Abstract
Angiosperm plastid genomes are generally conserved in gene content and order with rates of nucleotide substitutions for protein-coding genes lower than for nuclear protein-coding genes. A few groups have experienced genomic change, and extreme changes in gene content and order are found within the flowering plant family Geraniaceae. The complete plastid genome sequence of Pelargonium X hortorum (Geraniaceae) reveals the largest and most rearranged plastid genome identified to date. Highly elevated rates of sequence evolution in Geraniaceae mitochondrial genomes have been reported, but rates in Geraniaceae plastid genomes have not been characterized. Analysis of nucleotide substitution rates for 72 plastid genes for 47 angiosperm taxa, including nine Geraniaceae, show that values of dN are highly accelerated in ribosomal protein and RNA polymerase genes throughout the family. Furthermore, dN/dS is significantly elevated in the same two classes of plastid genes as well as in ATPase genes. A relatively high dN/dS ratio could be interpreted as evidence of two phenomena, namely positive or relaxed selection, neither of which is consistent with our current understanding of plastid genome evolution in photosynthetic plants. These analyses are the first to use protein-coding sequences from complete plastid genomes to characterize rates and patterns of sequence evolution for a broad sampling of photosynthetic angiosperms, and they reveal unprecedented accumulation of nucleotide substitutions in Geraniaceae. To explain these remarkable substitution patterns in the highly rearranged Geraniaceae plastid genomes, we propose a model of aberrant DNA repair coupled with altered gene expression.
Keywords: comparative genomics, genome evolution, plastid genome
Angiosperm plastid genomes are generally highly conserved in gene order, gene content, and organization (1). Whereas the rates of nucleotide substitutions are highly variable in protein-coding genes of angiosperm nuclear genomes, rates in plastid genes are generally lower (2). Rates of nonsynonomous substitutions (dN), those that cause an amino acid change, are substantially lower than rates of synonymous substitutions (dS), those that do not cause an amino acid change. Aside from a recent report describing elevated dN for a single gene in Oenothera and lineages within Caryophyllaceae (3), plastid genes of photosynthetic plants are under strong purifying selection and the rapid accumulation of either dN or dS has not been described.
The plastid genomes of nonphotosynthetic plants reveal accelerated rates of nucleotide substitutions in many protein-coding genes; furthermore, these genomes exhibit extensive gene loss and genome rearrangement (4–6). However, analyses involving either few genes or few taxa for photosynthetic angiosperm plastid genomes generally reveal that modest rate variation is locus- and lineage-specific. A few groups of angiosperms have experienced lineage-specific rate variation, including the lineages leading to the grasses (7), pea (2), Gnetum (8), and Welwitschia (9). The largest degree of locus-specific variation has been shown for genes encoding RNA polymerases, ATPases, and ribosomal proteins (7, 10). In addition, location within the quadripartite plastid genome affects rate variation; genes encoded in the two single-copy regions exhibit higher rates than genes duplicated in the large inverted repeat (IR) regions (11).
Geraniaceae organelle genomes exhibit two unusual features. First, mitochondrial genomes show multiple, major shifts in dS, especially in the genus Pelargonium (12). Similar rapid increases have only been documented in one other plant lineage, Plantago (Plantaginaceae) (13). These studies showed that rates were not affected in nuclear or plastid genes, but few genes were examined. A correlation between rates of sequence evolution and genomic rearrangement has not been documented for plant mitochondrial genomes, but this relationship was been shown in animal mitochondrial genomes (14, 15). Second, Geraniaceae plastid genomes are structurally unusual. The plastid genome of Pelargonium X hortorum exhibits unprecedented levels of change in both gene order and content (16). This genome also contains the largest IR, along with numerous dispersed repeats contributing to increased genome size and potential rearrangement hotspots. In addition, restriction site mapping studies revealed extensive rearrangements and gene/intron loss throughout the family, and in Erodium and Monsonia the loss or severe reduction of the IR (17).
A recent phylogenetic analysis of 81 plastid genes from 64 seed plants described a positive correlation between changes in gene order, gene/intron loss, and lineage-specific rate acceleration (18). Geraniaceae are an ideal group to study plastid genome evolution, because they are highly rearranged (16, 17). In this study, we examine rates and patterns of sequence evolution across Geraniaceae and provide evidence for major locus- and lineage-specific substitution rate increases. We also show a strong correlation between gene function and rates of nucleotide substitutions. We examine plastid sequence evolution at a genome-wide scale and show major increases in dN for protein-coding genes in the typically conserved plastid genomes of photosynthetic angiosperms.
Results
Alignment and annotation of Geraniaceae gene sequences relative to sequences from other angiosperms revealed a number of genomic changes, including gene/intron loss, alternative start codons (see data matrix http://www.biosci.utexas.edu/ib/faculty/jansen/lab/research/data_files/index.htm), and accelerated rates of sequence evolution. Our analyses included 72 genes [supporting information (SI) Table S1] from 47 angiosperms (Tables S2 and S3).
