Abstract
Proteomics has progressed radically in the last 5 years and is now on par with most genomic technologies in throughput and comprehensiveness. Analyzing peptide mixtures by liquid chromatography coupled to high-resolution mass spectrometry (LC-MS) has emerged as the main technology for in-depth proteome analysis whereas two-dimensional gel electrophoresis, low-resolution MALDI, and protein arrays are playing niche roles. MS-based proteomics is rapidly becoming quantitative through both label-free and stable isotope labeling technologies. The latest generation of mass spectrometers combines extremely high resolving power, mass accuracy, and very high sequencing speed in routine proteomic applications. Peptide fragmentation is mostly performed in low-resolution but very sensitive and fast linear ion traps. However, alternative fragmentation methods and high-resolution fragment analysis are becoming much more practical. Recent advances in computational proteomics are removing the data analysis bottleneck. Thus, in a few specialized laboratories, “precision proteomics” can now identify and quantify almost all fragmented peptide peaks. Huge challenges and opportunities remain in technology development for proteomics; thus, this is not “the beginning of the end” but surely “the end of the beginning.”
Analysis of individual proteins by classical methods and by mass spectrometry (MS) has been an indispensable cornerstone of biochemistry for many decades. Large-scale analysis of the whole protein complement of cells, tissues, and body fluids (proteomics) would additionally enable the unbiased comparison of different cellular states in biology and medicine at a “systems-wide” level. However, technological challenges associated with proteomics have long prevented its widespread adoption. Two-dimensional (2D) gel electrophoresis was conceived more than 30 years ago (1). This technology has been useful for low-complexity protein mixtures but never matured into a comprehensive and accurate proteomics technology. The introduction of high-sensitivity protein identification by MS at first seemed to help 2D gel analysis, but in fact it revealed that the thousands of spots seen in the gel maps are actually variants of a few hundred of the most abundant proteins (2). Recently, it has also become clear that quantitation of even these proteins is far from accurate because of spot overlap (3). Accordingly, “biomarkers” found by these technologies tend to be the same regardless of the system under investigation (4).
In principle, protein arrays might be applicable to proteomics in a similar way that gene chips have been to the measurement of RNA. However, the challenge associated with expressing thousands of full-length proteins and immobilizing them in a native state on a chip is daunting (5, 6). In practice, the role of protein arrays has been limited, and the literature contains few examples of their successful use. MS technology with low resolving power, especially in the form of the so-called SELDI method (7), caught the imagination of clinicians a few years ago. This approach involves measuring a MALDI spectrum of proteins from the body fluid of a patient and then employs machine learning to differentiate disease and healthy states. However, from a mass-spectrometric point of view, SELDI boils down to simple MALDI spectra of very complex mixtures and would be expected to only yield a subset of the most abundant low-mass peptides and protein fragments. Such species could still have proven sufficient to classify patient samples. However, as the scientific community demanded identification of the peaks comprising the SELDI patterns, these usually turned out to belong to the same nonspecific proteins unlikely to be directly associated with the disease.
In contrast to the above approaches, which were discussed as promising proteomics technologies as late as a few years ago, MS-based proteomics has taken great strides in development. MS-based protein science has always been extremely useful in studies focused on individual proteins, but large-scale proteomics is increasingly realizing its turn-of-the-millennium promises, too. In particular, technological improvements in the last 5 years have dramatically increased the routine availability of extremely high-performance MS. In many but not all cases, these technologies already existed but could only be applied in specialized situations by expert laboratories and at low throughput. The main purpose of this perspective is to show that MS techniques with high accuracy can and should now be applied routinely in most proteomics contexts, and that there is no penalty for their use. In fact, we argue that precise and comprehensive analysis of complex proteomes is best achieved by using high-resolution proteomics technologies. There are many other important aspects of MS-based proteomics that have been the subjects of recent reviews and that will not serve as focal points here. For example, the remarkable inroads of proteomics strategies into the quantitative analysis of posttranslational modifications (8), the determination of protein interactions (9), and the ongoing integration of MS technology with other powerful tools of molecular biology (10) are not discussed here.
The Importance of Being Highly Resolved
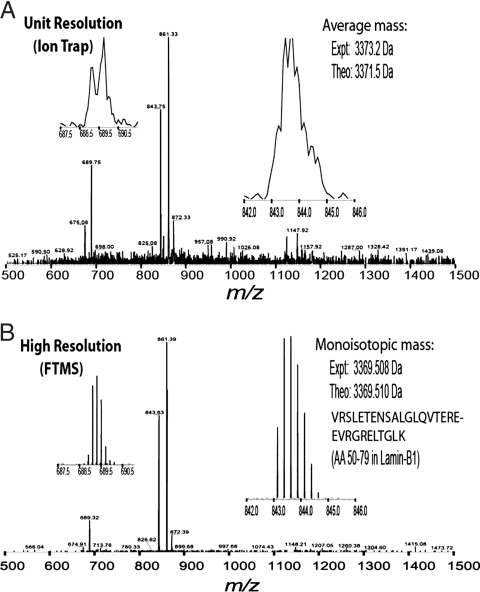
The current mainstream format in large-scale proteomics involves the analysis of very complex peptide mixtures. In this “shotgun approach” (11), tens of thousands of peptides with very large dynamic range (i.e., the concentration difference between the most and least abundant peptides) have to be analyzed in several chromatographic runs. If these mixtures are measured with ion traps or other MS instruments of lower resolving power, coeluting peptides with similar m/z ratios frequently overlap. This precludes accurate mass analysis, accurate charge state determination, and accurate quantitation. Fig. 1 shows a mass spectrometric scan of a relatively simple peptide mixture at high resolution and low resolution. From a theoretical point of view, there is no clear cut-off for desired mass resolving power, but in our experience 100,000 [full width at half height (FWHH)] is both practical and desirable for complex mixture analysis.
