Abstract
Oxidoreduction in ferritin protein nanocages occurs at sites that bind two Fe(II) substrate ions and O2, releasing Fe(III)2–O products, the biomineral precursors. Diferric peroxo intermediates form in ferritins and in the related diiron cofactor oxygenases. Cofactor iron is retained at diiron sites throughout catalysis, contrasting with ferritin. Four of the 6 active site residues are the same in ferritins and diiron oxygenases; ferritin-specific Gln137 and variable Asp/Ser/Ala140 substitute for Glu and His, respectively, in diiron cofactor active sites. To understand the selective functions of diiron substrate and diiron cofactor active site residues, we compared oxidoreductase activity in ferritin with diiron cofactor residues, Gln137 → Glu and Asp140 → His, to ferritin with natural diiron substrate site variations, Asp140, Ser140, or Ala140. In Gln137 → Glu ferritin, diferric peroxo intermediates were undetectable; an altered Fe(III)–O product formed, ΔA350 = 50% of wild type. In Asp140 → His ferritin, diferric peroxo intermediates were also undetectable, and Fe(II) oxidation rates decreased 40-fold. Ferritin with Asp140, Ser140, or Ala140 formed diferric peroxo intermediates with variable kinetic stabilities and rates: t1/2 varied 1- to 10-fold; kcat varied approximately 2- to 3-fold. Thus, relatively small differences in diiron protein catalytic sites determine whether, and for how long, diferric peroxo intermediates form, and whether the Fe–active site bonds persist throughout the reaction cycle (diiron cofactors) or break to release Fe(III)2–O products (diiron substrates). The results and the coding similarities for cofactor and substrate site residues—e.g., Glu/Gln and His/Asp pairs share 2 of 3 nucleotides—illustrate the potential simplicity of evolving active sites for diiron cofactors or diiron substrates.
Keywords: diiron protein catalysts, dioxygen, iron biominerals, protein nanocages, Differic peroxo
Ferritins are protein nanocages with multiple sites that catalyze the oxidoreduction of Fe(II) and O2 to produce diferric mineral precursors; the mineral precursors migrate into a central cavity 8 nm in diameter and form Fe(III) hydrated oxo mineral. Iron minerals in ferritins serve to concentrate iron to the high levels needed for the synthesis of iron-containing cofactors. The Fe(II) oxidation reactions in ferritins consume dioxygen or hydrogen peroxide, thereby minimizing dangerous Fenton chemistry (1). Ferritin protein nanocages self-assemble from 24 or 12, 4-α-helix-bundle, subunits in maxiferritin or miniferritin, respectively; miniferritins were named, historically, Dps (DNA protection during starvation) proteins and, to date, are known only in bacteria and archaea; heme-containing ferritins in bacteria are called bacterioferritins (Bfr) (2). The biological importance of ferritin is reflected by wide distribution in most cells of humans, other animals, bacteria, and archaea, by the lethality of gene deletion in mice (3), by mutation effects on the central nervous system (4), and by protection during oxidant stress (5). In addition, the unusual physical stability of ferritin nanocages to temperature and chaotropes has application to materials and protein engineering (6, 7).
Oxidoreductase/ferroxidase (Fox) catalytic centers with diferrous binding sites, Fe1 and Fe2, occur in each ferritin subunit except in animals, where a second gene encodes a catalytically inactive subunit (ferritin L) that co-assembles with the active subunits in various ratios that depend on the tissue. The ferritin oxidoreductase sites share properties with diiron cofactor catalysts that include the first detectable reaction intermediate in ferritin, a diferric peroxo (DFP) complex, characterized by UV/visible (UV/vis), resonance Raman, Mössbauer, and extended X-ray absorption fine structure (EXAFS) spectroscopies (8–14). The reaction in ferritins is shown in Scheme 1.
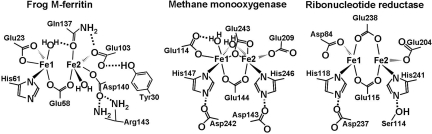
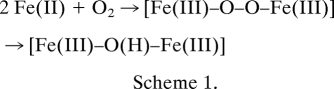 |
DFP decays to Fe(III) oxo or hydroxo dimers or multimers (15), representing reaction products from multiple sites, that move across the protein cage to the cavity where the Fe(III) oxide mineral forms. Parallel formation of diferric dimers and multimers (15), the effects on reaction rates of covalent crosslinks that decrease conformational flexibility (16), and the properties of Fe(II) binding measured directly by circular dichroism/magnetic circular dichroism (CD/MCD) (17) all suggest interactions among the polypeptide subunits during the reactions at the multiple catalytic centers in ferritin protein cages. The active site of a single ferritin polypeptide subunit, based on protein co-crystals with the Fe(II) analogue Mg(II) (18) and recent CD/MCD analysis (17) is shown in Fig. 1. Similar results for identification of the metal-binding amino acid residues, or ligands, were obtained with a Ca(II) co-crystal of ferritin (19) and with a solution study of an L ferritin [no Fe(II)/O2 oxidoreductase catalytic center] where the putative catalytic center ligands replaced the wild-type (WT) residues (20). A distinguishing feature of the ferritin diiron substrate catalytic centers is the weaker Fe(II) ligands in the Fe2 site that contrast with diiron cofactor catalytic centers, in proteins such as methane monooxygenase (MMO), ribonucleotide reductase (RNR) and Δ9-stearoyl-acyl carrier protein desaturase (Δ9 desaturase) (21–23) and the heme-containing Bfr (2). Ferritin diiron substrate centers also lack one of the two carboxylate ligands of the diiron cofactor sites, which, like iron in the ferritin substrate sites, react with O2 to form a DFP intermediate (8–10).
Fig. 1.
Comparisons of diiron sites for Fe substrate (ferritin) and cofactors reveal significant differences in iron ligands and second-shell residues. The ferritin Fe2 site ligands are Glu103, Gln137, and Asp140, with a bridging carboxylate, Glu58. The distinctive ferritin Gln137 is compared with Glu243 in methane monooxygenase (MMO) and Glu238 in ribonucleotide reductase (RNR). Multiple histidine bonding interactions in diiron cofactor centers are absent from ferritin diiron substrate centers. The figure uses the frog M ferritin model [Protein Data Bank (PDB) file 1MFR, a crystal structure with Mg(II) as an Fe(II) homologue (18)] and CD/MCD analysis of diferrous binding (17). Diiron cofactor site structures include diferrous, as in MMO (1FYZ) (21) and RNR (1PFR) (22).
