The biosphere is brimming with examples of symbiotic interactions between distantly related organisms that have evolved mutually beneficial relationships to ensure their survival. One might assume that there are many examples of syntrophic interactions among microorganisms because of their diversity and abundance (1, 2). Remarkably few cases between 2 or more microbial species have been described, however, and even fewer have been characterized in detail (3–6). With incomplete evidence to support the mechanisms that are involved in the emergence of symbiosis, a central question remains unanswered: how do different microbes become mutually agreeable neighbors? An understanding of the mechanisms that underlie this question will play a fundamental role in our understanding of ecology, evolution, and microbiology and may provide a starting point and a set of design principles for engineering “synthetic” multicellular communities that have new structure and function. In this issue of PNAS, Kim et al. (7) break a barrier along the pathway to engineering synthetic communities of bacteria by demonstrating how their spatial organization is involved in the development of syntrophy.
The creation of syntrophic populations of microbes may be perceived as being as simple as mixing together strains that have capabilities that the other strains lack, which are required for their survival (e.g., the ability to degrade antimicrobial compounds in an antibiotic-rich environment). It is reasonable to assume that if each organism encodes a phenotype that is beneficial to the community, the consortium will reach a stable population in which every strain survives. This assumption is a gross undersimplification, however, and in practice the creation of stable, mixed populations of n bacterial strains is remarkably difficult to achieve, particularly when n ≥ 3 (8–10). This situation arises for several reasons. (i) The rates of growth of the different species are typically not balanced and the consumption and production of metabolites are not matched. (ii) The fluctuation of physicochemical factors produced by the quorum biases the growth and survival of some strains (e.g., some species form biofilms) and is bacteriostatic or even bactericidal to other strains (e.g., changes in pH or osmolality). (iii) Microbial warfare kills organisms that are perceived as competitors through the production of toxic secondary metabolites. The combination of these and other characteristics produce systems that are intrinsically nonlinear. Until recently it appeared that models of these systems, which would make it possible to predict and design syntrophic communities, would be characterized by a series of differential equations that include variables that are currently unknown or are just beginning to emerge. The article by Kim et al. (7) describes the important of “space” and defines how this variable affects the growth and homeostasis of a population of unrelated bacteria.
The role of spatial organization in multicellular populations is not new. Other groups have demonstrated that spatial dynamics are important for multicellular interactions (11, 12). For example, the spatial colocalization of 2 engineered strains of Escherichia coli produces an engineered consortium in which a consensus gene expression response arises (13). Programmed pattern formation has been observed between engineered strains of E. coli in which 1 strain sends a chemical signal and the other acts as a receiver (14). The relationship between microscale spatial structure and syntrophic communities of bacteria, however, has not been characterized in detail and is the focus of the article by Kim et al. (7).
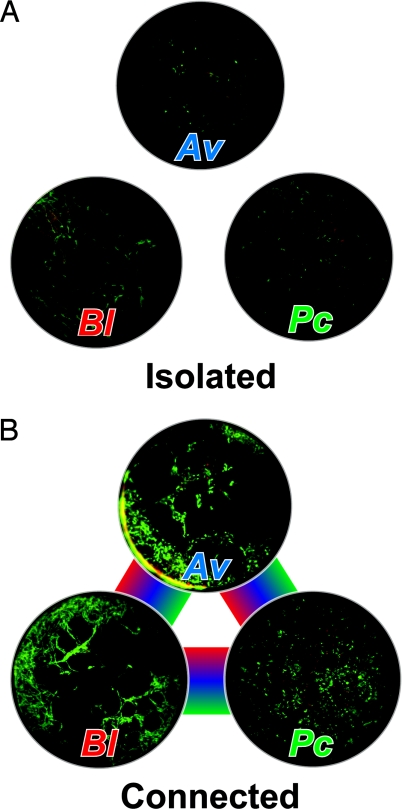
To study the affect of spatial structure on multispecies communities of bacteria, Kim et al. (7) developed a microfluidic system (15) to grow populations of 3 unrelated colonies of WT soil bacteria that are not known to interact in nature: Azotobacter vinelandii fixes nitrogen gas into amino acids; Bacillus licheniformis degrades penicillin G by hydrolyzing the β-lactam ring; and Paenibacillus curdlanolyticus hydrolyzes carboxymethyl cellulose to d-glucose. The microfluidic system provides a physical barrier (the equivalent of a “fence”) between monoclonal liquid cultures of the 3 organisms but leaves them in fluidic contact as the floor of the compartments consists of a porous membrane with nanoscopic holes through which the cells cannot penetrate. Below the membrane is a channel filled with liquid that connects the colonies together and provides a conduit for the transport of small molecules and ions between the cultures. By controlling the distance between the 3 organisms, Kim et al. demonstrate the emergence of reciprocal syntrophy during growth in nutrient conditions in which the organisms are forced to cooperate for their survival (Fig. 1).