Phylogenetic analyses were performed on the 47-taxa, 72-gene dataset of 58,998 aligned nucleotide positions. Maximum parsimony (MP) and maximum likelihood (ML) methods were used to infer relationships (Geraniaceae only, Fig. 1; 47-taxa tree, Fig. S1]. A single tree was inferred using MP with a length of 109,664 steps, a consistency index (CI; excluding uninformative characters) of 0.3785, and a retention index (RI) of 0.5897. Four ML analyses inferred the same tree topology (-lnL scores: μ = 607633.5555, σ = 0.001). The poorly supported clade including Geraniales and Myrtales is the sister group to the eurosids II. Although tree topologies were identical in MP and ML analyses and consistent with a recent angiosperm phylogeny (18), relationships among Geraniales, Myrtales, eurosids I, and eurosids II are generally poorly understood (18–20). ML analysis was performed for a smaller dataset including 27 slowly evolving genes to confirm that relationships were not incorrectly inferred because of rapidly evolving genes, and this analysis resulted in a tree identical to full MP and ML analyses (Fig. S2). Relationships within Geraniaceae are consistent with well-accepted phylogenies (17, 21, 22), and branches are resolved and strongly supported (Fig. 1).
Fig. 1.
ML tree of Geraniaceae taken from 72-gene, 47-taxa analysis (Fig. S1). All nodes had 100% ML and MP bootstrap support.
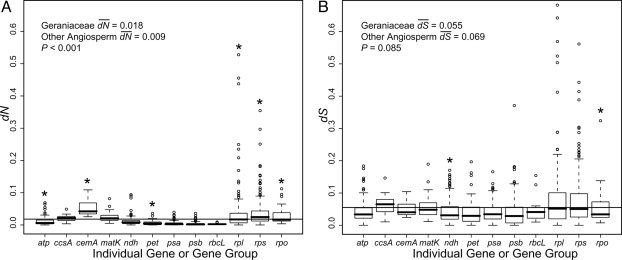
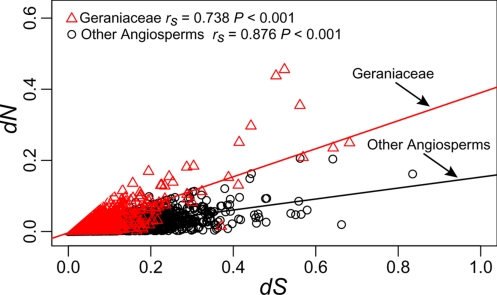
Values of dN and dS were compared within Geraniaceae and between Geraniaceae and other angiosperms to reveal patterns of sequence evolution. Mean dN is higher in Geraniaceae relative to other angiosperms, and Wilcoxon rank sum tests show that values of dN are significantly different in Geraniaceae relative to other angiosperms (P < 0.001; Fig. 2). Values of dS are not significantly different (P = 0.085; Fig. 2). dN and dS are more strongly correlated (P < 0.001) in other angiosperms (Spearman's rank correlation, rS = 0.876) than in Geraniaceae (rS = 0.738; Fig. 3). Rates for each gene group were compared to reveal patterns of substitutions (Fig. 2; post-hoc statistics in Tables S4 and S5). In general, values of dN are higher in Geraniaceae in comparison to other angiosperms, and comparisons revealed that ribosomal protein and RNA polymerase genes are the most significantly different relative to genes involved in photosynthesis. Values of dS exhibit similar yet weaker patterns of variation; large and small subunit ribosomal protein genes (rpl and rps, respectively) and psb genes being the most significantly different (both P < 0.001). Phylogenetic trees for rpl- and rps-genes with the highest values of dN and dS are shown in Fig. 4 A–D (labeled trees shown in Fig. S3), and the long branches in the Geraniaceae clade (shaded) show the extreme rate acceleration. psaI and psaC have the highest values of dN and dS for photosystem I genes, respectively, and branch lengths show that these genes are not evolving faster relative to other angiosperms (Fig. 4E and labeled trees shown in Fig. S3). Nucleotide substitutions were compared for Geraniaceae branches, and post hoc comparisons tested the significance of branch acceleration (Fig. S4 and Tables S6 and S7). These tests revealed a strong lineage-specific pattern of increases in dN and dS for the branches leading to Geraniaceae (branch 1) and the terminal taxon E. chrysanthum (branch 10). However, these results also show that values of dN are greater overall for Geraniaceae branches relative to other angiosperms.
Fig. 2.
Boxplots of the number of (A) nonsynonymous (dN) and (B) synonymous (dS) substitutions for individual Geraniaceae genes or groups of genes (see Materials and Methods). Wilcoxon rank sum tests were used to show that values for Geraniaceae dN were significantly higher than for other angiosperms (P < 0.001), whereas values of dS were not significantly different (P = 0.085). The horizontal line is the mean value of dN or dS in Geraniaceae. For each gene group, the box represents values between quartiles, dotted lines extend to minimum and maximum values, outliers are shown as circles, and the thick, black lines show median values. Asterisks show values for gene groups that are statistically different (P < 0.05 after Bonferroni correction; data not shown) than the values for same gene group in other angiosperms. See Tables S4 and S5 for post hoc statistics.
Fig. 3.
Correlation between dN and dS for the Geraniaceae (red triangles) and other angiosperms (black circles). Spearman's rank correlation rho (rS) is significant (P < 0.001) for both Geraniaceae and angiosperms. Correlation coefficients were compared by using Fisher's Z transformation to show that there is a stronger correlation between dN and dS in the other angiosperms compared with Geraniaceae (P < 0.001).
Fig. 4.
ML trees for the fastest evolving large subunit ribosomal (A and B), small subunit ribosomal (C and D), and photosystem I genes (E and F). Labeled trees are shown in Fig. S3. Branch length is defined as the number of nucleotide substitutions per codon. The Geraniaceae clade is highlighted within each tree to show locus- and lineage-specific rate acceleration. Asterisks show the branch leading to grasses, a group known for lineage-specific rate acceleration (7).
