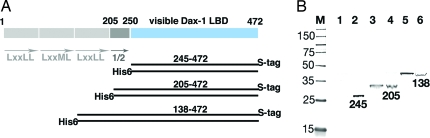
The trimeric structure of the complex suggested that Dax-1 may form a homodimer. To probe the existence of a Dax-1 dimer, we coexpressed three pairs of Dax-1 deletion mutants tagged with either His6 or S tags (Fig. 2A). Analysis of purified His6-tagged proteins revealed that the Dax-1 fragment 205–472 used in our structural analysis is a dimer in solution, as determined by its association with the S-tagged Dax-1 (Fig. 2B). This finding was confirmed by size exclusion chromatography revealing the presence of a single protein species with molecular mass of ≈60 kDa in a purified Dax-1 205–472 protein sample (Fig. S5A). This value agrees with the calculated molecular mass for the Dax-1 dimer. In contrast, a shorter fragment of Dax-1 corresponding to the visible Dax-1 LBD (aa 245–472) is a monomer, as determined by Western blot and size exclusion chromatography analyses (Figs. 2 and S5B). We conclude that 40 residues (aa 205–245) N-terminal to the visible Dax-1 LBD are sufficient to form and maintain a stable Dax-1 homodimer in vitro. Therefore, the presence of the second Dax-1 LBD in the (Dax-1)2:LRH-1 heterotrimer is a direct consequence of the dimeric state of Dax-1 (Fig. S5C).
Fig. 2.
Structural determinants of Dax-1 dimerization. (A) Schematic representation of Dax-1. The N-terminal domain of Dax-1 (gray) has 3 and 1/2 structural repeats (indicated by arrows); the 1/2 repeat is the N-terminal to the visible Dax-1 LBD (blue). Pairs of coexpressed Dax-1 fragments 245–472, 205–472, and 138–472 with either His6 or S tags are shown as black bars. (B) Western blot analysis showing formation of Dax-1 homodimers. Coexpressed fragments were purified using Ni-NTA matrix and analyzed by Western blot using the S-tagged antibody. Lanes 2, 4, and 6 correspond to the soluble bacterial extracts; lanes 1, 3, and 5 correspond to the purified Dax-1 proteins.
Specific Structural Features of the Dax-1 LBD.
The dimerization domain of Dax-1 was not visualized on electron density maps of the (Dax-1)2:LRH-1 trimer, likely because of its flexible nature (Fig. S5C). The residues constituting the dimerization domain of Dax-1 precede helix H3 and correspond to helices H1 and H2 in other nuclear receptors. The absence of H1 and H2 in Dax-1 is consistent with secondary structure predictions suggesting that the N-terminal region of Dax-1 LBD lacks the expected signatures for these helices. Notably, the N- and C-terminal boundaries of the visible structure of Dax-1 (aa 250–472) match the minimal transcriptional silencing domain of Dax-1 defined in mutational and cellular studies (18). A second disordered region in Dax-1 LBD (aa 314–352) corresponds to the loop connecting helices H5 and H7. The length of this loop varies among Dax-1 LBDs from different species, and large deletions in this region do not alter either the subcellular localization or the corepressor function of Dax-1 (46, 47). With the exception of these two regions, the rest of the Dax-1 LBD folds into a well-defined, compact structure.
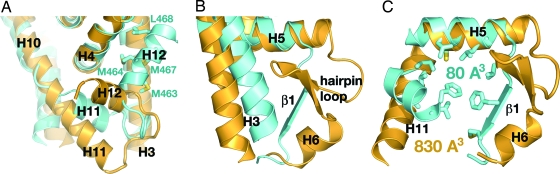
Remarkably, the ligand-binding pocket, which usually forms the hydrophobic core of nuclear receptor LBDs, is absent in Dax-1. Three structural elements of Dax-1—helices H11, H5, and the short β-strand following H5—reroute in a short cut to eliminate the pocket (Fig. 3 A and B). The resulting volume of the ligand-binding cavity is about 80 Å3 (compared with 830 Å3 in mLRH-1) (33), which is insufficient to accommodate even a small ligand (Fig. 3C). Mass spectroscopy analysis of the purified (Dax-1)2:LRH-1 complex confirms the absence of any ligand bound to the Dax-1 LBD.
Fig. 3.
Structural comparison of LRH-1 (yellow) and Dax-1 (blue) LBDs. (A) The AF-2 sites of LRH-1 and Dax-1. Helix H12 of LRH-1 is in an “active” conformation, whereas helix H12 of Dax-1 is in an “inactive” conformation and is docked in the coactivator groove; the residues participating in the docking interactions are indicated. Helix H11 of Dax-1 is rotated by ≈45° compared with its counterpart in LRH-1. (B) Helices H3 and H5-H6 of LRH-1 and Dax-1. Helix H3 of Dax-1 is shifted toward the core of the LBD. Helix H5 of Dax-1 is shorter and is followed by a short β-strand (indicated). (C) The ligand-binding pockets of LRH-1 and Dax-1. Residues filling the putative ligand-binding cavity of Dax-1 are shown as stick models.
Functional Conformations of Dax-1 and LRH-1 LBDs.
