Abstract
Sex-lethal (Sxl), the master regulatory gene of Drosophila somatic sex determination, is stably maintained in an on or an off state by autoregulatory control of Sxl premRNA processing. Establishment of the correct Sxl splicing pattern requires the coordinate regulation of two Sxl promoters. The first of these promoters, SxlPe, responds to the female dose of two X chromosomes to produce a pulse of Sxl protein that acts on the premRNA products from the second promoter, SxlPm, to establish the splicing loop. SxlPm is active in both sexes throughout most of development, but nothing is known about how SxlPm is expressed during the transition from X signal assessment to maintenance splicing. We found that SxlPm is activated earlier in females than in males in a range of Drosophila species, and that its expression overlaps briefly with that of SxlPe during the syncytial blastoderm stage. Activation of SxlPm depends on the scute, daughterless, and runt transcription factors, which communicate X chromosome dose to SxlPe, but is independent of the X signal element sisA and the maternal co-repressor groucho. We show that DNA sequences regulating the response of SxlPe to the X chromosome dose also control the sex-differential response of SxlPm. We propose that co-expression of Sxl protein and its premRNA substrate facilitates the transition from transcriptional to splicing control, and that delayed activation of SxlPm in males buffers against the inappropriate activation of Sxl by fluctuations in the strength of the X chromosome signal.
Keywords: D. virilis, D. yakuba, X:A ratio, XSE, genetic switch
Cell fate decisions often are controlled by regulatory genes or pathways that respond to small quantitative differences in the concentrations of signaling molecules. The mechanisms that maintain such genes or pathways in stable states often differ from those that initially signal the responses. Such is the case in the somatic sex determination system of Drosophila. Throughout nearly all of the fly life cycle, sexual identity is maintained via a positive feedback mechanism that controls the splicing of the transcripts of the switch gene, Sex-lethal (Sxl), so that they encode active Sxl proteins in females and truncated, inactive proteins in males (1–3). This occurs because transcripts from the “maintenance” promoter, SxlPm, are processed into functional mRNA species only in the presence of Sxl protein (4–6). Whereas positive autoregulatory splicing of SxlPm-derived transcripts constitutes a self-sustaining loop, a different mode of regulation is required to initiate the process. The key event is the transient activation of the “establishment” promoter, SxlPe, in response to the female dose of two X chromosomes (7–9). Transcripts from SxlPe, unlike those from SxlPm, are spliced by default so as to produce functional Sxl mRNA and protein (7, 10, 11). Thus, the burst of SxlPe activity in XX embryos supplies the protein that initiates female-specific splicing from SxlPm. In contrast, the failure to activate SxlPe in XY embryos leaves the SxlPm-derived RNAs to be spliced in the nonfunctional male state.
Whereas the transient female-specific activation of SxlPe in response to X chromosome dose has been the target of much experimental scrutiny, little is known about the control of SxlPm. The standard view is that SxlPm is a “housekeeping” promoter active in both sexes from around the time of gastrulation through adulthood. Analysis of Sxl RNA by Northern blot or RNase protection assays (4, 7, 12, 13) has suggested a time lag of 1–2 h between the cessation of SxlPe activity in early nuclear cycle 14 and the onset of SxlPm expression (14), supporting the concept that the two promoters are expressed independently. On the other hand, Barbash and Cline (15) detected SxlPm-derived transcripts during cycle 14, and Keyes (16) noted that SxlPm appeared to be expressed earlier in XX embryos than in XY embryos, raising the possibility of a direct regulatory connection between SxlPm and SxlPe.
To gain insight into the switch between the two Sxl promoters, we analyzed the time course of SxlPm activity during and immediately after syncytial cycles 12, 13, and early 14, when the X chromosome dose is assessed at SxlPe (9, 17, 18). Remarkably, we found that the activities of the two promoters overlap briefly in female embryos and that there is a short, but distinct, delay in the appearance of SxlPm transcripts in males. This female-first pattern of maintenance promoter activity is conserved in Drosophila species, suggesting that SxlPm also responds to the number of X chromosomes. Here we show, using genetic and transgenic analyses, that the earlier onset of SxlPm in females is not simply a consequence of the female-specific activation of the downstream SxlPe, but that the two promoters share a common enhancer that responds to X chromosome dose. We report that some, but not all, of the X-linked signal element (XSE) proteins that regulate SxlPe (8, 9, 19) are needed for the earliest expression from SxlPm, demonstrating an unexpected complexity in the X-counting mechanism. We propose that the early overlapping activities of the two promoters facilitates the transition to stable splicing control, and that the association between X chromosome dose and SxlPm further amplifies the XSE signal and thus contributes to the robustness of the on-or-off control of Sxl.