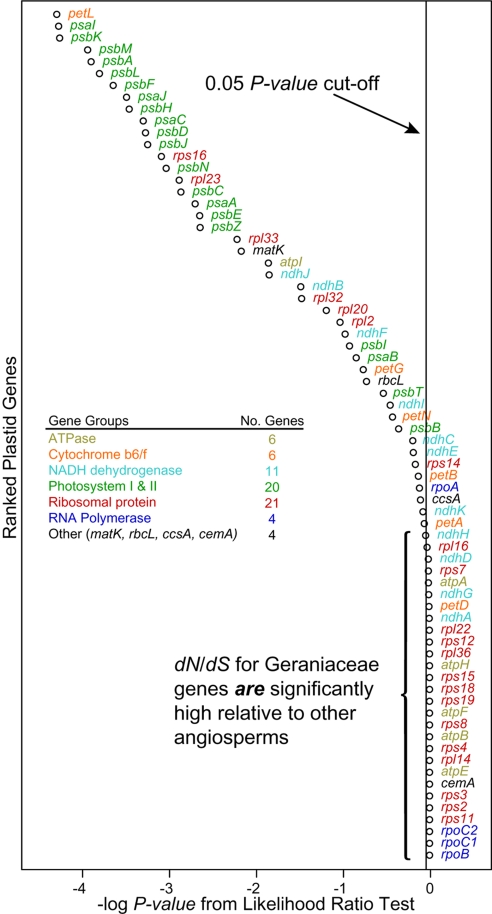
Likelihood ratio tests were used to compare the fit of two models; H0, where values of dN/dS are not significantly different among angiosperms, and HA, where values of dN/dS are greater in Geraniaceae relative to other angiosperms (Table S8). Overall, HA is significantly different from H0 (P < 0.001; data not shown). Values of dN/dS for photosystem I and II genes are not significantly different between Geraniaceae and other angiosperms; however, values of dN/dS for ribosomal protein and RNA polymerase genes are in general significantly higher in Geraniaceae (Fig. 5). Three of four RNA polymerase genes (rpoB, rpoC1, and rpoC2) are the most significantly different, that is, the P-values are the lowest, showing that HA fit these genes the best. The dN/dS value for the gene rpoA, although absent in Pelargonium, is not significantly different in the remaining Geraniaceae relative to other angiosperms.
Fig. 5.
Plot of ranked log P-values for each gene from likelihood ratio tests (LRT). P-values were determined by comparing two models H0 and HA. Corrected P-values were ranked and plotted. The vertical line shows a P value cut-off, and genes appearing to the right of the vertical line have a significantly higher dN/dS in Geraniaceae relative to other angiosperms. Genes that encode subunits of a functional complex were grouped according to Matsuoka et al. (56) and Chang et al. (57).
Discussion
Accelerated Rates of Sequence Evolution in Geraniaceae.
Although photosynthetic angiosperm plastid genomes are generally conserved in gene order, content, and rates of sequence evolution for protein-coding genes, we report extensive nucleotide rate variation in the flowering plant family Geraniaceae. We use complete plastid genomes. This is the first genome-wide analysis to characterize rates and patterns of sequence evolution throughout photosynthetic angiosperm plastids. Accelerated rates of sequence evolution have rarely been reported in angiosperm plastid genomes, and elevated dN has only been shown for the plastid gene clpP (3). We chose not to include clpP in our analyses, because we were unable to align either amino acid or nucleotide sequences, and determining intron/exon boundaries proved difficult. Misalignment would lead to erroneous inference of substitutions rates (23).
Our study also provides evidence for locus- and lineage-specific patterns of rate variation in plastid protein-coding genes. To explain this extreme rate variation, we describe a model that involves faulty DNA repair and variable levels of gene expression. We argue that each of these factors alone cannot explain the rates and patterns of nucleotide substitutions. If faulty DNA repair alone explained the accumulation of substitutions, we would expect rate increases for all genes in these rearranged plastid genomes. Only Geraniaceae rpl-, rps-, and rpo-genes are accumulating substitutions at a significantly higher rate relative to other angiosperms. Moreover, if gene expression level alone explained substitution patterns, we would expect that rapidly evolving genes in Geraniaceae would not evolve any faster than the same genes in other angiosperms. Our data show that rpl-, rps-, and rpo-genes have higher values of dN, and values are significantly different compared with values for the same genes in other angiosperms.
Plastid DNA Repair.
Pelargonium X hortorum contains the most highly rearranged angiosperm plastid genome sequenced to date (16), and based on restriction site mapping studies, plastid genomes in other Geraniaceae are also highly rearranged (17). A significant positive correlation between rates of nucleotide substitutions and genome rearrangements was recently shown for plastid genomes (18). Additionally, in highly rearranged arthropod mitochondrial genomes increased levels of genomic rearrangement are positively correlated with higher rates of sequence evolution (14, 15). We characterized rates of nucleotide substitutions by estimating dN, dS, and dN/dS in Geraniaceae plastid genomes relative to other angiosperms. Our analyses show that both dN (Fig. 2A) and dN/dS are significantly elevated in protein-coding genes, and this suggests that either positive selection or relaxed selection at nucleotide sites is acting on these genomes. We also show that there is a positive correlation between dN and dS in Geraniaceae and other angiosperm plant genomes (Fig. 3). These results suggest that selective constraints are coupled for both types of substitutions; however, there is a weaker constraint in Geraniaceae. To examine lineage-specific rate acceleration, we conducted tests to determine those branches within Geraniaceae that evolve at a faster rate. These data generally show that the branches leading to the most recent common ancestor of Geraniaceae and to E. chrysanthum are accumulating significantly higher values of dN and dS (Fig. S4). Although we only have complete genome sequence data for the P. hortorum genome (16) and data from restriction site mapping studies (17), there appears to be a strong correlation between genome rearrangements and rates of nucleotide substitutions. Completion and analysis of highly rearranged plastid genomes, including such groups as the Campanulaceae, Passifloraceae, Goodeniaceae, and additional Geraniaceae, will likely further support this correlation.