Similar to the structure of mLRH-1 alone (33), the mLRH-1 LBD in the trimer is without hormone and in an active conformation, as defined by the position of helix H12 (Fig. 1). Furthermore, superimposition of the complexed LRH-1 with free LRH-1 (33) shows nearly identical LBD structures (with a rmsd for 242 Cα atoms of 0.9 Å), demonstrating that LRH-1 does not assume a different conformation when inactivated by Dax-1. The trimer also can form with mLRH-1 mutant A268W, a functional receptor with an occluded ligand-binding cavity (33), indicating that Dax-1 does not require an LRH-1 hormone for binding.
Both Dax-1 molecules in the complex are found in an identical conformation (with an rmsd for 183 Cα of 0.7 Å), which differs from that of the LRH-1 LBD. Each is characterized by docking of helix H12 into its own coactivator-binding groove (Figs. 1 and 3A). In other nuclear receptors, this groove is known to bind the nuclear receptor coactivator boxes containing the LXXLL motif (43, 48, 49). In Dax-1, the conserved M464, M467, and L468 from H12 (MXXML; Fig. 3A) mimic the Leu residues from the LXXLL motif. The docked position of H12 in Dax-1 is consistent with its function as a repressor that does not need to bind coactivator.
Architecture of the (Dax-1)2:LRH-1 Heterotrimer.
The two bound molecules of Dax-1 are curiously positioned in the complex (Fig. 1). The first Dax-1 LBD is docked into the coactivator groove of LRH-1, forming a large binding interface of ≈1350 Å3. Surprisingly, the core structural element used by Dax-1 for binding to LRH-1 is a compact loop connecting helices H3 and H4. This loop forms a short helix, termed a “repression helix” (RH) (Fig. 4 A and B), bordered by two conserved Pro residues, P275 and P281, which introduce two turns into the Dax-1 polypeptide chain. The helical conformation of the RH site is very stable and is supported by multiple intramolecular contacts (Fig. 4B). This property is rather uncommon for accessible surface loops, which often are flexible. Another unusual property of the RH is the presence of a cluster of solvent-exposed hydrophobic residues (C276, I279, and L280; Fig. 4B), which suggests that this site might have evolved for intermolecular protein interactions.
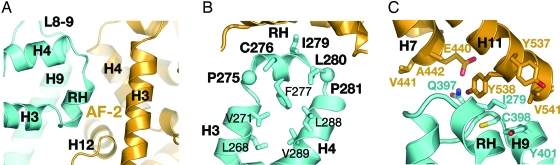
Fig. 4.
Binding interactions between Dax-1 (blue) and LRH-1 (yellow). (A) A magnified view of the primary Dax-1-LRH-1 interface indicating the structural elements involved in binding. (B) Architecture of the RH site. The repressor helix is bordered by two conserved Pro residues, P275 and P281 (shown as spheres). The helical conformation of the RH site is supported by multiple intramolecular interactions. Residues participating in binding with LRH-1 are shown in bold. (C) A magnified view of the secondary Dax-1-LRH-1 interface indicating the residues participating in binding interactions.
Superimposition of the (Dax-1)2:LRH-1 complex with the structures of hLRH-1 bound to the GRIP-1 peptide (34, 37) shows that the position and conformation of the RH in the complex are similar to those of the docked regulatory peptide. However, the RH site of mDax-1 does not have the LXXLL motif present in nuclear receptor coactivators. Instead, the RH site includes a sequence motif, 275-PCFXXLP-281, conserved among all members of the NR0B1 subfamily. Thus, the exposed hydrophobic residues C276, I279, and L280 from the Dax-1 repressor helix (Fig. 4B) substitute for the three Leu residues from the LXXLL motif of the coactivators. The N-terminal region of Dax-1 helix H9 with preceding loop L8–9 (Fig. 4A) extends this interface with LRH-1 to ≈1350 Å2.
The second Dax-1 is positioned in the vicinity of the first Dax-1 LBD, just outside the ligand-binding pocket of LRH-1 (Fig. 1). Curiously, for its interaction with LRH-1, the second Dax-1 uses the same site, consisting of the RH and the N-terminal part of helix H9 (Fig. 4C). The second Dax-1-LRH-1 interface includes the N- and C-terminal parts of LRH-1 helices H7 and H11, respectively, and is less extensive (≈700 Å2). Interestingly, the second Dax-1 LBD places the side chain of semiconserved Q397 in loop L8–9 at a distance of direct contact with residues at the mouth of the LRH-1 pocket. In LRH-1 from all species except rodents, this is the coordination site of the phosphate group of the bound phospholipid (Fig. S6) (34). Thus, a bound LRH-1 ligand might influence the second Dax-1 in vivo. We observe a weak binding interface (≈200 Å2; Fig. 1) between the visible structures of Dax-1 LBDs in the heterotrimer, formed between residues positioned on the exposed faces of helix H9 of the two Dax-1 LBDs. This observation is consistent with the dimeric state of Dax-1 (Fig. 2).
Mutational Analysis of the Dax-1-LRH-1 Interfaces.