Results
To define when SxlPm is activated, we developed an in situ hybridization assay using an intron-derived probe (Fig. 1) that enabled us to identify nascent SxlPm-derived transcripts as focused dots of staining in embryonic nuclei. Because Sxl is located on the X chromosome, we could differentiate between male (XY) and female (XX) embryos based on the number of dots visible in the nuclei. Progression through cycle 14 was monitored using two different parameters: the ratio between the length and width of the surface nuclei and the extent of cell membrane invagination during the cellularization process (20, 21).
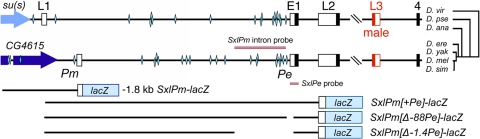
Fig. 1.
Map of Sxl locus and SxlPm-lacZ fusions. (Top) Structure of Sxl exons L1, E1, L2, L3, and 4 in D. pseudoobcura and D. melanogaster. Exons 5–10 are omitted. SxlPm and Pe promoters are marked. Female splice patterns are E1 to 4 and L1 to L2 to 4. The male splice pattern is L1–L2–L3–4. Diamonds represent known or predicted Sc/Da binding sites. Filled portions of Sxl exons represent coding regions. A chromosomal rearrangement exchanged the ancestral upstream su(s) gene for CG4615 after divergence of the D. ananassae and the D. erecta, D. yakuba, D. melanogaster, and D. simulans lineages. D. virilis diverged from the other species about 40 million years ago. (Bottom) SxlPm-lacZ transgenes. Genomic fragments extended 1.8 kb or 0.5 kb upstream of Sxl exon L1. Internal deletions from -88 to + 85 or -1,452 to + 85 bp relative to exon E1 removed the SxlPe promoter and regulatory sequences.
Sxl Maintenance Promoter Is Activated Earlier in Females than in Males.
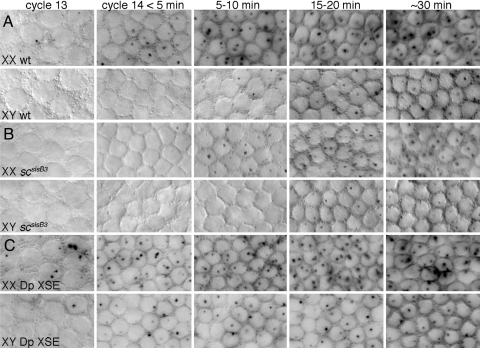
Inspection of embryos after hybridization with SxlPm-specific probes revealed that SxlPm is expressed in both sexes from early in the cellularization cycle until the completion of embryonic development (Fig. 2A and data not shown). Analysis of early embryos showed that the initial expression of SxlPm was sexually dimorphic. Transcripts from SxlPm first appeared in females during nuclear cycle 13 [Fig. 2A; supporting information (SI) Table S1]. Initially, only about 15% of the female nuclei expressed SxlPm, and many nuclei expressed it from only one of the two X chromosomes, suggesting that activation of SxlPm is a stochastic process occurring independently on each X chromosome. During the first minutes of cycle 14, the proportion of expressing nuclei, as well as the number of nuclei expressing both alleles, increased, until by 10–15 min, every female nucleus expressed both copies of SxlPm. In the male embryos, expression from SxlPm was delayed by about 10 min relative to that in the female embryos (Fig. 2A; Table S1). Nascent transcripts from SxlPm were first seen in the XY embryos as very faint dots in scattered nuclei in early cycle 14. As cycle 14 progressed, the proportion of XY nuclei expressing SxlPm and the staining intensity of the nuclear dots increased, until by 20–30 min, every male nucleus was transcribed from SxlPm. Once Sxl was fully active, both sexes maintained expression from SxlPm throughout embryogenesis (data not shown).
Fig. 2.