Geraniaceae plastid genomes appear exceptional with respect to rates of evolution and to genomic change; we hypothesize that these phenomena reflect altered organellar DNA repair mechanisms throughout the family. Moreover, both plastid and mitochondrial genomes are affected. Generally, rates of sequence evolution in plant mitochondrial genomes are low relative to rates in plastid and nuclear genomes (2, 24), but mitochondrial genes within Plantago and Geraniaceae show up to 4,000-fold rate increases in dS (12, 13). These studies compared few plastid genes within these lineages. Based on Geraniaceae- and genome-wide analyses, our data indicate numerous instances of accelerated rates that were not detected in earlier studies.
Several mechanisms explaining the correlation between genome rearrangements and rates of sequence evolution have been previously described, and it is possible that these mechanisms are responsible for rearrangement and accelerated rates in Geraniaceae plastid genomes. One mechanism involves homologs to the eubacterial recA gene. In E. coli, this gene is involved in homologous recombination and strand exchange and reduces deleterious mutations through efficient DNA repair. Extensive BLAST searches showed that recA homologs are found in plant and algal nuclear genomes (25), and the products of these genes are targeted to plastids (26) and mitochondria (27) in Arabidopsis. It is now well accepted that plastids and mitochondria arose through endosymbiosis between eukaryotic cells and eubacterial cyanobacteria and proteobacteria, respectively (28). Furthermore, the transfer of organellar genes to the nucleus has been shown, and products of some of these genes are targeted back to plastids and mitochondria (28). The role of recA homologs in plant organellar genomes has not been determined. recA appears to be lost in animal and fungal genomes, and Lin et al. (25) suggest that plant plastid and mitochondrial genomes are large relative to animal and fungal mitochondrial genomes because of recA-mediated recombination. Notably, Geraniaceae plastid genomes are highly variable in size (16, 17), and improper repair may lead to such extreme size variation. In addition to recA, other nuclear-encoded genes are involved in DNA repair (29, 30), and their expression has been shown in plastids (31). For example, mutation of recQ14A resulted in increased homologous recombination and genomic rearrangements in Arabidopsis nuclear genomes (32). The presence and function of rec-genes in highly rearranged plastid genomes has not been tested, and mutations in organellar-targeted genes could result in improper homologous recombination or strand exchange leading to both genome rearrangements and increased substitution rates.
Expression of Plastid Genes.
Our data show that ribosomal protein and RNA polymerase genes exhibit significantly higher values of dN relative to genes involved in photosynthesis in Geraniaceae compared to other angiosperms (Figs. 2, 4, and 5). Within Geraniaceae, comparisons revealed patterns of rate variation across gene groups (Tables S4 and S5). Values of dN are broadly accelerated. On the other hand, only 3% of dS comparisons are significantly different, and these tests indicate that values of rpl- and rps-genes are significantly different relative to psb-genes.
We suggest that a second factor, variable levels of gene expression, affects rates of sequence evolution in Geraniaceae plastid genomes. It has been shown that variation in nucleotide substitutions is correlated with expression levels, where genes that are highly expressed evolve at a lower rate, such as those reported in E. coli, S. cerevisiae, and Drosophila (33–35). Moreover, for plastid genes of the liverwort Marchantia polymorpha and the angiosperm Nicotiana tabacum, Morton (36) described a correlation between expression levels and increased dN and dS. The expression levels of Geraniaceae plastid genes have not been quantified, but it is known that proteins accumulate to different levels in plastids. Available data gathered from barley seedlings showed that there was accumulation of Rubisco, photosystem I and II, and α and β ATPase proteins (37, 38). Notably, genes encoding these highly accumulated proteins correspond to low values of dN, dS, and dN/dS in our analyses (see Figs. 2, 4, and 5), and it appears that expression level and rates of sequence evolution are also negatively correlated in Geraniaceae plastid genes. Genome-wide estimation of expression levels in Geraniaceae genes are needed to better understand the relationship between expression levels and rates of nucleotide substitutions.
Transcription of plastid genes in most land plants is shared by two RNA polymerases, the plastid-encoded polymerase (PEP) and the imported nuclear-encoded polymerase (NEP). Early models suggested that transcription of photosynthetic genes was controlled by PEP, whereas housekeeping genes were transcribed by NEP (39). More recent work has challenged this model, and plastid maturity, plant developmental stage, and posttranscriptional regulation are implicated in a biologically complex pattern of gene transcription (40–43). Nonetheless, if gene expression is correlated with gene transcription, differences in rates of sequence evolution could be due to variable transcription by these polymerases. It is notable that one genus of Geraniaceae, Pelargonium, has lost a functional copy of the PEP gene rpoA; however, rpoA-like ORFs were found dispersed throughout the plastid genome (16). rpoA was transferred from the plastid genome to the nuclear genome in a moss (44) and in the nonphotosynthetic angiosperm Cuscuta (42), but little is known about rpoA in Pelargonium and other Geraniaceae (45–47). In the case of Cuscuta, loss of rpoA and rpoB results in a change from PEP- to NEP-based promoter sequences in the rrn16 gene (42). Control of transcription was taken over by the nucleus, and it is possible that this has also occurred in Pelargonium.