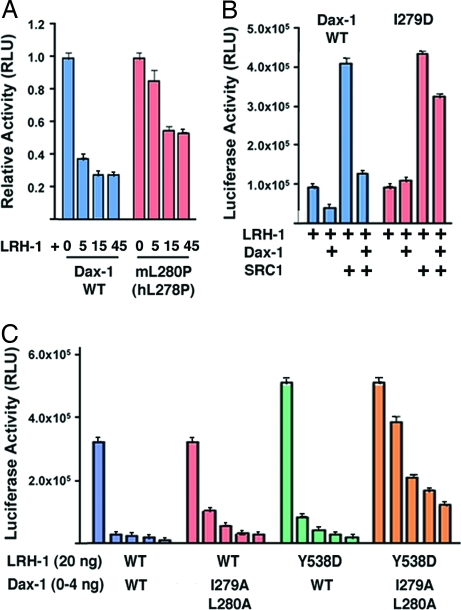
To date, more than 30 human Dax-1 missense mutations have been associated with adrenal disease (3). All mutations affect evolutionarily conserved residues positioned within the visible structure of Dax-1, suggesting that this structure corresponds to a critical functional domain. Most mutations affect buried Dax-1 residues and are predicted to destabilize the structural core of the LBD (50); however, one described mutation in hDax-1 affects a solvent-exposed hydrophobic residue (L278P in hDax-1 (3); L280P in mDax-1). Strikingly, L280 in mDax-1 is one of the conserved residues involved in the binding of Dax-1 to LRH-1 (Fig. 4B). Substitution of L278 for Pro in hDax-1 likely would distort the helical conformation of the RH site, thereby weakening the binding interactions of Dax-1 and LRH-1. Consistent with this prediction, equivalent mutation L280P in mDax-1 resulted in diminished repression of LRH-1 by Dax-1 (Fig. 5A). Mutating the adjacent hydrophobic I279 to negatively charged Asp eliminated the binding of Dax-1 to LRH-1 (Fig. S7) and produced a variant that no longer overpowered the coactivator (I279D in Fig. 5B). Consistent with these results, double-mutant I279A/L280A (Fig. 5C) attenuated the ability of Dax-1 to act as a potent corepressor of LRH-1 in cellular reporter assays. Taken together with structural data, these findings confirm the functional importance of the discovered RH and its conserved motif PCFXXLP for regulatory interactions of Dax-1 with LRH-1.
Fig. 5.
Regulatory interactions between Dax-1 and LRH-1. (A) Repression of LRH-1 by Dax-1 in HepG2 cells. Luciferase activity was measured after cotransfection of vectors encoding the Aro-Luc reporter, mLRH-1, and either wild-type or mutant mDax-1. Mutation L280P in the RH site of mDax-1 (L278P in hDax-1) resulted in diminished repression by Dax-1. (B) Repression of LRH-1 by Dax-1 in the presence of coactivator SRC-1. LRH-1 and Dax-1 variants were cotransfected into HepG2 cells with or without SRC-1, as indicated by “+.” Dax-1 is a potent repressor of LRH-1 that overpowers activation by SRC-1; however, a single mutation I279D in the RH site abrogates repression by Dax-1. (C) Diminished repression of Dax-1 by parallel mutations at the Dax-1 RH site and at the secondary binding site of LRH-1. Either wild-type or I279A/L280A Dax-1 RH mutant was cotransfected into HepG2 cells with either wild-type or mutant Y538D LRH-1.
Complementary to the described mutations in Dax-1 RH site, LRH-1 mutations R380E and E553R in the primary AF-2 site eliminated Dax-1 binding to the receptor (Fig. S7). In contrast to these AF-2 mutants, mutations at the secondary site of LRH-1 (Y538D, V541D, and Y538D/V541D; Fig. 4C) exhibited no effect on Dax-1 binding (Fig. S7). These results suggest that binding of the second Dax-1 to LRH-1 is mediated by the first Dax-1 docked into the AF-2 groove, likely through direct Dax-1-Dax-1 interactions that we see in part in the crystal structure. These data also are consistent with the hypothesis that Dax-1 binds to LRH-1 as a homodimer, with determining interactions at the primary AF-2 binding site. Although neither of the secondary site LRH-1 mutants produced noticeable effects on the complex formation in in vitro binding experiments, these mutants did attenuate the inhibitory function of Dax-1 in cell-based transcription assays (Fig. 5C). An effect of these mutants was apparent when they were paired with the RH site mutant I279A/L280A, suggesting that the presence of the second Dax-1 in the complex contributes to the repressor efficiency.
Discussion
Mechanism of Transcriptional Inhibition by Dax-1.
The data described in this work demonstrate that the Dax-1 LBD interacts directly with the LBD of LRH-1. Our finding that Dax-1 binds in the coactivator groove of LRH-1 is consistent with previous data showing that repression by Dax-1 could be through competitive inhibition (9). A novel twist that our analysis introduces to this hypothesis is that the repression site is located in the Dax-1 LBD and contains the subfamily conserved sequence motif PCFXXLP (Fig. 4B). The observed novel mode of interaction does not contradict the previous assignment for the N-terminal repeats as the region responsible for nuclear localization of Dax-1 (17). Furthermore, the observed docking of Dax-1 LBD into the coactivator groove of LRH-1 is consistent with the recently demonstrated mode of repression of nuclear receptors Nur77 and ERRγ by Dax-1, for which Dax-1 LBD was shown to be sufficient (7, 8).