Time course of nascent transcripts from SxlPm determined by in situ hybridizations with a probe specific for SxlPm-derived premRNA. Surface views of syncytial nuclei. (A) Wild-type XX and XY embryos. (B) scsisB3 mutant XX and XY embryos. (C) XX and XY embryos with two extra copies of the XSEs sc+ and sisA+. Embryos in (B) were progeny of: y scsisB3/y scsisB3 SxlM4 sn females and y scsisB3/Y males. Embryos in (C) were progeny of: w1118 females and y w cm Sxlf1 ct6/Y; 2X P(mini-w+,sisA+)and 2X P(mini-w+,scsisB+)/CyO males.
Our findings demonstrate that SxlPm becomes active earlier than was previously estimated from Northern blot and RNase protection analyses (12, 13). They also indicate that the periods of SxlPe and SxlPm expression overlap in females during the first 10–20 min of cycle 14 (9, 18, 22). To determine whether the sexually dimorphic pattern of SxlPm activation is conserved in other Drosophila species, we examined D. virilis, D. yakuba, and D. simulans (Fig. 1) using in situ hybridization. We found that all three species expressed SxlPm similarly to D. melanogaster (Fig. S1 and A.N.G. and J.W.E., unpublished data), suggesting that the female-first pattern of maintenance promoter activation is, like the female-specific activation of SxlPe (22), an ancient response to the number of X chromosomes.
The XSEs scute and runt and Maternal daughterless Regulate SxlPm.
SxlPe is activated during nuclear cycle 12 and expressed through the first 10–20 min of cycle 14 in response to the two X chromosome dose of XSEs (9, 15, 18). To determine whether the same XSEs that control the on-or-off response of SxlPe also regulate SxlPm, we analyzed mutations in several XSEs and co-factors to determine whether they affected transcription from the maintenance promoter.
The XSE scute encodes a dose-sensitive bHLH transcription factor that dimerizes with the maternally supplied daughterless protein to directly activate SxlPe (23). We found that loss of zygotic scute (sc) or maternal daughterless (da) also affects SxlPm. In scsisB3 and maternal da1, mutant progeny expression of SxlPm was delayed in both sexes by about 5–10 min compared with wild type (Fig. 2B and Fig. S2A). In XX embryos, no expression was observed during cycle 13, and only a fraction of nuclei showed expression by 5 min in cycle 14. Thereafter the proportion of expressing nuclei increased, however, until by about 20 min in cycle 14, all female zygotic nuclei expressed SxlPm in a manner indistinguishable from wild type. Expression in scsisB3 and da1 males was similarly delayed. About one-half of the XY nuclei expressed SxlPm at 15 min into cycle 14, and all stably activated the maintenance promoter by 30 min into the cellularization process.
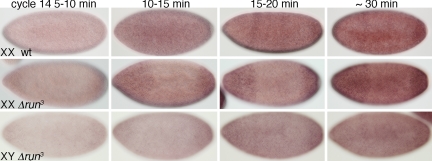
We observed similar results with the XSE runt, which is required to activate SxlPe in the central region of the embryo (24). In homozygous Δrunt3 females, the number of nuclei expressing SxlPm, and the staining intensity of the individual dots, decreased noticeably between 5 and 30 min in cycle 14 (Fig. 3). This caused a diminution of overall embryo staining intensity in central regions relative to the poles during early cycle 14. Similar effects were seen in Δrunt3 males, but the lower contrast resulting from the decreased expression level of their single Sxl allele made these effects more difficult to document photographically (Fig. 3).
Fig. 3.
The XSE runt controls expression from SxlPm, as determined by in situ hybridizations with a SxlPm-specific intron probe. (Top) Wild-type females at the indicated times in nuclear cycle 14. (Middle) Homozygous run3 mutant XX progeny. (Bottom) Hemizygous run3 XY embryos. Mutant XX embryos were progeny of w f run3/FM7c females and w f run3/Yy+, mal+ males. One-half of the XX embryos exhibited the abnormal staining pattern. The XY embryos were progeny of w f run3/FM7c females and FM7c/Y males. Wild-type females were from w1118 parents.
Our findings demonstrate that Sc/Da and Runt regulate the onset of SxlPm expression in both sexes, but that none of these three proteins is required for maintenance promoter activity during the rest of embryonic development. This implies that at least two temporally distinct mechanisms control SxlPm activity: one mechanism regulating the sexually dimorphic onset of transcription in response to X chromosome dose and the other conferring constitutive expression throughout the remainder of the life cycle.