Fig. 1.
Spatial organization of bacteria and syntrophy of a community. (A) Isolated cultures of A. vinelandii (Av), B. licheniformis (Bl), and P. curdlanolyticus (Pc). The bacteria were grown for 36 h in minimal media supplemented with penicillin G and containing carboxymethyl cellulose as the carbon source. Live cells appear green and dead cells appear red. (B) The strains were grown in the same nutrient conditions in a microfluidic system that controls the distance between the bacteria and permits their chemical communication with each other. The rainbow lines depict fluidic contact between the populations of bacteria and the sharing of metabolites. The connected cultures form a stable syntrophic community.
The mathematical model that emerges from their study illustrates how spatial structure balances the supportive and competitive interactions within a multispecies community of bacteria. The model demonstrates that stability is not achieved when the distance between the inhabitants is small because mutual consumption of metabolites exceeds the production of secondary metabolites. As the distance between the organisms is increased the consumption rate decreases and the system reaches a stable steady state. At even larger distances the system is incapable of reaching a stable steady state that balances the consumption and production of metabolites.
Three classes of bacterial communities are predicted from the models of nonlinear behavior described by Kim et al. (7). Class I communities are stable in a well-mixed environment and at intermediate distances between the inhabitants. This population is characterized by rates of production of essential secondary metabolites that can accommodate the consumption by all 3 organisms. Class I communities exist in nature and have been described before (7, 8), but many of these interactions have not been studied in the laboratory.
Class II communities are unstable when the organisms are positioned too close to each other. In these cases the consumption of metabolites exceeds their production. As the members of the community are separated, interspecific competition is reduced and the population becomes stable. These communities require spatial structure for their stability and are characteristic of the syntrophic system of A. vinelandii, B. licheniformis, and P. curdlanolyticus (10). Other synthetic class II communities should be possible by using strains of bacteria that possess complementary phenotypes. It is currently unclear whether natural communities of bacteria or other microorganisms belong to the class II category, because they would not be culturable with traditional microbiological methods. These communities require techniques for controlling their spatial organization (e.g., geometry and distance) to achieve stability. One way to impose this organization is by using a microfluidic device (16, 17), such as the system described by Kim et al. (7), or other techniques for controlling the position of bacteria on surfaces (18, 19).
Spatial structure balances the supportive and competitive interactions within a multispecies community of bacteria.
A class III community never attains stability because of the intrinsic mismatch of the consumption and production of metabolites by the bacterial strains.
The view that emerges from the work by Kim et al. (7) is that control over the spatial organization of a multispecies community of microbes makes it possible to override differences in the rate of growth and metabolism of organisms to achieve a stable, reciprocal syntrophic community. The principles that underlie the article by Kim et al. may find application in synthetic biology, the identification of new microbial communities in the biosphere, the study of the evolution of symbiosis, and the creation or possibly the recapitulation of the emergence of symbiotic relationships (e.g., mitochondria, chloroplasts). One of the most intriguing applications of Kim et al.'s article may be the isolation, growth, and identification of “unculturable” species of microbes that may require factors for their growth that can be provided by class II communities (20). Robert Frost, in his poem Mending Wall, described 2 neighbors strengthening a friendship while repairing the barrier between their properties. Kim et al. demonstrate that bacteria, growing collaboratively yet while demanding ideal length scales of separation, concur with the conclusion that “good fences make good neighbors,” whether they be stone walls or microfluidic channels.
Footnotes
The author declares no conflict of interest.
See companion article on page 18188.