Our work suggests two reasons why Dax-1 functions as an efficient transcriptional inhibitor that can overpower coactivators. First, the binding interactions of Dax-1 at the AF-2 site are more extensive than those of a coactivator with a single LXXLL motif. The repression site of Dax-1 is expanded by residues presented by the well-ordered helix H9. Second, the RH site of Dax-1 is well structured and stabilized through extensive intramolecular interactions. The stability of the RH site contrasts with the dynamic nature of LXXLL motifs of nuclear receptor coactivators that are followed by flexible loops (43), which require prestructuring for binding to nuclear receptors. We conclude that the increased interaction surface and the preordered state of the RH site underlie the efficiency of Dax-1 as a transcriptional repressor.
Dimerization of Dax-1 and Possible Functional Role of the Dax-1 Dimer.
Dax-1 binds to primary and secondary sites on LRH-1. What are these sites' contributions to the trimer assembly? As discussed earlier, LRH-1 mutations in the primary AF-2 site block formation of the complex. In contrast, LRH-1 mutants with altered secondary sites behave similarly to wild-type protein. In the context of the structure, our data indicate that Dax-1 binds LRH-1 as a dimer with determining interactions at the primary AF-2 site.
What is the structure of the Dax-1 dimer? Although our group and others (51) have observed formation of Dax-1 dimers, the (Dax-1)2:LRH-1 structure does not reveal an obvious configuration for the Dax-1 homodimer. A previously proposed model suggests that Dax-1 molecules might dimerize through docking of the N-terminal LXXLL motifs into the coactivator grooves of their respective partners (51). Our results argue against this model, because the structure of Dax-1 reveals occupation of the AF-2 site by helix H12. Our findings also demonstrate that Dax-1 can dimerize in the absence of the LXXLL motifs, and that the presence of 40 residues preceding helix H3 is sufficient for Dax-1 dimerization. The dimerization domain of Dax-1 is not seen in the (Dax-1)2:LRH-1 trimer, likely because of its flexible linkage to the visible Dax-1 LBDs.
In this work, we show Dax-1 to be a dimeric repressor with one repression site present on each of the two LBDs. Its binding partner LRH-1 has only one AF-2 site. Why is the Dax-1 dimer needed to bind to the LRH-1 monomer? What could be the role of the second Dax-1 in the regulatory complex? One possibility is that the second Dax-1 LBD might enhance the repressive power, providing more contacts to LRH-1 and thus ensuring repression and domination of Dax-1 over coactivators. Consistent with this idea, binding experiments showed that the dimeric Dax-1 binds LRH-1 with higher affinity (Kd = 0.9 ± 0.1 μM; Fig. S3C) compared with the truncated monomeric Dax-1 LBD (aa 245–472; Kd = 4.8 ± 0.2 μM; Fig. S8). In addition, the flexible association between the two Dax-1 molecules might facilitate the binding of different nuclear receptors with variable molecular surfaces. The weakly ordered contact relating the two Dax-1 LBDs in the trimer may be a functional feature that allows the pair to adjust and perform a “claw-like” clasping, to accommodate different nuclear receptor LBDs. We also note that the placement of the second Dax-1 at the entrance to the LRH-1 ligand-binding pocket makes the second Dax-1 LBD a candidate to serve as a “sensor” of the receptor's ligand state. An alternative possibility is that the second Dax-1 LBD might simply provide an additional binding interface and link the regulatory assembly to other components of the transcriptional machinery. The dimerization of Dax-1 in vivo might depend on its concentration in cells, be controlled by specific co-regulatory proteins, or be triggered by specific signaling events; therefore, both monomeric and dimeric forms of Dax-1 might exist and function in different cellular contexts. Clearly, gaining further insight into the dynamics of Dax-1 dimerization and identifying relevant partners of Dax-1 will help translate this unusual structure into physiology.
In summary, we describe the first three-dimensional structure of Dax-1 in association with one of its physiological targets, the nuclear receptor LRH-1. The RH in the Dax-1 LBD, which includes a subfamily conserved sequence motif, 275-PCFXXLP-281, mediates this association. Our findings demonstrate that Dax-1 functions as a ligand-independent nuclear receptor and explain its function as a competitive transcriptional corepressor. The three-dimensional structure of Dax-1 also rationalizes the naturally occurring missense mutations in the human Dax-1 leading to congenital X-linked adrenal hypoplasia. This work should aid in the design of further experiments aimed at clarifying the function of Dax-1 as a potent transcriptional regulator.
Materials and Methods
GST Pull-Down Assay.
The GST and the GST protein fusions with LRH-1 and SF-1 LBDs were expressed in Escherichia coli and immobilized on glutathione-agarose beads (Sigma). 35S-radiolabelled full-length mDax-1 was prepared in vitro in a reticulocyte lysate using the TNT T7 quick-coupled transcription-translation system (Promega) according to the manufacturer's protocol. The immobilized GST proteins with specifically bound mDax-1 were eluted, resolved by SDS-PAGE, and visualized by autoradiography.