Fig. 1.
A direct comparison of peptides detected at two different resolving powers. A modest mixture of peptides was measured on an ion trap instrument at unit resolution (A) vs. on a Fourier transform instrument at high resolution (B). (Insets) Expanded spectral regions for two individual peptides, with Insets at right highlighting an unmodified Lys-C peptide that is 30 residues long. Note that the natural isotopes mainly due to 13C are clearly resolved at high resolution but not resolved at unit resolution. The spectra shown were obtained during a chromatographic separation, and the theoretical masses shown are for the average (A) and the monoisotopic (B) molecular weight values.
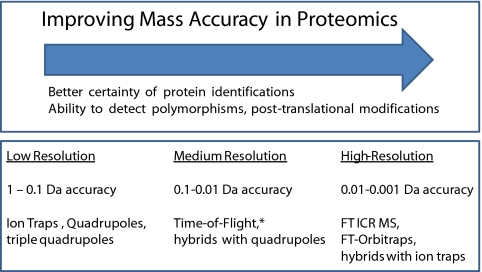
Until a few years ago, proteomics researchers had to choose between 3D ion traps, with high sequencing speed, high sensitivity, and very robust performance but low resolving power (≈300) or time-of-flight (TOF) instruments with higher resolving power (≈10,000) but less sensitivity and robustness (Fig. 2). The evolution of TOF instruments and even more so the introduction of hybrid linear ion trap Fourier-transform (FT) instruments have now made high-resolution MS broadly available. This development has been an unalloyed boon to proteomics, leading to much higher quality datasets and dramatically reduced false positive peptide identifications. High-resolving-power MS (≫50,000) has in fact been available in FT ion cyclotron resonance (ICR) instruments for several decades (12). However, FT ICR instruments were not routinely used in proteomics before the commercial introduction of the LTQ-FT in 2004 (13). This instrument consists of a linear ion trap as the front end, which was itself a great improvement over the 3D ion trap in terms of ion capacity, scan speed, and mass resolving power (14). Defined numbers of ions are pulsed into the FT part of the instrument, which analyzes them at high resolution (≈100,000 at m/z 400 for a 1-s scan; mass resolving power depends inversely on m/z in the FT-ICR instrument). During this time, the linear ion trap sequences the most prominent ions determined in the mass spectrum. Several years later, the LTQ-Orbitrap was introduced, in which the FT ICR back-end of the LTQ-FT with its superconducting magnet is replaced by an orbitrap. This analyzer also uses the FT principle; however, ions are confined in purely electric fields and the device is significantly smaller (15–17). The LTQ-Orbitrap has proven to be a tremendous advance for shotgun proteomics, combining high resolving power, mass accuracy, and reliability in a relatively compact form.
Fig. 2.
Overview of the impact that improving mass accuracy has in contemporary proteomics (Upper) along with various flavors of MS instruments and their approximate mass accuracy for tryptic peptides (Lower). The asterisk indicates that the latest models of TOF instruments are approaching the lower end of the resolution range of FTMS instruments.
Other analyzer configurations include the hybrid quadrupole TOF instruments. They achieve medium mass resolving power and good mass accuracy. In principle, they can be much faster than scanning instruments, but so far they are in practice slower and less sensitive than the LTQ-Orbitrap. In one innovative approach, a quadrupole TOF instrument is used without peptide ion selection to fragment all precursor ions at once (18). Product and precursor ions are then correlated by chromatographic elution time. One potential limitation of this approach is the limited dynamic range in the fragmentation mode, because highly abundant and low-abundance peptides are fragmented together. A multiplexed approach to fragmentation has also been performed in a linear ion trap (19).
The triple quadrupole was the classic analyzer at the beginning of protein MS. The “triple quad” is currently experiencing a renaissance for a specific use in proteomics because it allows the monitoring of targeted peptide masses and fragment combinations in an experiment called multiple reaction monitoring (MRM). In MRM, the triple quadrupole constantly monitors specific, preprogrammed fragmentation reactions, leading to high quantitative accuracy of targeted peptides. Even in very complex mixtures, the MRM experiment quantifies many peptides per LC injection by using the abundance of one or more of its fragment ions, and makes use of the large dynamic range of triple quadrupoles (20).
MALDI is used predominantly in single gel band or gel spot analysis but can also be coupled to LC separation via a spotting plate. Peptide mass fingerprints are generally not accepted as sufficient evidence for protein identification anymore, making fragmentation capability a near prerequisite of MALDI instruments for proteomics, especially when working in eukaryotic systems. Current MALDI/TOF instruments have medium mass resolving power (<15,000) for both intact MS and fragmentation spectra acquired on the tandem TOF/TOF instruments (21), except for the combination of MALDI with the LTQ-Orbitrap.
How Accurate Is Accurate Enough?