Geraniaceae RNA polymerase genes rpoB, rpoC1, and rpoC2 are accumulating an unusual amount of nonsynonymous substitutions, indicating either positive or relaxed selection (Fig. 5). Aside from Pelargonium, other Geraniaceae genomes appear to contain a functional plastid-encoded rpoA gene, but the other PEP subunits are rapidly accumulating nonsynonymous substitutions. It is possible that rpoA was transferred to the nucleus early in the evolution of Geraniaceae. Models of organellar gene loss require gene duplication and transfer before loss. Functional loss of rpoA may have only occurred in Pelargonium, but a nuclear transfer event in the evolution of Geraniaceae could result in strong selective pressure and high levels of substitutions in the other subunits of the plastid-encoded polymerase. Substitutions in a nuclear-encoded rpoA in Geraniaceae would then incur compensatory substitutions in rpoB, rpoC1, and rpoC2 to maintain the function of the polymerase. Indeed in the nuclear-encoded rpoA of the moss Physcomitrella patens, substitutions are numerous and show a strong bias toward nuclear codon use (44). Alternatively, we cannot exclude the possibility that in the plastid genome of Pelargonium the rpoA-like ORFs identified by Chumley et al. (16) are actually highly divergent functional genes, and compensatory substitutions in other subunits of the polymerase result. Notably, expression data from plastids suggests that rpoA may be functional (P. Kuhlman and J. D. Palmer, personal communication).
This study provides evidence for unprecedented increases in nucleotide substitutions in angiosperm plastid genomes, and we use protein-coding sequences from complete plastid genomes to characterize rates and patterns of sequence evolution in the plastids of photosynthetic angiosperms. We report extreme accumulation of dN in ribosomal protein and RNA polymerase genes, and these data provide evidence for relaxed selection or positive selection that is unique among photosynthetic angiosperm plastid genomes. In addition, this study provides evidence that angiosperm plastid genomes contain a high degree of locus- and lineage-specific rate variation, and plastid genomes are far more dynamic than previously described. Our observations are consistent with earlier models of plastid sequence evolution (48), which indicated that rates of both dS and dN vary across lineages, rates of dS are relatively homogenous across loci, and rates of dN vary extensively across plastid genomes. However, our observations are the first to show the magnitude of rate variation among plastid loci and angiosperm lineages.
Geraniaceae plastid genomes provide the rare opportunity to examine anomalous plastid sequence evolution and to model genomic changes. Our model involves improper DNA repair leading to genome rearrangement and increased nucleotide substitutions and possible transcriptional control of plastid genes by the nucleus leading to altered expression and increased nucleotide substitutions. Completed sequences for additional highly rearranged Geraniaceae and angiosperm plastid genomes and characterization of genes involved in DNA repair in Pelargonium and other Geraniaceae are needed to better understand the highly accelerated substitution patterns in this family. In addition, expression data are needed to correlate rates of nucleotide substitutions with levels of gene transcription.
Materials and Methods
Gene Sequencing.
To maximize sampling in major angiosperm clades, 47 taxa were chosen, including one previously published Geraniaceae plastid genome (16) (Table S2). Nearly complete draft sequences are available for eight additional Geraniaceae taxa (see Table S3 for genome sequence status). Protocols for plastid isolation, RCA amplification, sequencing, and sequence assembly are previously described (49). Protein-coding sequences for 72 genes were used with several exclusions (Table S1).
Gene Annotation and Alignment.
Genes were annotated by using DOGMA (50). Start and stop codons were determined based on similarity with known sequences following the bacterial and plastid genetic code. For highly divergent genes adjacent start/stop codons were used. Amino acid sequences were aligned by using MSWAT (http://mswat.ccbb.utexas.edu), manually adjusted, and used to constrain the nucleotide alignment.
Phylogenetic Analyses.
MP analyses were performed by using 100 random addition replicates and TBR branch swapping with the Multrees option using PAUP* Version 4.0b10 (51). Four independent ML analyses were performed by using GARLI and default settings (52). Bootstrap values were generated by using PAUP* and GARLI with 100 replicates and the above settings. ML analysis were also performed on a set of 27 slowly evolving genes (including the individual gene rbcL and subunits of pet-, psa, and psb-genes).
Evolutionary Rate Estimation.
Using the codon frequencies model F3 × 4, PAML's codeml (53) was used to estimate dN, dS, and dN/dS. Gapped regions were excluded using cleandata = 1 to avoid spurious rate inference. The phylogenetic tree generated using MP and ML was used as a constraint tree, but branch lengths were inferred by using PAML. Using the method described by Yang (54), a null model (H0; branch model = 0), where one dN/dS ratio was fixed across angiosperms, was compared with an alternative model (HA; branch model = 2), where the Geraniaceae clade was allowed to have a different dN/dS. Likelihood ratio tests (LRT) were used to test model fit. P-values were transformed according to Sokal and Rohlf (55) [-log(P value + 1)], ranked, and plotted. Genes that encode subunits of a functional complex were grouped according to Matsuoka et al. (56) and Chang et al. (57). Statistical analyses were conducted by using the R software package (http://www.r-project.org), and Bonferroni correction was used.