Preparation, Crystallization, and Crystallographic Analysis of the Dax-1:LRH-1 Complex.
A DNA fragment encoding mLRH-1 LBD (aa 313–560) with His6 tags and a cleavage site for tobacco etch virus (TEV) protease was cloned into a pACYCDuet-1 vector; five different DNA fragments encoding mDax-1 residues 1–208, 205–472, 138–472, 70–472, and 1–472 were cloned into pETDuet-1 plasmid (Novagen). For coexpression of Dax-1 with LRH-1, BL21(DE3)Star cells (Invitrogen) were cotransformed with the corresponding plasmids. Coexpression was induced with 0.1 mM isopropyl β-d-1-thiogalactopyranoside (IPTG), and cells were grown for 12 h at 15 °C. Cell lysates were loaded onto Ni-NTA matrix, and specifically bound proteins were eluted with 200 mM imidazole. These experiments showed that only the Dax-1 LBD forms a stable complex with LRH-1. After the LRH-1 His6-tag was cleaved by TEV protease, the complex was purified using size-exclusion chromatography (Superdex 75, Amersham Pharmacia). The Dax-1:LRH-1 complex was crystallized by the vapor diffusion method, using a reservoir solution containing 0.1 M Hepes (pH 7.5), 20% PEG 8000, and 1 mM Deoxy-BigChap. Before data collection, crystals were flash-frozen in liquid nitrogen. X-ray diffraction data were obtained at −180 °C to 3.0 Å at the Advanced Light Source beamline 8.3.1 (λ = 1.1 Å) using a single crystal. Data were integrated and scaled using DENZO and SCALEPACK. The crystal was of the space group P43 with cell dimensions of a = b = 103.4 Å and c = 117.5 Å, and it contained one (Dax-1)2:LRH-1 heterotrimer in the asymmetric unit. The structure of the complex was determined by the molecular replacement method using the CNS algorithms with a starting model derived from the atomic coordinates for mLRH-1 (PDB ID 1PK5). Electron-density maps based on coefficients 2Fo − Fc were calculated from the phases of the initial model. Subsequent rounds of model building and refinement were performed using the QUANTA and CNS programs, respectively. At later stages of refinement, the structure was checked using simulated annealing composite omit maps.
Preparation of His6- and S-Tagged Dax-1 Fragments for Western Blot Analysis.
Three DNA fragments encoding mDax-1 residues 245–472, 205–472, and 138–472 with either N-terminal His6 tags or C-terminal S tags were cloned into pETDuet-1 and pACYCDuet-1 vectors, respectively (Novagen). For coexpression of the His6- and S-tagged fragments, BL21(DE3)Star cells (Invitrogen) were cotransformed with the corresponding plasmids. Coexpression was induced with 0.1 mM IPTG, and cells were grown for 12 h at 15 °C. Cell lysates were loaded onto Ni-NTA matrix, and specifically bound proteins were eluted with 200 mM imidazole. The proteins were resolved by SDS-PAGE and analyzed by Western blot, using mouse anti-S-tag monoclonal antibody and goat anti-mouse IgG alkaline phosphatase conjugate (Novagen), and then developed with colorimetric detection reagents.
Surface Plasmon Resonance (SPR) Assay.
The His6-tagged mLRH-1 LBD and Dax-1 fragments 205–472 and 245–472 were expressed and purified using affinity and size-exclusion chromatography, and the purified LRH-1 LBD was immobilized to an Ni2+-tri-NTA chip (52). For SPR assays using anti-GST chips, a DNA fragment encoding mLRH-1 LBD was cloned into a pGEX-4T-1 vector (Amersham Pharmacia). The GST-LRH-1 protein was purified using glutathione sepharose beads and size-exclusion chromatography and immobilized to an anti-GST chip, which was prepared by immobilizing goat anti-GST antibody to a CM5 chip. All measurements were performed using a Biacore T100 instrument. The purified LRH-1 proteins were immobilized to a surface density of about 200 resonance units, and the purified Dax-1 fragments were injected over immobilized LRH-1 and reference surfaces. The equilibrium dissociation constants (Kd) were determined using nonlinear regression analysis for the association and dissociation phases of the sensorgrams.
Plasmids, Site-Directed PCR Mutagenesis, and Transfection.
The Aro-Luc reporter vector pGL2 was purchased from Promega; pcDNA3, pcDNA3-FLAGmDax-1, and pSG5-SRC-1, pcineo-HAmLRH-1 plasmids were described earlier. Mutagenesis of all LRH-1 and Dax-1 constructs was performed using the QuikChange site-directed mutagenesis kit (Stratagene) according to the manufacturer's protocol. Before the transcriptional assays, expression of all mutant proteins was tested in Cos7 cells by Western blot analysis of nuclear and cytoplasmic extracts using the corresponding antibodies to epitope tags. HepG2 cells were transfected with FuGene 6 as specified by the manufacturer (Roche). In all transfection experiments, cells were grown on 24 well plates, and reporter activity was measured 48 h after transfection.