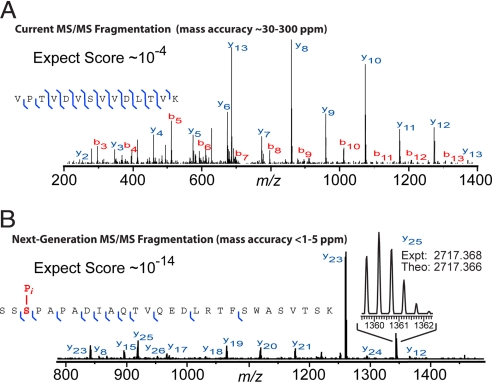
Surprisingly, the need for or even the desirability of high mass accuracy in proteomics has not been universally acknowledged (22, 23). However, the measured peptide mass acts as a filter that directly reduces the number of potential false positive assignments. With good scoring, higher mass accuracy proportionately increases certainty of identification, a concept that applies to intact peptides as well as their fragmentation products (vide infra). By using the new wave of LTQ-FT hybrids, obtaining low parts-per-million (ppm) numbers for intact peptides on a chromatographic time scale is now routine, with software that fully utilizes this accuracy now catching up with the intrinsic capabilities of the hardware. This is a huge advance compared with 3D ion trap measurements with mass uncertainty of several daltons, which corresponds to several thousand ppm. Intact peptides measured with accurate FTMS approaches by using either electrospray or MALDI have been shown to identify proteins in bacterial systems with an “accurate mass tag” approach to peptide mapping (24). However, the mass by itself, even when combined with the elution time, is normally not considered sufficient evidence for identification of the peptide. It is far more common to use ion trap-FT hybrids in a mode where mass spectra of the eluting peptides (i.e., “survey spectra” or MS1 spectra) are acquired at FT resolution and the MS2 (MS/MS) spectra are acquired at unit-resolution in the ion trap (Fig. 3A).
Fig. 3.
Fragmentation spectra for peptides interrogated by using two different workflows. (A) Fragmentation spectrum of a tryptic peptide recorded on a mainstream workflow in contemporary proteomics. Specifically, the intact peptides (not shown) are recorded at high resolution in a FT instrument, and the tandem mass spectrum (MS/MS) of a single tryptic peptide is recorded on a lower-resolution but faster linear quadrupole ion trap (LTQ) instrument during a LC-MS run. (B) Fragmentation spectrum recorded on a developmental workflow where fragment ions produced during MS/MS are recorded at FT resolution and <2 ppm mass accuracy. The high mass accuracy translates to high confidence in database retrieval, even when searching a complex database that includes modifications, polymorphisms, and alternative splicing. The high-resolution MS/MS data shown for a phosphopeptide in B was recorded on a Fourier-transform (LTQ FT) instrument during LC/MS/MS analysis of an endoproteinase Lys-C digestion of human cell line extract. The phosphorylation was stable through the standard MS/MS fragmentation process; it was readily detected and localized because it had been previously reported in the literature and actually stored in the database searched. (The actual phosphopeptide shown is the 230–257 segment from a Ras-GTPase activating protein, NCBI accession no. Q13283.)
How good should mass accuracy become? This question was answered in small-molecule MS a long time ago: it should be accurate enough to provide a unique chemical composition. Interestingly, recent high-resolution and high-accuracy proteomics studies have come within an order of magnitude of this goal, which requires a maximum mass deviation of ≈100 ppb for small tryptic peptides (23, 25). Grauman et al. (26) achieved ≈300 ppb as the average absolute mass deviation in a large-scale study of stem cells. For some of the smaller tryptic peptides, this already specifies the chemical composition. For larger tryptic peptides or those produced by more restrictive proteases (Figs. 1 and 3B), even higher mass accuracy is needed. However, the “database congestion” for larger peptides (>2 kDa) eases somewhat to often rule out all but one peptide sequence given the measured mass, at least for unmodified peptides. Model experiments have already indicated the potential for low ppb accuracy (27). Measurements around 30 ppb may soon become routine for well defined peaks. This would represent the advent of “quantized masses,” and would be a milestone achievement for MS-based proteomics. Note that even perfect mass accuracy does not prevent misidentification of related peptide sequences that differ by amino acid exchanges but leave the chemical composition unchanged (28). Importantly, peptide mass accuracy should be determined individually for each peptide to avoid degrading a high-accuracy instrument into a low-accuracy instrument in the process of database searching (23, 25).
Sequencing All Peptides: The Need for Speed
Ideally, all peptides visible to the mass spectrometer at its dynamic range should also be fragmented. This is still not the case, despite the subsecond fragmentation cycles of modern ion traps and TOF instruments (29), limiting comprehensiveness of analysis. It also causes part of the irreproducibility associated with shotgun proteomics because different subgroups of peptides are “picked for sequencing” in different LC-MS/MS analyses of the same sample. Furthermore, the acquisition software controlling the mass spectrometer does not exclusively target every eluting peptide for fragmentation once and only once. Instead, abundant peptides are fragmented multiple times, and low-level signals may never be targeted.
Currently, even the fastest mass spectrometers are incapable of comprehensively targeting each peptide signal for fragmentation. Replicate runs, especially with “exclusion lists” that prevent sequencing of previously fragmented peptides, partially alleviate this problem, albeit at the expense of measurement time. A combination of faster sequencing speed and more intelligent distribution of the available sequencing capacity should soon be able to target the great majority of peptides seen in shotgun experiments at the current dynamic range. Often, the peptides of interest are a small subgroup of all peptides. If these peptides can be targeted, the current sequencing speeds are already sufficient. This is enabled by duplicate analysis or—more elegantly—by the recently introduced “RePlay” technology. In RePlay, a part of the chromatographic effluent is split into a delay line allowing repeat analysis of the sample without decreasing signal levels or requiring additional sample material (30).