To test the notion that XSE dose specifies the timing of SxlPm activation, we investigated whether an increase in XSE copy number can cause earlier activation of the promoter (19). To increase the XSE dose, we crossed males heterozygous for a second chromosome carrying two transgenic copies each of sc+ and sisA+ with normal females (25, 26). This created a population consisting of XX embryos with four copies of sc+ and sisA+, XY embryos with three copies of sc+ and sisA+, and normal XX and XY embryos. We found that SxlPm was activated earlier in both sexes when the XSE dose was increased (Fig. 1C; Table S1). In females, the extra XSE genes caused about 70% of the nuclei to express SxlPm during cycle 13 and all XX nuclei to express SxlPm from the earliest stages of cycle 14. In males, the additional XSE copies caused nearly 30% of XY nuclei to express SxlPm ectopically during cycle 13 and nearly all to express SxlPm by 10 min into cycle 14.
Our findings demonstrate that the sexually dimorphic activation of SxlPm is controlled by some of the same determinants that signal the female-specific expression of SxlPe. They do not, however, allow us to distinguish whether the XSE proteins directly regulate SxlPm or whether their effects on SxlPm reflect indirect effects, due perhaps to local chromatin changes associated with the activation of the adjacent SxlPe (Fig. 1). As a first step toward answering the question of whether activation of SxlPm is linked in cis to that of SxlPe, we investigated whether we could identify conditions under which the activities of the two promoters could be uncoupled.
SxlPe Activity Is Not Needed for Proper Regulation of SxlPm.
Duplication of XSE activators leads to strong ectopic expression of SxlPe in male embryos. Reciprocally, elimination of the maternal co-repressor Groucho also causes strong ectopic Sxl expression in males by decreasing the threshold XSE concentrations necessary to activate SxlPe (17). We reasoned that if the initial female-specific response of SxlPm is coupled to the activation of the SxlPe, then a loss of groucho should result in premature expression from SxlPm. On the other hand, if Sc/Da and Runt activate the two promoters directly, then the sex-specific response of SxlPm may well be independent of groucho.
We found that embryos derived from mothers lacking groucho germline function expressed SxlPm in a wild-type pattern (Fig. S2C). Females first activated SxlPm in cycle 13 and fully expressed the promoter by 10–15 min into cycle 14. Males initiated expression early in cycle 14 and fully expressed SxlPm some 20–30 min later. Our observations are consistent with direct regulation of the two promoters by the XSEs scute and runt, a conclusion further supported by our finding that the XSE sisA does not regulate SxlPm.
The X-linked sisA gene encodes a bZIP transcription factor needed for the female-specific activation of SxlPe in all somatic cells of the embryo (26, 27). When we analyzed the strong loss-of-function allele sisA1, we found that neither homozygous sisA1 females nor hemizygous sisA1 males exhibited any delay in SxlPm activation or decreased staining of SxlPm-derived nascent transcripts compared with wild type (Fig. S2B). Taken together, our findings with sisA and groucho show that SxlPe activity can be blocked without affecting expression from SxlPm, and also that SxlPe activity can be induced ectopically without activating SxlPm. This strongly suggests that some, but not all, XSEs regulate the two promoters directly, but leaves open the question of whether the promoters share common enhancers or use independent cis regulatory elements.
SxlPm and SxlPe Share a Common Regulatory Element That Responds to X Chromosome Dose.
Our finding that SxlPm is activated earlier in XX embryos than in XY embryos in response to scute, da, and runt suggests that SxlPm, like SxlPe, responds directly to the number of X chromosomes present in the embryo. To determine whether the XSEs and other proteins regulate the two promoters through independent regulatory elements or whether they instead share a common enhancer, we analyzed the structure of SxlPm by creating a series of transgenes that fused different portions of the Sxl gene to a lacZ reporter.