Supplementary Material
Acknowledgments.
Thus work was supported by the National Institutes of Health (R.J.F. and H.A.I.). We thank Gregory M. Waitt (GlaxoSmithKline) for the mass spectrometry analysis of the purified Dax-1:LRH-1 complex.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3F5C).
This article contains supporting information online at www.pnas.org/cgi/content/full/0808936105/DCSupplemental.
References
- 1.Lalli E, Sassone-Corsi P. DAX-1, an unusual orphan nuclear receptor at the crossroads of steroidogenic function and sexual differentiation. Mol Endocrinol. 2003;17:1445–1453. doi: 10.1210/me.2003-0159. [DOI] [PubMed] [Google Scholar]
- 2.Bardoni B, et al. A dosage-sensitive locus at chromosome Xp21 is involved in male to female sex reversal. Nat Genet. 1994;7:497–501. doi: 10.1038/ng0894-497. [DOI] [PubMed] [Google Scholar]
- 3.Lin L, et al. Analysis of DAX1 (NR0B1) and steroidogenic factor-1 (NR5A1) in children and adults with primary adrenal failure: Ten years' experience. J Clin Endocrinol Metab. 2006;91:3048–3054. doi: 10.1210/jc.2006-0603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ito M, Yu R, Jameson JL. DAX-1 inhibits SF-1-mediated transactivation via a carboxy-terminal domain that is deleted in adrenal hypoplasia congenita. Mol Cell Biol. 1997;17:1476–1483. doi: 10.1128/mcb.17.3.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Crawford PA, Dorn C, Sadovsky Y, Milbrandt J. Nuclear receptor DAX-1 recruits nuclear receptor corepressor N-CoR to steroidogenic factor 1. Mol Cell Biol. 1998;18:2949–2956. doi: 10.1128/mcb.18.5.2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nachtigal MW, et al. Wilms' tumor 1 and Dax-1 modulate the orphan nuclear receptor SF-1 in sex-specific gene expression. Cell. 1998;93:445–454. doi: 10.1016/s0092-8674(00)81172-1. [DOI] [PubMed] [Google Scholar]
- 7.Song KH, et al. The atypical orphan nuclear receptor DAX1 interacts with orphan nuclear receptor Nur77 and represses its transactivation. Mol Endocrinol. 2004;18:1920–1940. doi: 10.1210/me.2004-0043. [DOI] [PubMed] [Google Scholar]
- 8.Park YY, et al. An autoregulatory loop controlling orphan nuclear receptor DAX-1 gene expression by orphan nuclear receptor ERRgamma. Nucleic Acids Res. 2005;33:6756–6768. doi: 10.1093/nar/gki976. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 9.Zhang H, Thomsen JS, Johansson L, Gustafsson JA, Treuter E. DAX-1 functions as an LXXLL-containing corepressor for activated estrogen receptors. J Biol Chem. 2000;275:39855–39859. doi: 10.1074/jbc.C000567200. [DOI] [PubMed] [Google Scholar]
- 10.Holter E, et al. Inhibition of androgen receptor (AR) function by the reproductive orphan nuclear receptor DAX1. Mol Endocrinol. 2002;16:515–528. doi: 10.1210/mend.16.3.0804. [DOI] [PubMed] [Google Scholar]
- 11.Agoulnik IU, et al. Repressors of androgen and progesterone receptor action. J Biol Chem. 2003;278:31136–31148. doi: 10.1074/jbc.M305153200. [DOI] [PubMed] [Google Scholar]
- 12.Suzuki T, Kasahara M, Yoshioka H, Morohashi K, Umesono K. LXXLL-related motifs in Dax-1 have target specificity for the orphan nuclear receptors Ad4BP/SF-1 and LRH-1. Mol Cell Biol. 2003;23:238–249. doi: 10.1128/MCB.23.1.238-249.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fayard E, Auwerx J, Schoonjans K. LRH-1: An orphan nuclear receptor involved in development, metabolism and steroidogenesis. Trends Cell Biol. 2004;14:250–260. doi: 10.1016/j.tcb.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 14.Niakan KK, et al. Novel role for the orphan nuclear receptor DAX1 in embryogenesis, different from steroidogenesis. Mol Gen Metabol. 2006;88:261–271. doi: 10.1016/j.ymgme.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 15.Wang J, et al. A protein interaction network for pluripotency of embryonic stem cells. Nature. 2006;444:364–368. doi: 10.1038/nature05284. [DOI] [PubMed] [Google Scholar]
- 16.Zanaria E, et al. An unusual member of the nuclear hormone receptor superfamily responsible for X-linked adrenal hypoplasia congenita. Nature. 1994;372:635–641. doi: 10.1038/372635a0. [DOI] [PubMed] [Google Scholar]
- 17.Kawajiri K, et al. Role of the LXXLL-motif and activation function 2 domain in subcellular localization of DAX-1 (dosage-sensitive sex reversal-adrenal hypoplasia congenita critical region on the X chromosome, gene 1) Mol Endocrinol. 2003;17:994–1004. doi: 10.1210/me.2002-0360. [DOI] [PubMed] [Google Scholar]