A Question of Sensitivity (and Dynamic Range)
Currently, peptide MS can achieve sensitivity in the attomole range and sometimes even below. Although ultimate sensitivity is important, dynamic range is even more crucial because many practical applications involve ample supplies of sample, but high-abundance proteins limit what can be observed with the given dynamic range of detection. For peptide detection in complex mixtures, this is currently in the range of 103 to 104. The dynamic range of protein detection is somewhat larger because different peptides from the same proteins have vastly different ionization efficiencies (31); therefore, very well detected peptides from low-abundance proteins sometimes make these low-level proteins detectable. The dynamic range needed to obtain a complete proteome has not yet been achieved for eukaryotic cells. A reasonable proteome coverage—approaching the coverage of DNA microarrays—is possible but presently only when combined with extensive fractionation of the proteome at the protein or peptide levels (32). As other areas of MS-based proteomics move closer to the desired performance, dynamic range in the mass spectrometer is clearly becoming the limiting factor. At least an order of magnitude improvement would be highly desirable, and two orders of magnitude (to 106) would be a major leap forward in proteomics technology. This would, for example, allow the identification of many posttranslationally modified peptides without specific enrichment—which in turn would enable the estimation of the stoichiometry of the modification, generally not possible for peptide-driven proteomics. Such additional dynamic range for peptide detection would increase the required speed for collecting fragmentation spectra by at least an order of magnitude. Alternatively, smarter mass spectrometers could direct their sequencing capacity toward peaks of particular interest (e.g., those indicating interesting abundance differences).
Accuracy in Protein Quantitation: The End of One-Size-Fits-All
One of the most important themes in current proteomics is the move toward quantitation (33). For almost all applications, proteomes need to be compared with each other—typically after one proteome is stimulated or otherwise perturbed with respect to the control proteome. As a half-way measure toward realizing true quantitation, “spectral counting” is currently widely applied (34–36). This concept relies on the aforementioned observation that peptides are targeted repeatedly for fragmentation by the mass spectrometer. The larger the number of detected peptides per protein and the more often they are fragmented, the more abundant the protein. A related concept, the “exponentially modified abundance index” (emPAI), divides the observed proteins by the observable peptides for each protein (i.e., peptides in the correct mass range for MS) (37) and does not depend on repeated targeting of the same peptide. These schemes are only an approximation to true quantitation, especially for low-abundance proteins with few peptides, for which differences become stochastic. This type of “label-free quantitation” separately quantifies the signal for each protein and compares these signals across separate experiments. It crucially depends on high mass resolving power and high mass accuracy, because peptides must not overlap in the mass spectra and the correct peptides must be matched to each other across experiments. If experimental conditions and purification procedures can be well controlled, label-free quantitation is an attractive strategy, because it requires no additional sample-preparation steps.
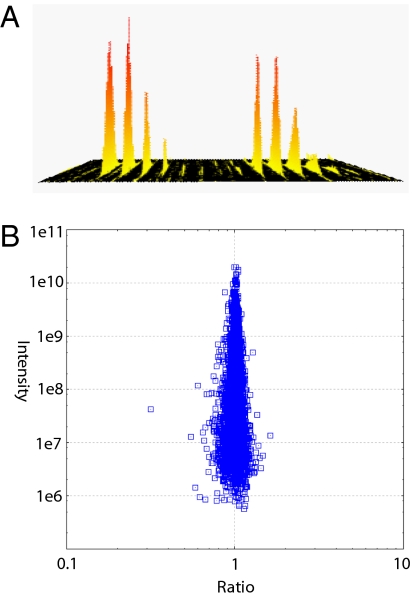
More precise quantitation can be achieved through the use of stable isotopes, introduced either chemically after sample collection or metabolically during cell or organismal growth. In chemical labeling, reactive groups are targeted with a reagent provided in either light or heavy isotope form. This has the advantage of being applicable to any protein source, and the original concept of comparing two proteomes has been multiplexed up to four or even eight. Disadvantages include chemical side-reactions and quantitation errors arising from separate processing of case and control proteomes. In metabolic labeling, dividing cells incorporate the label into their entire proteome. Of the two most common forms, 15N labeling is usually used for microorganisms or small metazoan (38, 39) whereas stable isotope labeling in cell culture (SILAC) is mostly used for mammalian cells (40, 41). SILAC labels one or two specific amino acids, making peptide pairs easy to identify by virtue of their known mass differences. In conjunction with high-resolution MS, SILAC quantitation of protein-expression ratios can be very accurate even in high-throughput experiments (Fig. 4).
Fig. 4.
Quantitative determination of a single peptide expression ratio (A), with a representation of thousands of such measurements (B) in a large-scale study of how embryonic stem cells differentiate. Two proteomes were mixed 1:1, and 4,668 proteins with at least three quantitation data points are plotted as a function of fold-change and summed peptide intensities (B). Excellent quantitation accuracy is observed with almost all proteins within 50% of ratio one. Significance of fold change is calculated by protein abundance. Data are from Graumann et al. (26). The quantitative distribution is narrower at 106 than at 107 because there are very few proteins in this abundance class, and these occupy the most probable states close to ratio one.
How accurate should MS-based quantitation be? Obviously, the more accurate the better; however, as in microarray experiments, biological replicates are often required, and these will introduce errors of their own. As a rule of thumb, MS-based proteomics should strive to be accurate within a 1.3- to 2-fold change, which is a cut-off often chosen for biological significance. Obviously, this depends on the experiment—for instance, a 2-fold accuracy is not sufficient for some biomarkers. Furthermore, more abundant proteins have more quantifiable peptides, and precision of quantitation is higher than for low-abundance proteins with few peptides. Thus, quantitation software should determine the significance of an observed fold-change in the context of absolute protein abundance.