We first assessed the function of the region upstream of the SxlPm transcription start site by fusing sequences from -1.8 kb to + 34 bp within exon L1 to lacZ (Fig. 1). We found that none of the four reporter lines tested expressed detectable lacZ mRNA in embryos, indicating that key regulatory elements necessary for SxlPm activity likely are located downstream of exon L1 (data not shown). Considering that the sequences upstream of Sxl are not conserved in all Drosophila species, having been exchanged by a chromosomal rearrangement some 10–15 million years ago, and that the 3′ ends of the upstream genes are located within about 200–500 bp of the SxlPm start site, we next tested a shorter upstream (−0.8 kb) and large downstream (+6.0 kb) segment encompassing the SxlPe regulatory elements for SxlPm function (Fig. 1). We found that these transgenes expressed lacZ mRNA in a manner consistent with normal SxlPe and SxlPm promoter activity (data not shown).
To analyze SxlPm independent of SxlPe activity, we created a modified version of the full-length transgene in which we removed a 171-bp segment that included the SxlPe basal promoter and part of the E1 exon (Fig. 1). This construct, designated SxlPm[Δ-88Pe]-lacZ, was expressed in a manner indistinguishable from the endogenous SxlPm promoter (Figs. 4A and S3A). Weak lacZ expression was detected in cycle 13 nuclei in XX embryos, and by 10–15 min into cycle 14, every nucleus appeared to express both copies of SxlPm[Δ-88Pe]-lacZ. Male embryos first expressed SxlPm[Δ-88Pe]-lacZ in cycle 14, with full activation occurring about 20 min later. Notably, XX embryos expressed SxlPm[Δ-88Pe]-lacZ mRNA at higher levels than XY embryos even when the transgenes were present in two copies in both sexes (Fig. 4A). This difference was maintained through cycle 14 and then gradually disappeared during gastrulation and germ band extension (data not shown). These results establish that all of the sequences necessary for normal expression of SxlPm lie between -0.8 and + 6.0 kb and confirm that a functional SxlPe is not required for the earlier onset of SxlPm activity in females.
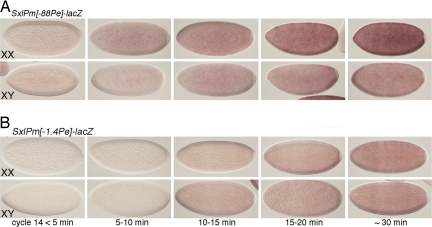
Fig. 4.
A 1.4-kb deletion of SxlPe regulatory DNA equalizes SxlPm activity in the sexes. In situ hybridization was used to detect steady-state lacZ mRNA levels. (A) Embryos carried two copies of an autosomal SxlPm[Δ-88Pe]-lacZ transgene deleted for the SxlPe basal promoter region. (B) Embryos with two copies of an autosomal SxlPm[Δ-1.4Pe]-lacZ transgene deleted for SxlPe and its regulatory sequences to -1.4 kb. Sex was determined by fluorescent detection of endogenous SxlPe-derived transcripts. Times after the onset of cycle 14 are indicated. Four independent lines of each transgene were examined, with indistinguishable results.
Normal sex-specific regulation of SxlPe requires sequences an extension to −1.4 kb upstream of exon E1 (28). Within these sequences, two regions spanning from +20 to −400 bp and from −800 to −1,400 bp have been identified as being crucial for full SxlPe activity. To determine whether these SxlPe regulatory sequences also regulate SxlPm, we created a modified SxlPm-lacZ fusion carrying an internal deletion spanning the region from -1452 to +85 relative to the start site of exon E1 (Fig. 1). We found that the onset of expression from the SxlPm[Δ-1.4Pe]-lacZ transgenes was delayed relative to that from the SxlPm[Δ-88Pe] lines, and also that there was no longer any difference in the timing or level of expression between the sexes (Fig. 4 and Fig. S3). Expression from SxlPm[Δ-1.4Pe]-lacZ lines was first seen in a few nuclei in both XX and XY embryos about 10–15 min into cycle 14 (Fig. S3). The number of expressing nuclei increased thereafter, reaching a maximum 15–20 min later. Mature transcripts accumulated over time, but XX and XY embryos expressed equal lacZ mRNA levels at all times evaluated. We conclude that the 1.4-kb region that controls the female-specific expression of SxlPe also is required for the sex differential expression of SxlPm. We also note that the effects of the [Δ-1.4Pe] mutation on SxlPm appeared to be stronger than those of sc, da, and runt mutations on the endogenous locus, because the mutations in the transacting regulators did not abolish male/female differences in SxlPm expression. This suggests that factors besides Sc/Da and Runt interact with the 1.4-kb region to control the initial activation of SxlPm.