Ferritins are isoforms that vary not only in overall polypeptide sequences but also in the sequence at the catalytic centers. The selective expression of the different ferritin isoforms in specific tissues or organelles indicates that the variations are physiologically relevant (24, 25). Four of the 6 iron ligands at the Fox catalytic centers of ferritins are present in diiron enzyme cofactor centers (Fig. 1), except in Bfr with all six diiron cofactor site ligands. The two divergent residues at the ferritin catalytic centers are both in the Fe2 site: a conserved glutamine at position 137* and a variable residue at position 140; in Fig. 1, residue 140 is aspartate from the extensively characterized frog M ferritin model.
We now report the results of investigating the function of the ferritin-specific residues in the Fe2 site of ferritin catalytic centers by comparing the catalytic properties of natural variants in residue 140, aspartate, serine, or alanine, in a single polypeptide sequence (frog M) to that in the natural polypeptide (frog M, Asp140; frog H, Ser140; and human H, Ala140). We also examined the diiron cofactor residue, histidine, in the ferritin context. We observed that kcat, and Kapp for DFP formation, and DFP decay (t1/2), determined from the progress curves of A650, were sensitive both to residue 140 and to the polypeptide backbone, suggesting coevolution of second-shell or distant residues, while histidine inhibited Fe(II) oxidation in the ferritin polypeptide background. Changing ferritin-specific invariant glutamine to glutamate inhibited DFP formation (A650) at either 20 °C or 4 °C but not Fe(II) oxidation, measured by A350, although the Fe(III)O product(s) had altered spectral properties. The absorbance at 350 nm is not a specific transition but is in the middle of the broad absorbance at 300–400 nm, due to Fe(III)O species that include DFP, diferric Fe(III) oxo/hydroxo mineral precursors, and Fe(III) oxide mineral with large absorbance of protein aromatic amino acids between 280 and 300 nm. The results illustrate the importance of ferritin Fe2 site-specific residue DFP formation and in directing DFP decay away from higher-valence iron species, such as Fe(IV), as in diiron cofactor proteins, and toward the diferric mineral precursors.
Results
Natural Asp140 Variants in Ferritin Fe2 Site Alter Catalytic Rates.
The variations at residue 140 in the Fe2 site of the catalytic centers (1, 20, 25–27) are inconsistent with the high conservation of the other residues as well as the conservation of histidine at the analogous position in the catalytic centers of related diiron oxygenases. The variations in residue 140 at the ferritin Fe2 sites appear to be physiologically relevant based on patterns of expression (24, 25); the physiological parameter driving the selection is unknown, but it could be temperature, oxygen concentration, or supramolecular interactions with other cellular components that are cell or organelle specific. To examine the effects of residue 140 variants, we compared the activity of proteins with ferritin Fe2 site Asp140Ala (Fe site2 with Glu103, Gln137, and Ala140; EQA) and Asp140Ser (EQS) in the same ferritin backbone, the frog M-ferritin subunit, to the WT, Asp140 (EQD).
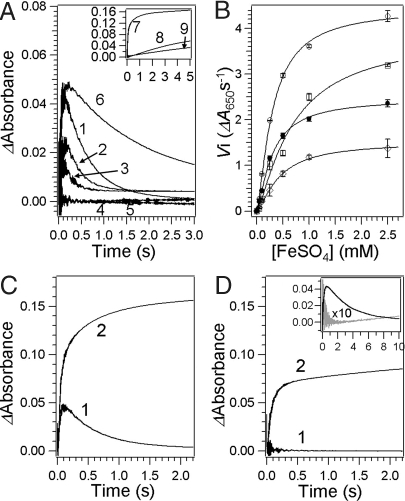
Rapid Fe(II) oxidation with DFP formation was observed with all natural residue 140 variants, measured either as the increase in A650, at the absorption maximum of ferritin DFP (8, 9, 15, 28) (Fig. 2A), or as the increase in A350, the less specific Fe(III)O species described above (Table 1). Fitting the data for initial rates at different concentrations of iron showed positive cooperativity (Fig. 2B), as previously observed with WT protein (20). Note that the Hill coefficient for DFP formation is smaller than for the binding of Fe(II) in the absence of O2 (n ∼ 3) (17), indicating that the basis for cooperativity is different for binding of the first substrate, Fe(II), to the protein in the absence of the second substrate, O2, compared with the reaction between Fe(II) and the second substrate, O2.
Fig. 2.
Effects of different Fe2 site residues with natural ferritin variants or with diiron oxygenase ligands, on rates in ferritin. (A) Progress curves of A650. Apoferritin (0.1 mM subunit) (4.2 μM nanocages) in 200 mM Mops, 200 mM NaCl at pH 7.0 was mixed with an equal volume of 0.2 mM ferrous sulfate (48 Fe per nanocage) in 1 mM HCl, at room temperature (20 °C) in a stopped-flow spectrophotometer (mixing time <5 ms). Traces are from the following: 1, WT frog M (EQD); 2, EQA; 3, EQS; 4, EQH; 5, EEH; and 6, WT human H (EQA). (Inset) Time course of A350 for the following: 7, WT frog M (EQD); 8, EQH; and 9, EEH. (B) The initial rate (Vi) for DFP formation as a function of iron concentration. The conditions were the same as for A. The curves represent the results of the Hill equation fitting. ○, WT frog M (EQD); □, EQA; ◇, EQS; and ●, WT human. (C and D) Effect of Gln137 → Glu substitution. The mixing conditions were the same as for A and B. (C) Progress curve for WT frog M (EQD). (D) Progress curve for EED. Traces shown here are 1, A650; and 2, A350. (Inset) Kinetic traces obtained at 4 °C for WT frog M (EQD) (black) and EED (gray). The trace for EED was multiplied by 10 for comparison.
Table 1.