References
- 1.Gans J, Wolinsky M, Dunbar J. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science. 2005;309:1387–1390. doi: 10.1126/science.1112665. [DOI] [PubMed] [Google Scholar]
- 2.Curtis TP, Sloan WT, Scannell JW. Estimating prokaryotic diversity and its limits. Proc Natl Acad Sci USA. 2002;99:10494–10499. doi: 10.1073/pnas.142680199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frostl JM, Overmann J. Phylogenetic affiliation of the bacteria that constitute phototrophic consortia. Arch Microbiol. 2000;174:50–58. doi: 10.1007/s002030000172. [DOI] [PubMed] [Google Scholar]
- 4.Glaeser J, Overmann J. The significance of organic carbon compounds for in situ metabolism and chemotaxis of phototrophic consortia. Environ Microbiol. 2003;5:1053–1063. doi: 10.1046/j.1462-2920.2003.00516.x. [DOI] [PubMed] [Google Scholar]
- 5.Glaeser J, Overmann J. Characterization and in situ carbon metabolism of phototrophic consortia. Appl Environ Microbiol. 2003;69:3739–3750. doi: 10.1128/AEM.69.7.3739-3750.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kanzler BEM, Pfannes KR, Vogl K, Overmann J. Molecular characterization of the nonphotosynthetic partner bacterium in the consortium Chlorochromatium aggregatum. Appl Environ Microbiol. 2005;71:7434–7441. doi: 10.1128/AEM.71.11.7434-7441.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim HJ, Boedicker JQ, Choi JW, Ismagilov RF. Defined spatial structure stabilizes a synthetic multi-species bacterial community. Proc Natl Acad Sci USA. 2008;105:18188–18193. doi: 10.1073/pnas.0807935105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kato S, Haruta S, Cui Z, Ishii M, Igarishi Y. Stable coexistence of five bacterial strains as a cellulose-degrading community. Appl Environ Microbiol. 2005;71:7099–7106. doi: 10.1128/AEM.71.11.7099-7106.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kato S, Haruta S, Cui Z, Ishii M, Igarishi Y. Network relationships of bacteria in a stable mixed culture. Microb Ecol. 2008;56:403–411. doi: 10.1007/s00248-007-9357-4. [DOI] [PubMed] [Google Scholar]
- 10.Haruta S, et al. Construction of a stable microbial community with high cellulose-degradation ability. Appl Microbiol Biotechnol. 2002;59:529–534. doi: 10.1007/s00253-002-1026-4. [DOI] [PubMed] [Google Scholar]
- 11.Hassell MP, Comins HN, May RM. Species coexistence and self-organizing spatial dynamics. Nature. 1994;370:290–292. [Google Scholar]
- 12.Hansen SK, Rainey PB, Haagensen JAJ, Molin S. Evolution of species interactions in a biofilm community. Nature. 2007;445:533–536. doi: 10.1038/nature05514. [DOI] [PubMed] [Google Scholar]
- 13.Brenner K, Karig DK, Weiss R, Arnold FH. Engineered bidirectional communication mediates a consensus in a microbial biofilm consortium. Proc Natl Acad Sci USA. 2007;104:17300–17304. doi: 10.1073/pnas.0704256104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Basu S, Gerchman Y, Collins CH, Arnold FH, Weiss R. A synthetic multicellular system for programmed pattern formation. Nature. 2005;434:1130–1134. doi: 10.1038/nature03461. [DOI] [PubMed] [Google Scholar]
- 15.Weibel DB, DiLuzio WR, Whitesides GM. Microfabrication meets microbiology. Nat Rev Microbiol. 2007;5:209–218. doi: 10.1038/nrmicro1616. [DOI] [PubMed] [Google Scholar]
- 16.Ingham CJ, et al. The micro-Petri dish, a million-well growth chip for the culture and high-throughput screening of microorganisms. Proc Natl Acad Sci USA. 2007;104:18217–18222. doi: 10.1073/pnas.0701693104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Abhyankar VV, Beebe DJ. Spatiotemporal micropatterning of cells on arbitrary substrates. Bacterial printing press that regenerates its ink: Contact-printing bacteria using hydrogel stamps. Anal Chem. 2007;79:4066–4-73. doi: 10.1021/ac062371p. [DOI] [PubMed] [Google Scholar]
- 18.Weibel DB, et al. Bacterial printing press that regenerates its ink: Contact-printing bacteria using hydrogel stamps. Langmuir. 2005;21:6436–6442. doi: 10.1021/la047173c. [DOI] [PubMed] [Google Scholar]
- 19.Rozhok S, et al. Methods for fabricating microarrays of motile bacteria. Small. 2005;1:445–451. doi: 10.1002/smll.200400072. [DOI] [PubMed] [Google Scholar]
- 20.Risenfeld CS, Schloss PD, Handelsman J. Metagenomics: Genomic analysis of microbial communities. Annu Rev Genet. 2004;38:525–552. doi: 10.1146/annurev.genet.38.072902.091216. [DOI] [PubMed] [Google Scholar]