Discussion
The Drosophila sex determination pathway elegantly illustrates the use of premRNA splicing control in development; however, the establishment of sex-specific splicing ultimately depends on the coordinated activities of two promoters for the master regulatory gene Sxl. In this work, we show that the switch from the initial assessment of X chromosome dose at SxlPe to the stable autoregulatory control of Sxl premRNA splicing exploits an unexpected level of transcriptional control of the Sxl maintenance promoter. We demonstrate that, contrary to the prevailing view, SxlPm responds to the X chromosome dose, and that it does so by sharing common X-signal elements and a common enhancer with SxlPe. The switch between Sxl promoters thus serves as a tractable model for exploring the logical circuitry and molecular mechanisms that control the fidelity of developmental switches and coordinate the uses of multiple promoters for a single gene (29).
Why Is SxlPm Regulated?
A priori, a female embryo must do two things to establish and then remember its sex: It must produce a pulse of Sxl protein by transiently activating SxlPe in response to the XX signal, and it must activate SxlPm, so that its transcripts can be spliced to produce yet more Sxl protein. A male embryo needs only to keep SxlPe off so that no Sxl protein is present when SxlPm is active. The system would seem to impose no requirement for sexually dimorphic expression from SxlPm or even for a temporal overlap in transcription from the two promoters (14), yet both features are conserved across the breadth of Drosophila species. We suggest that the resolution of this paradox lies in recognizing that the transition to stable autoregulatory Sxl splicing requires the presence of substantial amounts of Sxl protein (4, 14) rather than being driven by trace quantities of Sxl protein (30, 31). Given this, we propose that overlapping expression from the two promoters ensures that XX cells rapidly engage autoregulatory Sxl splicing, whereas the delayed activation of SxlPm in XY cells buffers against improper Sxl activation due to random variations in regulatory protein concentrations. In effect, we suggest that robustness is conferred on the system by rapid reinforcement of correct decisions. In XX embryos, strong induction of SxlPe, coupled with early activation of SxlPm, ensures that high levels of Sxl protein and its premRNA substrate are present during the transition to splicing control. In XY embryos, chance fluctuations in XSE or inhibitor concentrations that caused low-level activation of SxlPe would not persist to activate SxlPm, thus preventing amplification of rare mistakes into the fully on state. We note that a logically similar, two-target control process operates in the primary sex determination of Caenorhabditis elegans. There, four XSE proteins exert primary control of the master regulator xol-1 at the level of transcription, and a fifth XSE acts posttranscriptionally to ensure the fidelity of X chromosome counting (32, 33). The inclusion of multiple regulatory steps may prove to be a general mechanism for conferring robustness on dose-sensitive regulatory switches.
How Is SxlPm Regulated?
SxlPm appears to be equally active in both sexes after the onset of gastrulation. Before that time, SxlPm is expressed in a graded fashion, becoming active earlier and being expressed more strongly in XX embryos than in XY embryos. Sequences governing the early sexually dimorphic expression of SxlPm are included in the same 1.4-kb DNA segment that controls the on-or-off regulation of SxlPe (28). Importantly, the 1.4-kb region must work as an enhancer for SxlPm rather than exert an indirect effect in cis via activation of SxlPe, because deletion of the SxlPe core promoter has no effect on SxlPm activity. This, combined with the involvement of the XSEs scute and runt in SxlPm regulation, suggests that SxlPm, like SxlPe, responds directly to the number of X chromosomes present in the embryo. However, the fact that neither loss of the strong XSE sisA nor loss of the potent maternal co-repressor Groucho affects SxlPm suggests that the mechanism of X-counting at SxlPm differs from that at SxlPe, despite their shared common cis- and trans-acting components. We suspect that additional transcription factors contribute to both early SxlPm activation and the female/male differences in timing.