Effects of altering Fe2 sites in ferritin Fe(II)/O2 oxidoreductase catalysis
| Protein |
Single turnover |
Multiple turnover (48–480 Fe per protein cage) |
|||||
|---|---|---|---|---|---|---|---|
| DFP (A650) |
DFP (A650) |
Fe3+O formation (A350) |
|||||
| Fe2 site and backbone* | t1/2 (ln2/kdecay),†s | kcat,§ s−1 | Kapp, mM | n | Max Vi,‖ Δ A350·s−1 | Kapp, mM | n |
| EQD-Fr M | 0.37 ± 0.02 | 89 ± 2 | 0.20 ± 0.02 | 1.4 ± 0.1 | 7.4 ± 0.2 | 0.13 ± 0.01 | 1.6 ± 0.1 |
| EQS-Fr M | 0.15 ± 0.01 | 30 ± 2 | 0.32 ± 0.12 | 1.4 ± 0.2 | 5.8 ± 0.2 | 0.55 ± 0.08 | 1.4 ± 0.1 |
| EQA-Fr M | 0.21 ± 0.02 | 77 ± 7 | 0.64 ± 0.17 | 1.3 ± 0.2 | 7.5 ± 0.2 | 0.62 ± 0.05 | 1.3 ± 0.1 |
| EQS-Fr H¶ | — | 25 ± 6 | 0.20 ± 0.03 | 1.6 ± 0.1 | — | — | — |
| EQA-Hu H | 1.5 ± 0.2 | 49 ± 7 | 0.20 ± 0.02 | 1.3 ± 0.1 | 4.1 ± 0.1 | 0.18 ± 0.03 | 1.5 ± 0.1 |
| EQH-Fr M | — | — | — | — | 0.33 ± 0.10 | 1.4 ± 1.3 | 0.72 ± 0.29 |
| EEH-Fr M | — | — | — | — | 0.20 ± 0.04 | 0.81 ± 0.56 | 0.71 ± 0.19 |
| EED-Fr M | — | — | — | — | 8.6 ± 0.2 | 0.37 ± 0.06 | 1.2 ± 0.1 |
*Single-letter amino acid code for Fe2 sites in ferritins with polypeptide backbone as: human (Hu) or frog (Fr). Boldface letters indicate amino acids present in the Fe2 site of ferritins and also in diiron cofactor active sites.
†t1/2 values were obtained by single-exponential fitting to DFP decay process of A650; 48 Fe/protein cage (2Fe/active site). The results compare, within experimental error, to double exponential fitting (data not shown) for all the proteins except EQS-Fr M and EQA-FrM where t1/2, using double exponential fitting, is 0.069 ± 0.04 and 0.14 ± 0.04, respectively but the fitting quality, judged by nonrandom residuals, is poor. Regardless of the model used to fit the data, t1/2 was significantly different (P<0.01) in the mutants and wild type Hu H compared to Fr M.
§The value for kcat is reported/site (24 sites/protein nanocage) for comparison to single site diiron oxygenases, but contrasting with values per ferritin nanocage reported previously (20), which are 24-fold higher. The values presented were computed assuming a constant 1,000 M−1·cm−1 (9) as before (20). Using double exponential fitting as in (9) to compute ε, the values were within experimental of the reported value, ε = 1040 ± 80, 960 ± 140 and 1060 ± 80 M−1·cm−1 for frog M WT, D140A and human H WT, respectively, except for EQS where ε = 720 ± 120; non-random residuals indicated that the goodness of fits were poorer for the double exponential fitting model. Nevertheless, kcat values results with each fitting model were within 10% of each other except for EQS where the vaues differed by 30%. All the computed values of kcat were significantly different (P<0.01) in the mutants, EQS FrM, EQA, Fr M, and in EQS-Fr H and EQA-Hu H compared to EQD-Fr M.
‖Maximum Vi (ΔA350 nm·s−1) is used instead of Vmax because the spectra of the Fe(III)O species (DFP, diferric product, mineral) have not, to date, been spectroscopically resolved.
¶Data from ref. 20.
The initial rates and the kinetic parameters such as kcat, Kapp, and decay of DFP (t1/2) were dependent on the Fe2 residue 140 (Fig. 2B and Table 1). Asp140 was the most efficient among natural variants in residue 140 with frog M backbone, which is the WT combination. DFP kinetic stability, t1/2 values, varied only slightly among the proteins with variable residues at position 140 (Fig. 2A and Table 1), Asp140, 0.34 ± 0.02 s; Ala140, 0.21 ± 0.02 s; and Ser140, 0.15 ± 0.01 s. The kcat value in the Ala140 ferritin was comparable to WT (Asp140), but kcat for Ser140 was decreased to less than 50% of WT. Among the three types of catalytic centers, the order of Kapp, Asp140 < Ser140 < Ala140, suggests that Fe(II)/O2 binding was stabilized by a ligand and/or longer side chain. Slower DFP formation, higher Kapp [lower affinity for Fe(II)], smaller kcat, and faster DFP decay result in lower accumulations of DFP and can explain the lower peak absorbance in Ala140 or Ser140 than Asp140 (Fig. 2A).
When the same measurements were made in proteins where residue 140 was matched to the homologous polypeptide backbone, the results contrasted with those in the frog M polypeptide, illustrating the contribution of the polypeptide backbone. For example, Kapp was constant among WT ferritins, independent of residue 140 for frog M (Asp140), frog H (Ser140), and human H (Ala140), indicating that binding stability with weak ligands at the active centers could be compensated for by changes elsewhere in the protein structure (Table 1). In the Ala140 ferritins, in the human H and frog M polypeptide backbones, the kinetic differences were particularly large (Fig. 2A and Table 1). For example, Ala140 in the frog M polypeptide context had a 7-fold lower DFP kinetic stability, and a 3-fold larger Kapp than in Ala140 in the homologous human H polypeptide. In addition, Ser140 in the frog M ferritin polypeptide, which predominates in liver, had a significantly higher Kapp than in the homologous frog H ferritin polypeptide, which predominates in red blood cells (Table 1). These observations, which may reflect ferritin physiology at the different temperatures of warm-blooded or cold-blooded animals, or subcellular supramolecular interactions, emphasize the role of residues beyond the active site in ferritin catalysis.
His140, Required for Diiron Cofactor Oxidation, Eliminates Fe(II) Oxidation in Ferritin.