Winds of Change: High-Resolution Tandem Mass Spectrometry
A few laboratories have started to acquire large-scale MS/MS data in addition to MS data with the ultrahigh mass accuracy afforded by FTMS (42, 43). This simple switch in collecting both intact MS and MS/MS fragmentation data in the FTMS has not been compared directly with the more widespread mode of “FT/ion trap” data acquisition—or with TOF instruments. Fig. 3 offers a glimpse at the current tradeoffs between sensitivity (speed) and resolving power. It is clear that the ion trap is faster for MS2 data acquisition but can lead to many recorded spectra without confident peptide identifications. In a technique called “higher energy dissociation,” ions are fragmented in the intermediate “C-trap” between linear ion trap and orbitrap (44). This dissociation mode does not have a low mass cut-off and fragments are analyzed at high resolution in the orbitrap. Another fragmentation method, electron transfer dissociation (ETD) (45)—a relative of electron capture dissociation (ECD) (46)—provides a fragmentation pattern complementary to the normal ion trap dissociation and has also now been coupled to high-resolution “read out” in the orbitrap.
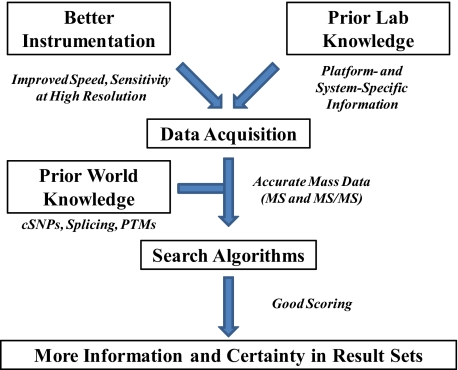
The great virtue of high-resolution tandem mass spectra is the orders-of-magnitude better specificity for searching databases (Fig. 3B), even those containing known modifications (vide infra). Software is increasingly able to harness the high-resolution fragmentation data now generated by using “bottom-up” (tryptic digestion) or “top-down” (intact protein) strategies. The use of accurate mass MS/MS data will allow better error-tolerant searching, more reliable determination of diverse modifications, and reliable “multiplexing” of identifications (i.e., identifying more than one peptide in a MS/MS spectrum). Better determination and tracking of MS1 peptide masses are also needed to capture the full information content of complex LC-MS/MS analyses in the context of large-scale proteome projects. Thus, top-down and proteolysis-driven approaches using high resolution, coupled with improving software for data acquisition and processing, are ongoing trends for evolution of the field (see Fig. 5).
Fig. 5.
Conceptual model for the ongoing development of MS-based proteomics. Along with improving hardware, smarter software at run time and embedding known information on protein variation into database searching are all trends for the future.
Sequencing Larger Peptides and Top-Down Proteomics
Driven by interest in detecting combinations of posttranslational modifications and improving instrumentation (16, 47), momentum is building to increase the size of peptides or small proteins analyzed by LC-MS/MS. This “middle-down” concept combines aspects of both top-down and bottom-up strategies and can be as simple as changing the protease from trypsin to Lys-C or Glu-C (e.g., Fig. 3 A vs. B). Here, peptides longer than ≈20 residues (48, 49) or large endogenous peptides (42) are sought for analysis to increase the sequence coverage provided by each successfully identified peptide.
The accurate mass MS/MS approach has been mostly associated with the rise of top-down MS. Given that intact proteins tend to have lower ion signals vs. the best responding small peptides, the combination of custom FTMS instruments with intact protein fragmentation was a good temporal match. With the speed and sensitivity of FTMS improving significantly in past years, top-down LC-MS/MS has recently become possible on complex mixtures of yeast (50) and human proteins (51). For top-down MS/MS, identification of 20–30 proteins ranging from 5–40 kDa during a single LC-MS/MS run is now possible using commercial instrumentation (50). The performance gap between the top-down approach and the bottom-up approach (which is able to identify many hundreds of unique peptides in a single LC/MS injection) will take some time to close. However, because the top-down approach does not involve proteolysis, the proteome is not obfuscated by the creation of exceedingly complicated peptide mixtures.
Computational Proteomics: The Move Toward Posttranslational Modifications
Although well over a decade has passed since the first demonstration of automated protein identification by database retrieval, many laboratories still encounter a bottleneck with this component of modern proteomics. No one search algorithm dominates. In fact, an entire January 2008 issue of the Journal of Proteome Research was dedicated to statistical and computational proteomics, with leading laboratories often having significant internal expertise.
Recently, a move to include known protein variation within databases has begun. Predicted in 2001 (52) and 2003 (53), large bioinformatic efforts are underway, including UniProt, which has established a hierarchical ontology for posttranslational modifications called “UniMOD,” based largely on the RESID database (54). Newer databases including HPRD [now called Proteopedia (55)] are also becoming available. However, incorporation of this knowledge into MS search engines is far less developed. Recently, cSNPs have been put into databases embedded within mass spectral search engines (56, 57). Some time ago, SONAR demonstrated identification of peptides that spanned exon–exon boundaries. Thousands of posttranslational modifications, polymorphisms, and alternative splicing patterns become known yearly. Software meant for primary searching of mass spectral data should be aware of such variations, and with each passing year this becomes a better approach toward comprehensive proteomics in model eukaryotes. As mass spectrometers become increasingly capable of acquiring MS data at low- to sub-ppm accuracy, appropriate software is maturing to capture the value of such data to interrogate eukaryotic proteins—in all of their complexity—with improved certainty (Fig. 5, bottom).