Supplementary Material
Acknowledgments.
We thank Lauren Ancel Meyers and Claus Wilke for help with statistics; Chris Blazier, Stephen Downie, Cynthia Londeore, and Elizabeth Ruck for careful review of the manuscript; Robin Parer at Geraniaceae.com for supplying plant material; Zhengqui Cai for access to MSWAT and for help submitting sequences to GenBank. Support for this work was provided by National Science Foundation Grant DEB-0717372 (to R.K.J.) and Integrative Graduate Education and Research Traineeship Grant DGE-0114387 (to M.M.G.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition footnote: The sequences in this paper have been deposited in the GenBank database. For a list of accession numbers, see Table S2.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806759105/DCSupplemental.
References
- 1.Raubeson LA, Jansen RK. In: Plant Diversity And Evolution: Genotypic And Phenotypic Variation In Higher Plants. Henry RJ, editor. Cambridge, MA: CABI Publishing; 2005. pp. 45–68. [Google Scholar]
- 2.Wolfe KH, Li WH, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci USA. 1987;84:9054–9058. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Erixon P, Oxelman B. Whole-Gene positive selection, elevated synonymous substitution rates, duplication, and indel evolution of the chloroplast clpP1 gene. PLoS ONE. 2008;3:e1386. doi: 10.1371/journal.pone.0001386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Funk HT, Berg S, Krupinska K, Maier UG, Krause K. Complete DNA sequences of the plastid genomes of two parasitic flowering plant species, Cuscuta reflexa and Cuscuta gronovii. BMC Plant Biol. 2007;7:45. doi: 10.1186/1471-2229-7-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McNeal JR, Arumugunathan K, Kuehl JV, Boore JL, dePamphilis CW. Systematics and plastid genome evolution of the cryptically photosynthetic parasitic plant genus Cuscuta (Convolvulaceae) BMC Biol. 2007;5:55. doi: 10.1186/1741-7007-5-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolfe KH, Morden CW, Palmer JD. Function and evolution of a minimal plastid genome from a nonphotosynthetic parasitic plant. Proc Natl Acad Sci USA. 1992;89:10648–10652. doi: 10.1073/pnas.89.22.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaut BS, Muse SV, Clegg MT. Relative rates of nucleotide substitution in the chloroplast genome. Mol Phylogenet Evol. 1993;2:89–96. doi: 10.1006/mpev.1993.1009. [DOI] [PubMed] [Google Scholar]
- 8.Wu CS, Wang YN, Liu SM, Chaw SM. Chloroplast genome (cpDNA) of Cycas taitungensis and 56 cp protein-coding genes of Gnetum parvifolium: Insights into cpDNA evolution and phylogeny of extant seed plants. Mol Biol Evol. 2007;24:1366–1379. doi: 10.1093/molbev/msm059. [DOI] [PubMed] [Google Scholar]
- 9.McCoy SR, Kuehl JV, Boore JL, Raubeson LA. The complete plastid genome sequence of Welwitschia mirabilis: An unusually compact plastome with accelerated divergence rates. BMC Evol Biol. 2008;8:130. doi: 10.1186/1471-2148-8-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Logacheva MD, Penin AA, Samigullin TH, Vallejo-Roman CM, Antonov AS. Phylogeny of flowering plants by the chloroplast genome sequences: In search of a “lucky gene”. Biochemistry (Mosc) 2007;72:1324–1330. doi: 10.1134/s0006297907120061. [DOI] [PubMed] [Google Scholar]
- 11.Perry AS, Wolfe KH. Nucleotide substitution rates in legume chloroplast DNA depend on the presence of the inverted repeat. J Mol Evol. 2002;55:501–508. doi: 10.1007/s00239-002-2333-y. [DOI] [PubMed] [Google Scholar]
- 12.Parkinson CL, et al. Multiple major increases and decreases in mitochondrial substitution rates in the plant family Geraniaceae. BMC Evol Biol. 2005;5:73. doi: 10.1186/1471-2148-5-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cho Y, Mower JP, Qiu YL, Palmer JD. Mitochondrial substitution rates are extraordinarily elevated and variable in a genus of flowering plants. Proc Natl Acad Sci USA. 2004;101:17741–17746. doi: 10.1073/pnas.0408302101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shao R, Dowton M, Murrell A, Barker SC. Rates of gene rearrangement and nucleotide substitution are correlated in the mitochondrial genomes of insects. Mol Biol Evol. 2003;20:1612–1619. doi: 10.1093/molbev/msg176. [DOI] [PubMed] [Google Scholar]
- 15.Xu W, Jameson D, Tang B, Higgs PG. The relationship between the rate of molecular evolution and the rate of genome rearrangement in animal mitochondrial genomes. J Mol Evol. 2006;63:375–392. doi: 10.1007/s00239-005-0246-5. [DOI] [PubMed] [Google Scholar]
- 16.Chumley TW, et al. The complete chloroplast genome sequence of Pelargonium x hortorum: Organization and evolution of the largest and most highly rearranged chloroplast genome of land plants. Mol Biol Evol. 2006;23:2175–2190. doi: 10.1093/molbev/msl089. [DOI] [PubMed] [Google Scholar]