To test the range of variability in residue 140 compatible with ferritin catalysis, given that all of the combinations of natural ferritin residues with various polypeptide backbones were kinetically competent (Table 1), we studied the effect of histidine at position 140 in the frog M polypeptide backbone. [Histidine is the WT residue at the equivalent position of the diiron oxygenase cofactor sites that form the same DFP intermediate from Fe(II) and O2 as ferritins.] All of the polypeptides with histidine substitutions at the catalytic centers assembled normally into the protein nanocages, based on gel filtration and iron mineralization properties. In the ferritin protein environment, histidine at position 140 prevented DFP formation (2 Fe per active site or 48 Fe per nanocage under standard, aerobic conditions) (Fig. 2A trace 4]. No DFP was detected, even with high concentrations of Fe(II) (up to 1,200 Fe per nanocage) (data not shown). Thus, to determine if Fe(II) was oxidized by the His140 protein, we measured the ΔA350. All Fe(III)O species, including DFP, Fe(III) oxo or hydroxo dimers, and Fe(III) oxide mineral have spectral changes in the region 300–400 nm. Fe(II) was oxidized at a very slow rate in the His140 protein, retarded by 40-fold compared with the frog M protein (Fig. 2A Inset). Moreover, the kinetic parameters such as maximum Vi and Kapp for Fe(II) oxidation, determined from the initial rate of ΔA350 at various concentrations of Fe(II), were greatly affected by His140 substitution. For example, the maximum Vi was decreased by 95%, and Kapp increased 10-fold (Table 1). A double mutant that replaced both ferritin-specific residues in the Fe2 substrate site with amino acids that completed the iron binding set of the cofactor sites in diiron oxygenases was as inactive as a single histidine substitution (Fig. 2A and Table 1) in the ferritin polypeptide context. Such data, although unexpected, show that even though histidine functions in very similar reactions between Fe(II) and O2 in the environment of diiron oxygenase centers, the variations in residue 140 compatible with reactivity in ferritins are restricted to aspartate, serine, alanine, or possibly another amino acid, but not histidine.
Gln137, Conserved in the Ferritin Fe2 Site, Controls the Diferric Peroxo Pathway.
Gln137 in the Fe2 site of all ferritins (Fig. 1) is a conservative substitution of an amide side chain for the absolutely conserved carboxylate side chain in glutamate at the equivalent position in diiron cofactor sites. A glutamine/glutamate substitution might produce an active protein with an activity distinct from either ferritin or a diiron oxygenase. Therefore, we created the Gln137 → Glu mutant of frog M that generates a ferritin Fe2 site with EED.
There were two effects of the Gln137 → Glu substitution: (i) No DFP detected; (ii) Fe(III)O product(s) with different spectral properties. We know of no other observation of a ferritin protein with such properties. The absence of DFP (Fig. 2D trace 1] could be caused by decreased kinetic stability of the intermediate or a change in the pathway of the reaction of Fe(II) with O2 (Fig. 2C trace 2 and Fig. 2D trace 2). For Fe(III)O product, the maximum initial rate, Vi, Kapp, and the Hill coefficient (n), determined from ΔA350, were comparable to the WT Fe2, EQD site (Table 1). When the reaction was measured at 4 °C, instead of the standard 20 °C, in an attempt to trap any reaction intermediates, we observed the appearance of a species (Fig. 2D Inset) with increasing absorbance from 350–700 nm, without the 650-nm absorbance maximum characteristic of the DFP complex of ferritin (data not shown). Even at 4 °C, the EED-specific, Fe(II) + O2 intermediate was only 1/15 as kinetically stable (t1/2: 0.15 ± 0.02 s) as DFP in the natural EQD site (t1/2: 2.0 ± 0.1 s). In addition, the maximum ΔA350 of the Fe(III)O products observed in Fe2 EED were only ∼50% that of WT Fe2 EQD (Fig. 2 C and D). Identification of the product of the Fe2 EED site in ferritin catalytic centers is outside the scope of this report, but existence of the product illustrates both a possible evolutionary intermediate between ferritin Fe(II)/O2 oxidoreductase sites and diiron cofactor oxygenase sites and the importance of Gln137 in control of the DFP reaction pathway for iron mineralization.
Discussion
Selectivity of Fe(II)/O2 chemistry at the diiron substrate sites in ferritin [Fe(II)/O2 oxidoreductase] catalytic centers, compared to the diiron cofactor centers in oxygenases that produce the same initial reaction intermediate Fe(III)–O–O–Fe(III), appears to depend on differences in only 2 of the 6 conserved residues at the diiron cofactor sites: (i) a conserved, glutamine in ferritins vs. glutamate in the cofactor sites, and (ii) a set of variable residues, aspartate/serine/alanine in ferritins vs. a conserved histidine in the cofactor sites. The two types of differences in the ferritin catalytic centers residues are associated with a weaker Fe(II) binding environment and a more open structure with a single carboxylate bridge between the two Fe atoms in structures of ferritin diiron substrate sites compared with the stronger binding environment and more closed structures in the oxygenase diiron cofactor sites (Fig. 1 and refs. 18 and 21–23). The kinetic studies presented here show that each natural variation among ferritin active sites alters kinetics of the Fe(II) + O2 reaction, but not the mechanism, and that each diiron catalytic center appears to have coevolved with the sequence of the polypeptide backbone, based on comparisons of the catalytic properties of natural variants in residue 140, aspartate, serine, or alanine in a single polypeptide sequence (frog M) with that in the natural polypeptide (frog M Asp140, frog H Ser140, and human H Ala140). However, substitution of the two Fe ligands from the diiron cofactor sites for the ferritin-specific residues effectively inactivated the ferritin oxidoreductase activity, which is facilitated by the same ligand set in the oxygenases, MMO, RNR, and Δ9 desaturase (11–14, 21–23).
Positive cooperativity, observed during DFP formation (Table 1), and in earlier studies, e.g., ref. 20, likely reflects interactions among the multiple catalytic sites of ferritin as recently observed during diferrous binding to the active sites in the absence of dioxygen (17). Local protein conformational changes, indicated by the differences among the Fe–Fe distances (EXAFS) of diferrous (3.4 Å), DFP (2.5 Å) and diferric mineral precursor (3.9 Å) (10), may also contribute to cooperativity.