High mass accuracy allows much more variability of proteins (vide supra) to be “locked down” during database searches (56). Current-generation FTMS instruments can record fragmentation data with better than 5 ppm mass accuracy routinely. This performance is adequate for searching complex databases containing modifications, polymorphisms, and their combinations. In high-resolution MS/MS measurements, the central issue is no longer mass measurement accuracy but rather detecting a high number of fragment ions (Fig. 3B) at the speed and sensitivity similar to that of ion traps (Fig. 3A). ProSight is the first high-throughput search environment to support error-tolerant proteomics based on resolved isotopic distributions for 3- to 40-kDa fragment ions, and uses a candidate expansion approach in a preconstructed database to expose known proteomic information for primary database searches (58). Such databases mimic the diversity of real proteomes by housing combinations of known protein information (from parsed Swiss-Prot flat-files), including posttranslational modifications, alternative splice forms, endogenous peptide cleavages, and polymorphisms.
Summary and Outlook
High-resolution measurements are becoming not only feasible but also necessary for confident, contemporary proteomics. Despite seemingly higher complexity and cost, precision proteomics can avoid errors that may otherwise lead to years of misdirected work. Decisive progress has been made in peptide identification, although continued progress on mass accuracy, peptide sequencing speed, and identification/scoring algorithms is still necessary. Dynamic range of measurement would greatly benefit from at least an order of magnitude advance. Increasingly, protein quantitation should move toward label free or stable isotope formats. A huge challenge will be to “roll out” the level of performance achieved in a few leading laboratories on a much broader scale. However, given the accelerating pace of the last few years, precision proteomics—deriving from core advances in mass spectrometry—will contribute to an overall increase in the efficiency of moving biological science forward. This reflects a general thread throughout this special issue of PNAS, which captures several examples of how measurement of one of the most fundamental properties—mass—drives the faster and better acquisition of new knowledge about the physical world around us.
Acknowledgments.
We thank Mingxi Li and Michael Boyne for assistance with figures and for enabling the research performed in N.L.K.'s laboratory, increasingly using mass spectrometry with high mass accuracy, and Fred McLafferty for the opportunity to contribute to this special issue. Jürgen Cox prepared Fig. 4 using MaxQuant software. This work was supported by National Institutes of Health Grant GM 067193-06 and the University of Illinois Institute for Genomic Biology (N.L.K.). Work in M.M.'s laboratory is partly supported by the Sixth Framework Programme of the European Union Commission (“Interaction Proteome” and HEROIC).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
References
- 1.O'Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250:4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 2.Fountoulakis M, Tsangaris G, Oh JE, Maris A, Lubec G. Protein profile of the HeLa cell line. J Chromatogr A. 2004;1038:247–265. doi: 10.1016/j.chroma.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 3.Campostrini N, et al. Spot overlapping in two-dimensional maps: A serious problem ignored for much too long. Proteomics. 2005;5:2385–2395. doi: 10.1002/pmic.200401253. [DOI] [PubMed] [Google Scholar]
- 4.Petrak J, et al. Deja vu in proteomics. A hit parade of repeatedly identified differentially expressed proteins. Proteomics. 2008;8:1744–1749. doi: 10.1002/pmic.200700919. [DOI] [PubMed] [Google Scholar]
- 5.Angenendt P, Kreutzberger J, Glokler J, Hoheisel JD. Generation of high density protein microarrays by cell-free in situ expression of unpurified PCR products. Mol Cell Proteomics. 2006;5:1658–1666. doi: 10.1074/mcp.T600024-MCP200. [DOI] [PubMed] [Google Scholar]
- 6.He M, Stoevesandt O, Taussig MJ. In situ synthesis of protein arrays. Curr Opin Biotechnol. 2008;19:4–9. doi: 10.1016/j.copbio.2007.11.009. [DOI] [PubMed] [Google Scholar]
- 7.Petricoin EF, Zoon KC, Kohn EC, Barrett JC, Liotta LA. Clinical proteomics: Translating benchside promise into bedside reality. Nat Rev Drug Discovery. 2002;1:683–695. doi: 10.1038/nrd891. [DOI] [PubMed] [Google Scholar]
- 8.Jensen ON. Interpreting the protein language using proteomics. Nat Rev Mol Cell Biol. 2006;7:391–403. doi: 10.1038/nrm1939. [DOI] [PubMed] [Google Scholar]
- 9.Kocher T, Superti-Furga G. Mass spectrometry-based functional proteomics: From molecular machines to protein networks. Nat Methods. 2007;4:807–815. doi: 10.1038/nmeth1093. [DOI] [PubMed] [Google Scholar]