- 17.Price RA, Calie PJ, Downie SR, Logsdon JM, Palmer JD. In: Vorster P, editor. The International Geraniaceae Symposium; Republic of South Africa: The University of Stellenbosch; 1990. pp. 237–244. [Google Scholar]
- 18.Jansen RK, et al. Analysis of 81 genes from 64 plastid genomes resolves relationships in angiosperms and identifies genome-scale evolutionary patterns. Proc Natl Acad Sci USA. 2007;104:19369–19374. doi: 10.1073/pnas.0709121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hilu KW, et al. Angiosperm phylogeny based on matK sequence information. Am J Bot. 2003;90:1758–1776. doi: 10.3732/ajb.90.12.1758. [DOI] [PubMed] [Google Scholar]
- 20.Zhu XY, et al. Mitochondrial matR sequences help to resolve deep phylogenetic relationships in rosids. BMC Evol Biol. 2007;7:217. doi: 10.1186/1471-2148-7-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fiz O, et al. Phylogeny and historical biogeography of Geraniaceae in relation to climate changes and pollination ecology. Syst Bot. 2008;33:326–342. [Google Scholar]
- 22.Price RA, Palmer JD. Phylogenetic relationships of the Geraniaceae and Geraniales from rbcL sequence comparisons. Ann Miss Bot Gard. 1993;80:661–671. [Google Scholar]
- 23.Wong KM, Suchard MA, Huelsenbeck JP. Alignment uncertainty and genomic analysis. Science. 2008;319:473–476. doi: 10.1126/science.1151532. [DOI] [PubMed] [Google Scholar]
- 24.Soria-Hernanz DF, Braverman JM, Hamilton MB. Parallel rate heterogeneity in chloroplast and mitochondrial genomes of Brazil nut trees (Lecythidaceae) is consistent with lineage effects. Mol Biol Evol. 2008;25:1282–1296. doi: 10.1093/molbev/msn074. [DOI] [PubMed] [Google Scholar]
- 25.Lin Z, Kong H, Nei M, Ma H. Origins and evolution of the recA/RAD51 gene family: Evidence for ancient gene duplication and endosymbiotic gene transfer. Proc Natl Acad Sci USA. 2006;103:10328–10333. doi: 10.1073/pnas.0604232103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cerutti H, Osman M, Grandoni P, Jagendorf AT. A homolog of Escherichia coli RecA protein in plastids of higher plants. Proc Natl Acad Sci USA. 1992;89:8068–8072. doi: 10.1073/pnas.89.17.8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khazi FR, Edmondson AC, Nielsen BL. An Arabidopsis homologue of bacterial RecA that complements an E. coli recA deletion is targeted to plant mitochondria. Mol Genet Genomics. 2003;269:454–463. doi: 10.1007/s00438-003-0859-6. [DOI] [PubMed] [Google Scholar]
- 28.Bock R, Timmis JN. Reconstructing evolution: Gene transfer from plastids to the nucleus. Bioessays. 2008;30:556–566. doi: 10.1002/bies.20761. [DOI] [PubMed] [Google Scholar]
- 29.Hartung F, Suer S, Puchta H. Two closely related RecQ helicases have antagonistic roles in homologous recombination and DNA repair in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2007;104:18836–18841. doi: 10.1073/pnas.0705998104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson-Schlitz D, Engels WR. Template disruptions and failure of double Holliday junction dissolution during double-strand break repair in Drosophila BLM mutants. Proc Natl Acad Sci USA. 2006;103:16840–16845. doi: 10.1073/pnas.0607904103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saotome A, et al. Characterization of four RecQ homologues from rice (Oryza sativa L. cv. Nipponbare) Biochem Biophys Res Commun. 2006;345:1283–1291. doi: 10.1016/j.bbrc.2006.04.134. [DOI] [PubMed] [Google Scholar]
- 32.Bagherieh-Najjar MB, Vries OMH, Hille J, Dijkwel PP. Arabidopsis RecQl4A suppresses homologous recombination and modulates DNA damage responses. Plant J. 2005;43:789–798. doi: 10.1111/j.1365-313X.2005.02501.x. [DOI] [PubMed] [Google Scholar]
- 33.Drummond DA, Raval A, Wilke CO. A single determinant dominates the rate of yeast protein evolution. Mol Biol Evol. 2006;23:327–337. doi: 10.1093/molbev/msj038. [DOI] [PubMed] [Google Scholar]
- 34.Sharp PM. Determinants of DNA sequence divergence between Escherichia coli and Salmonella typhimurium: Codon usage, map position, and concerted evolution. J Mol Evol. 1991;33:23–33. doi: 10.1007/BF02100192. [DOI] [PubMed] [Google Scholar]
- 35.Shields DC, Sharp PM, Higgins DG, Wright F. “Silent” sites in Drosophila genes are not neutral: Evidence of selection among synonymous codons. Mol Biol Evol. 1988;5:704–716. doi: 10.1093/oxfordjournals.molbev.a040525. [DOI] [PubMed] [Google Scholar]
- 36.Morton BR. Codon use and the rate of divergence of land plant chloroplast genes. Mol Biol Evol. 1994;11:231–238. doi: 10.1093/oxfordjournals.molbev.a040105. [DOI] [PubMed] [Google Scholar]