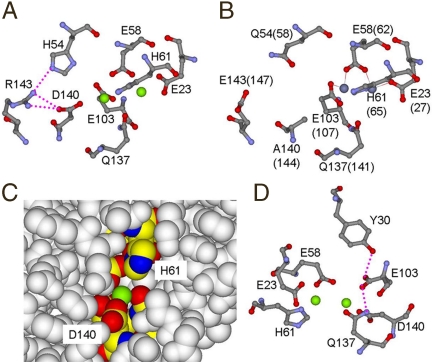
One of the residues at the ferritin-specific Fe2 site, residue 140, varies coincidentally with tissue-specific or organelle-specific ferritin expression (24, 25), without altering the reaction pathway (Scheme 1) (9, 10, 29). However, each ferritin active site in the homologous background had variable kcat and DFP kinetic stability, but constant Kapp (Table 1). In contrast, when the polypeptide sequence was the same, and just residue 140 was changed at the catalytic center by using the natural active center residues Ser and Ala as substitutions for Asp, the WT match with the polypeptide backbone, a different set of kinetic properties was observed (Table 1), indicating contributions of the global protein sequence to catalysis. Sequence and structure comparisons indicated that proximal residue 143, covarying with ferritin Fe2 site residue 140 (Fig. 3 A and B), might contribute to catalysis. However, changing residue 143 had no significant effects on DFP formation (T.T. and E.C.T., unpublished observations). Key residues matched to variations in residue 140 that modulate catalysis in ferritins are, thus, more likely to be some distance from the catalytic centers as discussed for other protein catalysts (reviewed in ref. 30).
Fig. 3.
Models of the environments around ferritin Fe2 site related to Fe(II)/O2 oxidoreductase activity. (A) The Fe2 environment with Asp at variable position 140. Arg143 hydrogen-bonded to Asp140 and His54, near Asp140, covary among ferritins. (B) The environment of variable residue with Ala140(144); same view as C. The hydrogen-bond network Arg143···His54 would be absent with Glu143 and Gln54. [The figure was created from PDB 1MFR, frog M-ferritin, for Asp140, His54, Arg 143 Mg(II) cocrystals (18) and PDB 2CEI, Ala140(144), Glu143(147), and Gln54(58), human H-ferritin Zn(II) cocrystals (35).] Dotted lines indicate presumed hydrogen bonds. (C) The inner cavity surface around the catalytic center. Fe2 site Asp140 is located at the inner cavity surface; Mg(II) (green) is exposed to cavity solvent. (D) A plausible hydrogen bonding network: the invariant ferritin Fe2 amide side chain of Gln137 hydrogen-bonds to the carboxylate group of conserved Glu103, which is hydrogen-bonded with the conserved Tyr30 phenoxyl.
Histidine at variable position 140 in the Fe2 site of the ferritin Fe(II)/O2 oxidoreductase centers marked a limit in the range of functional residues possible at Fe2 in ferritin catalytic centers, because His140 ferritins were ineffective not only in the coupling reaction that forms the DFP catalytic intermediate but also in Fe(II) oxidation itself (Fig. 2A and Table 1); histidine is conserved at the analogous site in diiron cofactors. Preliminary X-ray structural analysis (protein crystallography) on the Gln137 → Glu/Asp140 → His mutant showed that the active site was intact and the His140 configuration was similar to Asp140 in WT (T.T, H.-L. Ng, E.C.T., and T. Alber, unpublished observation). In addition, CD/MCD data indicate binding of 2Fe(II) in the catalytic center of Gln137 → Glu/Asp140 → His mutant (J. K. Schwartz, T.T, E.C.T., and E. I. Solomon, unpublished observation). Therefore, the activity loss of His140 ferritin is independent of Fe(II) binding or collapse of the active site structure. Several ferritin properties could explain the inactivation caused by inserting histidine at position 140. First, because residue 140 is exposed to nanocage cavity (Fig. 3C) it is in contact with the air-saturated, saline buffer in the cavity (31, 32), suggesting a role for residue 140 in active site interactions with small molecules such as O2 and H2O and possibly with the diferric oxo/hydroxo catalytic product; His140 might physically block transfer of product to the cavity. Second, the basicity of histidine is modulated by multiple protein side chain interactions in diiron cofactor sites (21–23) that are absent from WT ferritin around His61 in the Fe1 site (18) (Fig. 1) and may also be absent from the engineered His140 ferritin. A strong Fe–histidine interaction in the ferritin Fe2 site may impede O2 binding or electron transfer between Fe(II) and O2 or product release. Such effects illustrate the structural differences between the diiron cofactor centers in MMO, RNR, and Δ9 desaturase and the Fe(II)/O2 oxidoreductase centers in ferritin that coincide with different pathways from the common DFP catalytic intermediate.
Gln137, the conserved, ferritin-specific, active site residue with an amide side chain, contrasts with the glutamate carboxylate side chain at the equivalent site in the diiron cofactor oxygenases and controls DFP formation in ferritins, based on effects of glutamate substitution (Fig. 2D and Table 1), even though glutamate in the context of the diiron cofactor enzymes also forms DFP (11–14). Because the DNA code for glutamine and glutamate can differ by only one nucleotide (glutamine/glutamate codons can be CAG/GAG) (10, 18), ferritin with Glu137 could model an evolutionary intermediate on the path between diiron substrate and diiron cofactor catalytic centers. The important role of Gln137 in DFP-mediated iron mineralization by ferritin reflects the multiple interactions of Gln137 within the ferritin protein. First, crystal structures suggest a conserved hydrogen-bond network, Gln137···Glu103···Tyr30 (Fig. 3D). Gln137 → Glu catalysis is unusual in producing Fe(III)O products with a lower average absorbance than normal but with a normal rate of Fe(II) oxidation itself (Table 1), whereas substitutions for Glu103 or Tyr30 decrease Fe(II) oxidation (20, 33–35). The second Fe1–Fe2 bridge, formed by glutamate in the diiron oxygenases at the position analogous to Gln137 in ferritins (Fig. 1), not only is prevented by the amide but the side chain orientation itself differs because of constraints imposed by the hydrogen bond network. Second, in a diferrous model for the catalytic center ferritin developed with variable-temperature, variable-field magnetic circular dichroism (VTVH MCD) (17), the carbonyl O in Gln137 is coordinated to Fe(II) in the Fe2 site. Third, the VTVH MCD model (17) indicates bonding between the amide of Gln137 and H2O coordinated to Fe1 as shown in Fig. 1. Protonation at the DFP active site, required for decay of the intermediate and product release (15, 29), would be facilitated by a hydrogen bond between Gln137 and H2O coordinated to Fe1 (17). In addition, the oxo or hydroxo bridge suggested by the short Fe1–Fe2 distance (2.5 Å) in DFP (10) might also be facilitated by a hydrogen bond between Gln137 to H2O at Fe1. Thus, the particular orientation of Gln137 imposed by the hydrogen bond network could facilitate the proton transfer associated with DFP formation from Fe(II) + O2, or DFP decay to form Fe(III)–O(H)–Fe(III), or possibly the release of the catalytic product, Fe(III)–O(H)–Fe(III) during catalysis.