- 10.Cravatt BF, Simon GM, Yates JR., III The biological impact of mass-spectrometry-based proteomics. Nature. 2007;450:991–1000. doi: 10.1038/nature06525. [DOI] [PubMed] [Google Scholar]
- 11.Wolters DA, Washburn MP, Yates JR., III An automated multidimensional protein identification technology for shotgun proteomics. Anal Chem. 2001;73:5683–5690. doi: 10.1021/ac010617e. [DOI] [PubMed] [Google Scholar]
- 12.Comisarow MB, Marshall AG. The early development of Fourier transform ion cyclotron resonance (FT-ICR) spectroscopy. J Mass Spectrom. 1996;31:581–585. doi: 10.1002/(SICI)1096-9888(199606)31:6<581::AID-JMS369>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 13.Syka JE, et al. Novel linear quadrupole ion trap/FT mass spectrometer: Performance characterization and use in the comparative analysis of histone H3 post-translational modifications. J Proteome Res. 2004;3:621–626. doi: 10.1021/pr0499794. [DOI] [PubMed] [Google Scholar]
- 14.Schwartz JC, Senko MW, Syka JE. A two-dimensional quadrupole ion trap mass spectrometer. J Am Soc Mass Spectrom. 2002;13:659–669. doi: 10.1016/S1044-0305(02)00384-7. [DOI] [PubMed] [Google Scholar]
- 15.Makarov A. Electrostatic axially harmonic orbital trapping: A high-performance technique of mass analysis. Anal Chem. 2000;72:1156–1162. doi: 10.1021/ac991131p. [DOI] [PubMed] [Google Scholar]
- 16.Hu Q, et al. The Orbitrap: A new mass spectrometer. J Mass Spectrom. 2005;40:430–443. doi: 10.1002/jms.856. [DOI] [PubMed] [Google Scholar]
- 17.Makarov A, et al. Performance evaluation of a hybrid linear ion trap/orbitrap mass spectrometer. Anal Chem. 2006;78:2113–2120. doi: 10.1021/ac0518811. [DOI] [PubMed] [Google Scholar]
- 18.Purvine S, Eppel JT, Yi EC, Goodlett DR. Shotgun collision-induced dissociation of peptides using a time of flight mass analyzer. Proteomics. 2003;3:847–850. doi: 10.1002/pmic.200300362. [DOI] [PubMed] [Google Scholar]
- 19.Venable JD, Dong MQ, Wohlschlegel J, Dillin A, Yates JR. Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nat Methods. 2004;1:39–45. doi: 10.1038/nmeth705. [DOI] [PubMed] [Google Scholar]
- 20.Wolf-Yadlin A, Hautaniemi S, Lauffenburger DA, White FM. Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc Natl Acad Sci USA. 2007;104:5860–5865. doi: 10.1073/pnas.0608638104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Domon B, Aebersold R. Mass spectrometry and protein analysis. Science. 2006;312:212–217. doi: 10.1126/science.1124619. [DOI] [PubMed] [Google Scholar]
- 22.Haas W, et al. Optimization and use of peptide mass measurement accuracy in shotgun proteomics. Mol Cell Proteomics. 2006;5:1326–1337. doi: 10.1074/mcp.M500339-MCP200. [DOI] [PubMed] [Google Scholar]
- 23.Zubarev R, Mann M. On the proper use of mass accuracy in proteomics. Mol Cell Proteomics. 2007;6:377–381. doi: 10.1074/mcp.M600380-MCP200. [DOI] [PubMed] [Google Scholar]
- 24.Conrads TP, Anderson GA, Veenstra TD, Pasa-Tolic L, Smith RD. Utility of accurate mass tags for proteome-wide protein identification. Anal Chem. 2000;72:3349–3354. doi: 10.1021/ac0002386. [DOI] [PubMed] [Google Scholar]
- 25.Zubarev R, Harkansson P, Sundqvist B. Accuracy requirements for peptide characterization by monoisotopic molecular mass measurements. Anal Chem. 1996;68:4060–4063. [Google Scholar]
- 26.Graumann J, et al. Stable isotope labeling by amino acids in cell culture (SILAC) and proteome quantitation of mouse embryonic stem cells to a depth of 5,111 proteins. Mol Cell Proteomics. 2008;7:672–683. doi: 10.1074/mcp.M700460-MCP200. [DOI] [PubMed] [Google Scholar]
- 27.Schaub TM, et al. High-performance mass spectrometry: Fourier transform ion cyclotron resonance at 14.5 Tesla. Anal Chem. 2008;80:3985–3990. doi: 10.1021/ac800386h. [DOI] [PubMed] [Google Scholar]
- 28.He F, Emmett MR, Hakansson K, Hendrickson CL, Marshall AG. Theoretical and experimental prospects for protein identification based solely on accurate mass measurement. J Proteome Res. 2004;3:61–67. doi: 10.1021/pr034058z. [DOI] [PubMed] [Google Scholar]
- 29.Kuster B, Schirle M, Mallick P, Aebersold R. Scoring proteomes with proteotypic peptide probes. Nat Rev Mol Cell Biol. 2005;6:577–583. doi: 10.1038/nrm1683. [DOI] [PubMed] [Google Scholar]
- 30.Waanders LF, et al. A novel chromatographic method allows on-line reanalysis of the proteome. Mol Cell Proteomics. 2008;7:1452–1459. doi: 10.1074/mcp.M800141-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mallick P, et al. Computational prediction of proteotypic peptides for quantitative proteomics. Nat Biotechnol. 2007;25:125–131. doi: 10.1038/nbt1275. [DOI] [PubMed] [Google Scholar]
- 32.Cox J, Mann M. Is proteomics the new genomics? Cell. 2007;130:395–398. doi: 10.1016/j.cell.2007.07.032. [DOI] [PubMed] [Google Scholar]
- 33.Ong SE, Mann M. Mass spectrometry-based proteomics turns quantitative. Nat Chem Biol. 2005;1:252–262. doi: 10.1038/nchembio736. [DOI] [PubMed] [Google Scholar]