- 18.Lalli E, et al. A transcriptional silencing domain in DAX-1 whose mutation causes adrenal hypoplasia congenita. Mol Endocrinol. 1997;11:1950–1960. doi: 10.1210/mend.11.13.0038. [DOI] [PubMed] [Google Scholar]
- 19.Altincicek B, et al. Interaction of the corepressor Alien with DAX-1 is abrogated by mutations of DAX-1 involved in adrenal hypoplasia congenita. J Biol Chem. 2000;275:7662–7667. doi: 10.1074/jbc.275.11.7662. [DOI] [PubMed] [Google Scholar]
- 20.Zazopoulos E, Lalli E, Stocco DM, Sassone-Corsi P. DNA binding and transcriptional repression by DAX-1 blocks steroidogenesis. Nature. 1997;390:311–315. doi: 10.1038/36899. [DOI] [PubMed] [Google Scholar]
- 21.Nitta M, Ku S, Brown C, Okamoto AY, Shan B. CPF: An orphan nuclear receptor that regulates liver-specific expression of the human cholesterol 7alpha-hydroxylase gene. Proc Natl Acad Sci USA. 1999;96:6660–6665. doi: 10.1073/pnas.96.12.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.del Castillo-Olivares A, Gil G. Alpha 1-fetoprotein transcription factor is required for the expression of sterol 12alpha-hydroxylase, the specific enzyme for cholic acid synthesis: Potential role in the bile acid-mediated regulation of gene transcription. J Biol Chem. 2000;275:17793–17799. doi: 10.1074/jbc.M000996200. [DOI] [PubMed] [Google Scholar]
- 23.Luo Y, Liang CP, Tall AR. The orphan nuclear receptor LRH-1 potentiates the sterol-mediated induction of the human CETP gene by liver X receptor. J Biol Chem. 2001;276:24767–24773. doi: 10.1074/jbc.M100912200. [DOI] [PubMed] [Google Scholar]
- 24.Schoonjans K, et al. Liver receptor homolog 1 controls the expression of the scavenger receptor class B type I. EMBO Rep. 2002;3:1181–1187. doi: 10.1093/embo-reports/kvf238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clyne CD, Speed CJ, Zhou J, Simpson ER. Liver receptor homologue-1 (LRH-1) regulates expression of aromatase in preadipocytes. J Biol Chem. 2002;277:20591–20597. doi: 10.1074/jbc.M201117200. [DOI] [PubMed] [Google Scholar]
- 26.Liu DL, et al. Expression and functional analysis of liver receptor homologue-1 as a potential steroidogenic factor in rat ovary. Biol Reprod. 2003;69:508–517. doi: 10.1095/biolreprod.102.011767. [DOI] [PubMed] [Google Scholar]
- 27.Mendelson CR, Jiang B, Shelton JM, Richardson JA, Hinshelwood MM. Transcriptional regulation of aromatase in placenta and ovary. J Steroid Biochem Mol Biol. 2005;95:25–33. doi: 10.1016/j.jsbmb.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 28.Hinshelwood MM, Shelton JM, Richardson JA, Mendelson CR. Temporal and spatial expression of liver receptor homologue-1 (LRH-1) during embryogenesis suggests a potential role in gonadal development. Dev Dyn. 2005;234:159–168. doi: 10.1002/dvdy.20490. [DOI] [PubMed] [Google Scholar]
- 29.Gu P, et al. Orphan nuclear receptor LRH-1 is required to maintain Oct4 expression at the epiblast stage of embryonic development. Mol Cell Biol. 2005;25:3492–3505. doi: 10.1128/MCB.25.9.3492-3505.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ueda H, Sun GC, Murata T, Hirose S. A novel DNA-binding motif abuts the zinc finger domain of insect nuclear hormone receptor FTZ-F1 and mouse embryonal long-terminal repeat-binding protein. Mol Cell Biol. 1992;12:5667–5672. doi: 10.1128/mcb.12.12.5667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Solomon IH, et al. Crystal structure of the human LRH-1 DBD-DNA complex reveals Ftz-F1 domain positioning is required for receptor activity. J Mol Biol. 2005;354:1091–1102. doi: 10.1016/j.jmb.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 32.Val P, et al. Adrenocorticotropin/3′, 5′-cyclic AMP-mediated transcription of the scavenger akr1–b7 gene in adrenocortical cells is dependent on three functionally distinct steroidogenic factor-1-responsive elements. Endocrinology. 2004;145:508–518. doi: 10.1210/en.2003-1093. [DOI] [PubMed] [Google Scholar]
- 33.Sablin EP, Krylova IN, Fletterick RJ, Ingraham HA. A novel structural element accounts for the constitutive activity of the orphan nuclear receptor, LRH-1. Mol Cell. 2003;11:1575–1585. doi: 10.1016/s1097-2765(03)00236-3. [DOI] [PubMed] [Google Scholar]
- 34.Krylova IN, et al. Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell. 2005;120:343–355. doi: 10.1016/j.cell.2005.01.024. [DOI] [PubMed] [Google Scholar]