Catalysis at the ferritin Fe(II)/O2 oxidoreductase centers depends on combining ferritin-specific residues in the Fe2 site with residues common to diiron cofactor oxygenases. The reaction of Fe(II) with dioxygen that produces the DFP intermediate occurs for both diiron substrate and diiron cofactor protein catalysts. The ferritin-specific Fe2 site uses Glu103, also present in diiron cofactor sites, ferritin-specific conserved residue (Gln137), and a ferritin-specific set of ferritin-variable residue Asp140, Ser140, or Ala140, but cannot function with the cofactor residue, His140. Gln137 is required for DFP formation/decay and for the more open catalytic site structure (Fig. 1). The short Fe1–Fe2 distance (2.5 Å) and small Fe–O–O angle (8, 10) in the ferritin DFP compared to DFP in diiron cofactor enzymes, appears to facilitate peroxide release rather than the formation of high-valence reaction intermediate observed in diiron cofactor enzymes. The results reported here show that weaker iron binding at the ferritin-specific Fe2 site is required for the formation of DFP in ferritin and may contribute to a particular reaction pathway distinct from the heme-containing Bfr with an active site containing the diiron cofactor ligands (2) and a geometry of the ferritin DFP that is incompatible with the diiron cofactor ligand set. The two ferritin-specific ligands in the Fe2 site of the catalytic center, Gln137/Asp140, are related to Glu and His, the two diiron cofactor site ligands at equivalent positions, by 2 of 3 codon nucleotides in DNA. Thus, the results also illustrate the potential simplicity of evolving active sites for diiron cofactors or diiron substrates.
Materials and Methods
Cloning and Mutagenesis.
Site-directed mutagenesis used QuikChange Mutagenesis (Stratagene) on frog M-ferritin in pET-3a as previously described (20). Primers were Q137E (EED): forward, 5′-GAA TAT CTG GAG GAA GAG GTG AAG GAT ATT AAA CGC-3′; reverse, 5′-GCG TTT AAT ATC CTT CAC CTC TTC CTC CAG ATA TTC-3′; D140H (EQH): forward, 5′-G GAA CAG GTG AAG CAT ATT AAA CGC-3′; reverse, 5′-GCG TTT AAT ATG CTT CAC CTG TTC C-3′; Q137E/D140H (EEH): forward, 5′-GAA TAT CTG GAG GAA GAG GTG AAG CAT ATT AAA CGC-3′; reverse, 5′-GCG TTT AAT ATG CTT CAC CTC TTC CTC CAG ATA TTC-3′, D140A (EQA): forward, 5′-GAA TAT CTG GAG GAA CAG GTG AAG GCT ATT AAA CGC-3′; reverse, 5′-GCG TTT AAT AGC CTT CAC CTG TTC CTC CAG ATA TTC-3′, D140S (EQS): forward, 5′-GAA TAT CTG GAG GAA CAG GTG AAG TCT ATT AAA CGC-3′; reverse, 5′-GCG TTT AAT AGA CTT CAC CTG TTC CTC CAG ATA TTC-3′ (underlines indicate mutation sites). The human H-ferritin coding region, obtained by PCR amplification of Marathon-Ready HeLa cell cDNA (Clontech), was cloned in pUC19, followed by PCR to insert NdeI and BamHI sites, digestion with NdeI and BamHI, and cloning into pET-3a DNA, which was analyzed to confirm the sequence of human H-ferritin insert.
Protein Expression.
Recombinant ferritin samples, isolated as previously described (20, 36–38) from Escherichia coli BL21(DE3) incubated in LB medium, used 20 mM Bis-Tris-propane, pH 7.5, during sonication, coagulation of soluble nonferritin proteins (15 min, 65 °C), concentration by precipitation with 45% saturated (NH4)2SO4, ion-exchange fractionation (mono-Q column, Amersham Pharmacia) with a linear NaCl gradient of 0–1.0 M, and detection with SDS/PAGE. Pooled, homogenous, ferritin fractions were dialyzed against 100 mM Mops (pH 7.0), 100 mM NaCl, stored at 4 °C, and analyzed within 2 weeks.
Kinetic Analysis of Ferrous Oxidation and DFP Formation.
Fresh solutions of FeSO4 (0.1–5 mM in 1.0 mM HCl) were mixed with an equal volume of ferritin solutions [0.1 mM subunits in 200 mM Mops (pH 7.0), 200 mM NaCl] in a stopped-flow UV/vis spectrophotometer (Applied Photophysics) at 20 °C and 4 °C. Data were collected for A650, the absorbance maximum of DFP in ferritin (9), and A350, the midpoint of the Fe(III) oxy absorption, an unresolvable mixture of DFP, Fe(III) oxo/hydroxo dimer, and Fe(III) biomineral (20); the absorbance range 300–420 nm is often used in ferritin Fox analyses (20, 29, 33, 39–42). Initial rates (Vi) were calculated by linear fitting to the early time domain (∼50 ms) of A350 and A650 data. Vi values obtained at varied Fe concentrations were fitted to the Hill equation (20), Vi = Vmax[Fe(II)]n/(Kapp + [Fe(II)]n), where n is the Hill coefficient. The kcat values (= Vmax/[ferritin]) for DFP formation were estimated by two fitting methods. First, by computing the kcat value (= Vmax/[ferritin]) for DFP formation with Vmax and the DFP molar extinction coefficient (ε) of 1,000 M1·cm1 at 650 nm (9). Second, kcat was determined by using the extinction coefficient of each protein, under the conditions used here, computed from fitting the kinetic trace of A650 under single-turnover conditions (48 Fe per ferritin) with a double-exponential equation assuming two irreversible consecutive reactions; ΔA650 = A0kfwd × [exp(−kfwdt) − exp(−kdecayt)]/(kdecay − kfwd) (9), where A0 = ε × [ferritin]. The computed values for ε were within experimental error of 1,000 M1·cm1, ε = 1,040 ± 80, 960 ± 140, and 1,060 ± 80 M1·cm1 for frog M WT (EQD-Fr M), D140A, and human H WT, respectively, except for EQS-Fr M, where ε = 720 ± 120 M1·cm1; nonrandom residuals indicated that the goodness of fits were poorer for the double-exponential fitting model. The values of kcat were significantly (P < 0.01) different from the control WT frog M by either method. DFP decay, fit well by a single-exponential (20) and less well with the double-exponential equation (see above), gave similar results, using the equation t1/2 = ln2/kdecay. Fitting used Igor software (WaveMetrics). Except for EQA-Hu H, where only one protein preparation was studied, the data were from 3–5 protein preparations; each protein preparation was analyzed 2–7 times
Acknowledgments.