- 34.Liu H, Sadygov RG, Yates JR., III A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 35.Lu P, Vogel C, Wang R, Yao X, Marcotte EM. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol. 2007;25:117–124. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- 36.Sardiu ME, et al. Probabilistic assembly of human protein interaction networks from label-free quantitative proteomics. Proc Natl Acad Sci USA. 2008;105:1454–1459. doi: 10.1073/pnas.0706983105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ishihama Y, et al. Exponentially modified protein abundance index (emPAI) for estimation of absolute protein amount in proteomics by the number of sequenced peptides per protein. Mol Cell Proteomics. 2005;4:1265–1272. doi: 10.1074/mcp.M500061-MCP200. [DOI] [PubMed] [Google Scholar]
- 38.Krijgsveld J, et al. Metabolic labeling of C. elegans and D. melanogaster for quantitative proteomics. Nat Biotechnol. 2003;21:927–931. doi: 10.1038/nbt848. [DOI] [PubMed] [Google Scholar]
- 39.Li L, et al. Quantitative proteomic and microarray analysis of the archaeon Methanosarcina acetivorans grown with acetate versus methanol. J Proteome Res. 2007;6:759–771. doi: 10.1021/pr060383l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 41.Mann M. Functional and quantitative proteomics using SILAC. Nat Rev Mol Cell Biol. 2006;7:952–958. doi: 10.1038/nrm2067. [DOI] [PubMed] [Google Scholar]
- 42.Taylor S, et al. Efficient high-throughput discovery of large peptidic hormones and biomarkers. J Proteome Res. 2006;5:1776–1784. doi: 10.1021/pr0600982. [DOI] [PubMed] [Google Scholar]
- 43.Falth M, et al. SwedCAD, a database of annotated high-mass accuracy MS/MS spectra of tryptic peptides. J Proteome Res. 2007;6:4063–4067. doi: 10.1021/pr070345h. [DOI] [PubMed] [Google Scholar]
- 44.Olsen JV, et al. Higher-energy C-trap dissociation for peptide modification analysis. Nat Methods. 2007;4:709–712. doi: 10.1038/nmeth1060. [DOI] [PubMed] [Google Scholar]
- 45.Syka JE, Coon JJ, Schroeder MJ, Shabanowitz J, Hunt DF. Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc Natl Acad Sci USA. 2004;101:9528–9533. doi: 10.1073/pnas.0402700101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zubarev RA, et al. Electron capture dissociation for structural characterization of multiply charged protein cations. Anal Chem. 2000;72:563–573. doi: 10.1021/ac990811p. [DOI] [PubMed] [Google Scholar]
- 47.Olsen JV, et al. Parts per million mass accuracy on an Orbitrap mass spectrometer via lock mass injection into a C-trap. Mol Cell Proteomics. 2005;4:2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 48.Forbes A, Mazur M, Patel H, Walsh C, Kelleher N. Toward efficient analysis of >70 kDa proteins with 100% sequence coverage. Proteomics. 2001;1:927–933. doi: 10.1002/1615-9861(200108)1:8<927::AID-PROT927>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 49.Wu S, et al. Dynamic profiling of the post-translational modifications and interaction partners of epidermal growth factor receptor signaling after stimulation by epidermal growth factor using extended range proteomic analysis (ERPA) Mol Cell Proteomics. 2006;5:1610–1627. doi: 10.1074/mcp.M600105-MCP200. [DOI] [PubMed] [Google Scholar]
- 50.Parks BA, et al. Top-down proteomics on a chromatographic time scale using linear ion trap Fourier transform hybrid mass spectrometers. Anal Chem. 2007;79:7984–7991. doi: 10.1021/ac070553t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Roth M, Parks B, Ferguson J, Li M, Kelleher N. “Proteotyping”: Population proteomics of human leukocytes using top down mass spectrometry. Anal Chem. 2008;80:2857–2866. doi: 10.1021/ac800141g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Meng F, et al. Informatics and multiplexing of intact protein identification in bacteria and the archaea. Nat Biotechnol. 2001;19:952–957. doi: 10.1038/nbt1001-952. [DOI] [PubMed] [Google Scholar]
- 53.Mann M, Jensen ON. Proteomic analysis of post-translational modifications. Nat Biotechnol. 2003;21:255–261. doi: 10.1038/nbt0303-255. [DOI] [PubMed] [Google Scholar]
- 54.Garavelli JS. The RESID Database of Protein Modifications as a resource and annotation tool. Proteomics. 2004;4:1527–1533. doi: 10.1002/pmic.200300777. [DOI] [PubMed] [Google Scholar]
- 55.Mathivanan S, et al. Human Proteinpedia enables sharing of human protein data. Nat Biotechnol. 2008;26:164–167. doi: 10.1038/nbt0208-164. [DOI] [PubMed] [Google Scholar]
- 56.Roth MJ, et al. Precise and parallel characterization of coding polymorphisms, alternative splicing, and modifications in human proteins by mass spectrometry. Mol Cell Proteomics. 2005;4:1002–1008. doi: 10.1074/mcp.M500064-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schandorff S, et al. A mass spectrometry-friendly database for cSNP identification. Nat Methods. 2007;4:465–466. doi: 10.1038/nmeth0607-465. [DOI] [PubMed] [Google Scholar]
- 58.LeDuc RD, et al. Using ProSight PTM and related tools for targeted protein identification and characterization with high mass accuracy tandem MD data. Curr Prot Bioinf. 2007;13.6:1–28. doi: 10.1002/0471250953.bi1306s19. [DOI] [PubMed] [Google Scholar]