- 35.Li Y, et al. Structural and biochemical basis for selective repression of the orphan nuclear receptor liver receptor homolog 1 by small heterodimer partner. Proc Natl Acad Sci U S A. 2005;102:9505–9510. doi: 10.1073/pnas.0501204102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ortlund EA, et al. Modulation of human nuclear receptor LRH-1 activity by phospholipids and SHP. Nat Struct Mol Biol. 2005;12:357–363. doi: 10.1038/nsmb910. [DOI] [PubMed] [Google Scholar]
- 37.Wang W, et al. The crystal structures of human steroidogenic factor-1 and liver receptor homogue-1. Proc Natl Acad Sci USA. 2005;102:7505–7510. doi: 10.1073/pnas.0409482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee YK, Choi YH, Chua SS, Park YJ, Moore DD. Phosphorylation of the hinge domain of the nuclear hormone receptor LRH-1 stimulates transactivation. J Biol Chem. 2006;281:7850–7855. doi: 10.1074/jbc.M509115200. [DOI] [PubMed] [Google Scholar]
- 39.Yu FQ, et al. Activation of the p38MPAK pathway by follicle-stimulating hormone regulates steroidogenesis in granulosa cells differentially. J Endocrinol. 2005;186:85–96. doi: 10.1677/joe.1.05955. [DOI] [PubMed] [Google Scholar]
- 40.Kim JW, Peng N, Rainey WE, Carr BR, Attia GR. The role of the orphan nuclear receptor, liver receptor homologue-1, in the regulation of human corpus luteum 3b-hydroxysteroid dehydrogenase type II. J Clin Endocrinol Metab. 2004;89:3042–3047. doi: 10.1210/jc.2003-030880. [DOI] [PubMed] [Google Scholar]
- 41.Kim JW, Havelock JC, Carr BR, Attia GR. Liver receptor homolog-1 regulates the expression of steroidogenic acute regulatory protein in human granulosa cells. J Clin Endocrinol Metab. 2005;90:1678–1685. doi: 10.1210/jc.2003-031599. [DOI] [PubMed] [Google Scholar]
- 42.Peng N, Kim JW, Rainey WE, Carr BR, Attia G. The role of the orphan nuclear receptor, liver receptor homologue-1, in the regulation of human corpus luteum 3b-hydroxysteroid dehydrogenase type II. J Clin Endocrinol Metab. 2003;88:6020–6028. doi: 10.1210/jc.2003-030880. [DOI] [PubMed] [Google Scholar]
- 43.Darimont BD, et al. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Fujino T, et al. In vitro farnesoid X receptor ligand sensor assay using surface plasmon resonance and based on ligand-induced coactivator association. J Steroid Biochem Mol Biol. 2003;87:247–252. doi: 10.1016/j.jsbmb.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 45.Moore JM, et al. Quantitative proteomics of the thyroid hormone receptor-coregulator interactions. J Biol Chem. 2004;279:27584–27590. doi: 10.1074/jbc.M403453200. [DOI] [PubMed] [Google Scholar]
- 46.Lehmann SG, Sassone-Corsi P. X-linked adrenal hypoplasia congenita is caused by abnormal nuclear localization of the DAX-1 protein. Proc Natl Acad Sci USA. 2002;99:8225–8230. doi: 10.1073/pnas.122044099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Park YY, et al. Differential role of the loop region between helices H6 and H7 within the orphan nuclear receptors small heterodimer partner and DAX-1. Mol Endocrinol. 2004;18:1082–1095. doi: 10.1210/me.2003-0339. [DOI] [PubMed] [Google Scholar]
- 48.Shiau AK, et al. The structural basis of estrogen receptor/coactivator recognition and the antagonism of this interaction by tamoxifen. Cell. 1998;95:927–937. doi: 10.1016/s0092-8674(00)81717-1. [DOI] [PubMed] [Google Scholar]
- 49.Glass CK, Rosenfeld MG. The coregulator exchange in transcriptional functions of nuclear receptors. Genes Dev. 2000;14:121–141. [PubMed] [Google Scholar]
- 50.Lenmann S, Wurtz JM, Renaud JP, Sassone-Corsi P, Lalli E. Structure-function analyses reveal the molecular determinants of the impaired biological function of DAX-1 mutants in AHC patients. Hum Mol Genet. 2003;12:1063–1072. doi: 10.1093/hmg/ddg108. [DOI] [PubMed] [Google Scholar]
- 51.Iyer AK, Zhang YH, McCabe ER. Dosage-sensitive sex-reversal adrenal hypoplasia congenita critical region on the X chromosome, gene 1 (DAX1) (NR0B1) and small heterodimer partner (SHP) (NR0B2) form homodimers individually, as well as DAX1-SHP heterodimers. Mol Endocrinol. 2006;20:2326–2342. doi: 10.1210/me.2005-0383. [DOI] [PubMed] [Google Scholar]
- 52.Huang Z, Park JI, Watson DS, Hwang P, Szoka FC. Facile synthesis of multivalent nitrilotriacetic acid (NTA) and NTA conjugates for analytical and drug delivery applications. Bioconjug Chem. 2006;17:1592–1600. doi: 10.1021/bc0602228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.