We thank Dr. Xiaofeng S. Liu for helpful discussions. This work was supported by National Institutes of Health Grant DK20251 (E.C.T., M.R.H., and T.T.) and a Japan Society for the Promotion of Science postdoctoral fellowship for research abroad (T.T.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
The subunit residue numbers in the amino acid sequences used correspond to those for horse, frog, and soybean H, L, and M; to convert to the human H sequence add 4.
References
- 1.Liu X, Theil EC. Ferritins: Dynamic management of biological iron and oxygen chemistry. Acc Chem Res. 2005;38:167–175. doi: 10.1021/ar0302336. [DOI] [PubMed] [Google Scholar]
- 2.Frolow F, Kalb AJ, Yariv J. Structure of a unique twofold symmetric haem-binding site. Nat Struct Biol. 1994;1:453–460. doi: 10.1038/nsb0794-453. [DOI] [PubMed] [Google Scholar]
- 3.Ferreira C, et al. Early embryonic lethality of H ferritin gene deletion in mice. J Biol Chem. 2000;275:3021–3024. doi: 10.1074/jbc.275.5.3021. [DOI] [PubMed] [Google Scholar]
- 4.Curtis AR, et al. Mutation in the gene encoding ferritin light polypeptide causes dominant adult-onset basal ganglia disease. Nat Genet. 2001;28:350–354. doi: 10.1038/ng571. [DOI] [PubMed] [Google Scholar]
- 5.Touati D, Jacques M, Tardat B, Bouchard L, Despied S. Lethal oxidative damage and mutagenesis are generated by iron in delta fur mutants of Escherichia coli: Protective role of superoxide dismutase. J Bacteriol. 1995;177:2305–2314. doi: 10.1128/jb.177.9.2305-2314.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Douglas T, et al. Synthesis and structure of an iron(III) sulfide-ferritin bioinorganic nanocomposite. Science. 1995;269:54–57. doi: 10.1126/science.269.5220.54. [DOI] [PubMed] [Google Scholar]
- 7.Vriezema DM, et al. Self-assembled nanoreactors. Chem Rev. 2005;105:1445–1489. doi: 10.1021/cr0300688. [DOI] [PubMed] [Google Scholar]
- 8.Moenne-Loccoz P, et al. The ferroxidase reaction of ferritin reveals a diferric μ-1,2 bridging peroxide intermediate in common with other O2-activating nonheme diiron proteins. Biochemistry. 1999;38:5290–5295. doi: 10.1021/bi990095l. [DOI] [PubMed] [Google Scholar]
- 9.Pereira AS, et al. Direct spectroscopic and kinetic evidence for the involvement of a peroxodiferric intermediate during the ferroxidase reaction in fast ferritin mineralization. Biochemistry. 1998;37:9871–9876. doi: 10.1021/bi980847w. [DOI] [PubMed] [Google Scholar]
- 10.Hwang J, et al. A short Fe-Fe distance in peroxodiferric ferritin: Control of Fe substrate versus cofactor decay? Science. 2000;287:122–125. doi: 10.1126/science.287.5450.122. [DOI] [PubMed] [Google Scholar]
- 11.Moenne-Loccoz P, Baldwin J, Ley BA, Loehr TM, Bollinger JM., Jr O2 activation by nonheme diiron proteins: Identification of a symmetric μ-1,2-peroxide in a mutant of ribonucleotide reductase. Biochemistry. 1998;37:14659–14663. doi: 10.1021/bi981838q. [DOI] [PubMed] [Google Scholar]
- 12.Broadwater JA, Ai J, Loehr TM, Sanders-Loehr J, Fox BG. Peroxodiferric intermediate of stearoyl-acyl carrier protein Δ9 desaturase: Oxidase reactivity during single turnover and implications for the mechanism of desaturation. Biochemistry. 1998;37:14664–14671. doi: 10.1021/bi981839i. [DOI] [PubMed] [Google Scholar]
- 13.Shu L, et al. An FeIV2O2 diamond core structure for the key intermediate Q of methane monooxygenase. Science. 1997;275:515–518. doi: 10.1126/science.275.5299.515. [DOI] [PubMed] [Google Scholar]
- 14.Liu KE, et al. Characterization of a diiron(III) peroxo I intermediate in the reaction cycle of methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath) J Am Chem Soc. 1995;117:4997–4998. [Google Scholar]
- 15.Jameson GN, et al. Stoichiometric production of hydrogen peroxide and parallel formation of ferric multimers through decay of the diferric-peroxo complex, the first detectable intermediate in ferritin mineralization. Biochemistry. 2002;41:13435–13443. doi: 10.1021/bi026478s. [DOI] [PubMed] [Google Scholar]
- 16.Mertz JR, Theil EC. Subunit dimers in sheep spleen apoferritin: The effect on iron storage. J Biol Chem. 1983;258:11719–11726. [PubMed] [Google Scholar]
- 17.Schwartz JK, Liu XS, Tosha T, Theil EC, Solomon EI. Spectroscopic definition of the ferroxidase site in M ferritin: Comparison of binuclear substrate vs cofactor active sites. J Am Chem Soc. 2008;130:9441–9450. doi: 10.1021/ja801251q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ha Y, Shi D, Small GW, Theil EC, Allewell NM. Crystal structure of bullfrog M ferritin at 2.8 Å resolution: Analysis of subunit interactions and the binuclear metal center. J Biol Inorg Chem. 1999;4:243–256. doi: 10.1007/s007750050310. [DOI] [PubMed] [Google Scholar]
- 19.Hempstead PD, et al. Comparison of the three-dimensional structures of recombinant human H and horse L ferritins at high resolution. J Mol Biol. 1997;268:424–448. doi: 10.1006/jmbi.1997.0970. [DOI] [PubMed] [Google Scholar]
- 20.Liu X, Theil EC. Ferritin reactions: Direct identification of the site for the diferric peroxide reaction intermediate. Proc Natl Acad Sci USA. 2004;101:8557–8562. doi: 10.1073/pnas.0401146101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rosenzweig AC, Nordlund P, Takahara PM, Frederick CA, Lippard SJ. Geometry of the soluble methane monooxygenase catalytic diiron center in two oxidation states. Chem Biol. 1995;2:409–418. [PubMed] [Google Scholar]