- 37.Klein RR, Mason HS, Mullet JE. Light-regulated translation of chloroplast proteins. I. Transcripts of psaA-psaB, psbA, and rbcL are associated with polysomes in dark-grown and illuminated barley seedlings. J Cell Biol. 1988;106:289–301. doi: 10.1083/jcb.106.2.289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mullet JE. Chloroplast development and gene-expression. Ann Rev Plant Physiol Plant Mol Biol. 1988;39:475–502. [Google Scholar]
- 39.Hajdukiewicz PT, Allison LA, Maliga P. The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J. 1997;16:4041–4048. doi: 10.1093/emboj/16.13.4041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cahoon AB, Harris FM, Stern DB. Analysis of developing maize plastids reveals two mRNA stability classes correlating with RNA polymerase type. EMBO Rep. 2004;5:801–806. doi: 10.1038/sj.embor.7400202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hanaoka M, Kanamaru K, Fujiwara M, Takahashi H, Tanaka K. Glutamyl-tRNA mediates a switch in RNA polymerase use during chloroplast biogenesis. EMBO Rep. 2005;6:545–550. doi: 10.1038/sj.embor.7400411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Krause K, Berg S, Krupinska K. Plastid transcription in the holoparasitic plant genus Cuscuta: Parallel loss of the rrn16 PEP-promoter and of the rpoA and rpoB genes coding for the plastid-encoded RNA polymerase. Planta. 2003;216:815–823. doi: 10.1007/s00425-002-0933-z. [DOI] [PubMed] [Google Scholar]
- 43.Legen J, et al. Comparative analysis of plastid transcription profiles of entire plastid chromosomes from tobacco attributed to wild-type and PEP-deficient transcription machineries. Plant J. 2002;31:171–188. doi: 10.1046/j.1365-313x.2002.01349.x. [DOI] [PubMed] [Google Scholar]
- 44.Sugiura C, Kobayashi Y, Aoki S, Sugita C, Sugita M. Complete chloroplast DNA sequence of the moss Physcomitrella patens: Evidence for the loss and relocation of rpoA from the chloroplast to the nucleus. Nucleic Acids Res. 2003;31:5324–5331. doi: 10.1093/nar/gkg726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Downie SR, Katz-Downie DS, Wolfe KH, Calie PJ, Palmer JD. Structure and evolution of the largest chloroplast gene (ORF2280): Internal plasticity and multiple gene loss during angiosperm evolution. Curr Genet. 1994;25:367–378. doi: 10.1007/BF00351492. [DOI] [PubMed] [Google Scholar]
- 46.Palmer JD, et al. In: Current research in photosynthesis. Baltscheffsky M, editor. Amsterdam: Kluwer Academic Publishers; 1990. pp. 475–482. [Google Scholar]
- 47.Palmer JD, Baldauf SL, Calie PJ, dePamphilis CW. In: Molecular evolution. Clegg M, O'Brien S, editors. New York: Alan R. Liss, Inc.; 1990. pp. 97–106. [Google Scholar]
- 48.Muse SV, Gaut BS. A likelihood approach for comparing synonymous and nonsynonymous nucleotide substitution rates, with application to the chloroplast genome. Mol Biol Evol. 1994;11:715–724. doi: 10.1093/oxfordjournals.molbev.a040152. [DOI] [PubMed] [Google Scholar]
- 49.Jansen RK, et al. Methods for obtaining and analyzing whole chloroplast genome sequences. Methods Enzymol. 2005;395:348–384. doi: 10.1016/S0076-6879(05)95020-9. [DOI] [PubMed] [Google Scholar]
- 50.Wyman SK, Jansen RK, Boore JL. Automatic annotation of organellar genomes with DOGMA. Bioinformatics. 2004;20:3252–3255. doi: 10.1093/bioinformatics/bth352. [DOI] [PubMed] [Google Scholar]
- 51.Swofford D. Phylogenetic Analysis Using Parsimony (*and Other Methods). Version 4. Sunderland, MA: Sinauer Associates; 2003. PAUP*. [Google Scholar]
- 52.Zwickl DJ. GARLI: Genetic Algorithm for Rapid Likelihood Inference, Version 0.951. GARLI: Genetic Algorithm for Rapid Likelihood Inference, Version 0.951. 2006 Available at http://www.bio.utexas.edu/faculty/antisense/garli/Garli.html.
- 53.Yang Z. PAML 4: Phylogenetic Analysis by Maximum Likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 54.Yang Z. Likelihood ratio tests for detecting positive selection and application to primate lysozyme evolution. Mol Biol Evol. 1998;15:568–573. doi: 10.1093/oxfordjournals.molbev.a025957. [DOI] [PubMed] [Google Scholar]
- 55.Sokal RR, Rohlf FJ. Biometry : The Principles and Practice of Statistics in Biological Research. New York: W.H. Freeman; 1995. [Google Scholar]
- 56.Matsuoka Y, Yamazaki Y, Ogihara Y, Tsunewaki K. Whole chloroplast genome comparison of rice, maize, and wheat: Implications for chloroplast gene diversification and phylogeny of cereals. Mol Biol Evol. 2002;19:2084–2091. doi: 10.1093/oxfordjournals.molbev.a004033. [DOI] [PubMed] [Google Scholar]
- 57.Chang CC, Lin HC, Lin IP, Chow TY, et al. The chloroplast genome of Phalaenopsis aphrodite (Orchidaceae): Comparative analysis of evolutionary rate with that of grasses and its phylogenetic implications. Mol Biol Evol. 2006;23:279–291. doi: 10.1093/molbev/msj029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.