- 22.Logan DT, et al. Crystal structure of reduced protein R2 of ribonucleotide reductase: The structural basis for oxygen activation at a dinuclear iron site. Structure. 1996;4:1053–1064. doi: 10.1016/s0969-2126(96)00112-8. [DOI] [PubMed] [Google Scholar]
- 23.Lindqvist Y, Huang W, Schneider G, Shanklin J. Crystal structure of Δ9 stearoyl-acyl carrier protein desaturase from castor seed and its relationship to other di-iron proteins. EMBO J. 1996;15:4081–4092. [PMC free article] [PubMed] [Google Scholar]
- 24.Dickey LF, et al. Differences in the regulation of messenger RNA for housekeeping and specialized-cell ferritin. A comparison of three distinct ferritin complementary DNAs, the corresponding subunits, and identification of the first processed in Amphibia. J Biol Chem. 1987;262:7901–7907. [PubMed] [Google Scholar]
- 25.Levi S, et al. A human mitochondrial ferritin encoded by an intronless gene. J Biol Chem. 2001;276:24437–24440. doi: 10.1074/jbc.C100141200. [DOI] [PubMed] [Google Scholar]
- 26.Bou-Abdallah F, Santambrogio P, Levi S, Arosio P, Chasteen ND. Unique iron binding and oxidation properties of human mitochondrial ferritin: A comparative analysis with human H-chain ferritin. J Mol Biol. 2005;347:543–554. doi: 10.1016/j.jmb.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 27.Langlois d'Estaintot B, et al. Crystal structure and biochemical properties of the human mitochondrial ferritin and its mutant Ser144Ala. J Mol Biol. 2004;340:277–293. doi: 10.1016/j.jmb.2004.04.036. [DOI] [PubMed] [Google Scholar]
- 28.Bou-Abdallah F, et al. μ-1,2-Peroxobridged di-iron(III) dimer formation in human H-chain ferritin. Biochem J. 2002;364:57–63. doi: 10.1042/bj3640057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bou-Abdallah F, Zhao G, Mayne HR, Arosio P, Chasteen ND. Origin of the unusual kinetics of iron deposition in human H-chain ferritin. J Am Chem Soc. 2005;127:3885–3893. doi: 10.1021/ja044355k. [DOI] [PubMed] [Google Scholar]
- 30.Hammes-Schiffer S, Benkovic SJ. Relating protein motion to catalysis. Annu Rev Biochem. 2006;75:519–541. doi: 10.1146/annurev.biochem.75.103004.142800. [DOI] [PubMed] [Google Scholar]
- 31.Trikha J, Theil EC, Allewell NM. High resolution crystal structures of amphibian red-cell L ferritin: Potential roles for structural plasticity and solvation in function. J Mol Biol. 1995;248:949–967. doi: 10.1006/jmbi.1995.0274. [DOI] [PubMed] [Google Scholar]
- 32.Yang X, Chasteen ND. Molecular diffusion into horse spleen ferritin: A nitroxide radical spin probe study. Biophys J. 1996;71:1587–1595. doi: 10.1016/S0006-3495(96)79361-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Treffry A, Zhao Z, Quail MA, Guest JR, Harrison PM. Iron(II) oxidation by H chain ferritin: Evidence from site-directed mutagenesis that a transient blue species is formed at the dinuclear iron center. Biochemistry. 1995;34:15204–15213. doi: 10.1021/bi00046a028. [DOI] [PubMed] [Google Scholar]
- 34.Fetter J, Cohen J, Danger D, Sanders-Loehr J, Theil EC. The influence of conserved tyrosine 30 and tissue-dependent differences in sequence on ferritin function: use of blue and purple Fe(III) species as reporters of ferroxidation. J Biol Inorg Chem. 1997;2:652–661. [Google Scholar]
- 35.Toussaint L, Bertrand L, Hue L, Crichton RR, Declercq JP. High-resolution X-ray structures of human apoferritin H-chain mutants correlated with their activity and metal-binding sites. J Mol Biol. 2007;365:440–452. doi: 10.1016/j.jmb.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 36.Takagi H, Shi D, Ha Y, Allewell NM, Theil EC. Localized unfolding at the junction of three ferritin subunits. A mechanism for iron release? J Biol Chem. 1998;273:18685–18688. doi: 10.1074/jbc.273.30.18685. [DOI] [PubMed] [Google Scholar]
- 37.Jin W, Takagi H, Pancorbo NM, Theil EC. “Opening” the ferritin pore for iron release by mutation of conserved amino acids at interhelix and loop sites. Biochemistry. 2001;40:7525–7532. doi: 10.1021/bi002509c. [DOI] [PubMed] [Google Scholar]
- 38.Liu X, Jin W, Theil EC. Opening protein pores with chaotropes enhances Fe reduction and chelation of Fe from the ferritin biomineral. Proc Natl Acad Sci USA. 2003;100:3653–3658. doi: 10.1073/pnas.0636928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao G, et al. Multiple pathways for mineral core formation in mammalian apoferritin. The role of hydrogen peroxide. Biochemistry. 2003;42:3142–3150. doi: 10.1021/bi027357v. [DOI] [PubMed] [Google Scholar]
- 40.Levi S, et al. Construction of a ferroxidase center in human ferritin L-chain. J Biol Chem. 1994;269:30334–30339. [PubMed] [Google Scholar]
- 41.Bauminger ER, et al. Iron(II) oxidation and early intermediates of iron-core formation in recombinant human H-chain ferritin. Biochem J. 1993;296:709–719. doi: 10.1042/bj2960709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang X, Le Brun NE, Thomson AJ, Moore GR, Chasteen ND. The iron oxidation and hydrolysis chemistry of Escherichia coli bacterioferritin. Biochemistry. 2000;39:4915–4923. doi: 10.1021/bi992631f. [DOI] [PubMed] [Google Scholar]