The existence of a shared regulatory region between SxlPe and SxlPm raises the question of how enhancer activity is directed to the correct promoter at the appropriate time. The 1.4-kb region regulates SxlPe from cycle 12 through early cycle 14, yet the enhancer does not lead to significant expression from SxlPm until cycle 14. Expression from the two promoters overlaps briefly before SxlPe is silenced and SxlPm fully controls Sxl transcription. We imagine two general mechanisms that might explain how the enhancer can chose between the two promoters (34, 35). First, an insulator situated between the enhancer and the upstream promoter might block the 1.4-kb region from interacting with SxlPm until the insulating protein is removed from the DNA or its activity is overcome by additional positive signals. Second, promoter choice could be dictated by differences in the transcription machinery at the two promoters (34) or by a temporally restricted transcription factor that recruits the enhancer to one of the two Sxl promoters. The developmentally regulated competition between the promoters of the chicken ε-globin and β-globin genes for their common enhancer provides a precedent for the latter mechanism (36). The rapid changeover from SxlPe to SxlPm coincides with the Drosophila maternal-to-zygotic transition, when expression of the zygotic genome begins in earnest and numerous early mRNAs and proteins are eliminated from the embryo (37). It would not be surprising if the rapid changes at Sxl were directly connected to more general regulatory events occurring during this dynamic period of development.
Methods
P-Element Vectors and Transformation.
Sxl genomic fragments were made using the Expand Long-Template polymerase chain reaction system (Roche), cloned into pCRII-TOPO (Invitrogen), and ligated into P-element transformation vectors based on pCaSpeR-AUG-ßgal. Germline transformations were performed by Genetic Services, Inc, Cambridge, MA. Transgenes with internal deletions were cloned as upstream and downstream fragments and joined at primer-derived PacI sites. The -1.8-kb SxlPm-lacZ was made with primers 1.8Pm5′ and 1.8Pm3′; SxlPm[+Pe]-lacZ, with primers 1′ and 4′; SxlPm[Δ-88Pe]-lacZ, with primers 1′ and 2′/PacI and 3′PacI and 4′; and SxlPm[Δ-1.4Pe]-lacZ, with primers 1′ and 5′/PacI and 3′PacI and 4′. Control transgenes SxlPe[L2]-lacZ and SxlPe[Δ-88,L2]-lacZ were similar to SxlPm[+Pe]-lacZ and SxlPm[Δ-88Pe]-lacZ, except that sequences distal to 1.4 kb upstream of exon E1 were absent and the vector was pPelican. Two independent lines of SxlP[L2]-lacZ were expressed similarly to previous 1.4-kb SxlPe-lacZ lines (18, 28), but deletion of the core SxlPe promoter left both tested lines of SxlPe[Δ-88,L2]-lacZ inactive. Primer sequences were 1.8Pm5′-CTCACGCTAGAGAACACCGATCATTC; 1.8Pm3′-GACTTTCCTTCTTCGGCAAC; 1′-CCATCCGATCCGCGAGTCCA; 4′-GCACGCTCACTGTGCTTTCCTCTC; 2′/PacI-CCAttaattaaGGAGGCAAGGTGCGCGT; 3′/PacI-CCAttaattaaCGTAACTTTGTGATTATCCC; 5′/PacI-CCttaattaaATGCGAGCAGCGGAGAAGGG.
In Situ Hybridization.
Nonfluorescent in situ hybridization used digoxygenin or fluorescein-labeled probes (38). D. melanogaster and D. simulans SxlPm intron probes (1.4 kb) were transcribed from templates made using the following primers: Pm5′-CCCTTCTCCGCTGCTCGCAT and T3Pm-aattaaccctcactaaagggCCAGGTAGAAGATCGAAGGA. Templates for corresponding D. yakuba and D. virilis SxlPm probes were made with yakPm5′-CACCACCCCATTCCACCCG and T3Pm, or virPm5′-CGAGCCTTTCCGTAACTGTTCG and virT3Pm-aattaaccctcactaaagggTGCGCTACCTGTTGACAGTG. Probes for lacZ and exon E1 (SxlPe) have been described previously (9, 18, 26). Fluorescent detection of SxlPe transcripts was as detailed previously (see http://superfly.ucsd.edu/∼davek/). Nascent transcripts, visible as dots within stained nuclei, were seen with all probes but were more difficult to detect from lacZ transgenes. For X-linked genes, the number of nuclear dots indicates chromosomal sex. Times within cycle 14 were estimated by nuclear shape and length, as well as by the extent of membrane furrow invagination (20, 21). Specific developmental time estimates were based on the literature, but embryos grouped within specified time periods were staged as close as possible to one another.
Genetic Analysis.
The alleles sisA1, da1, and scsisB3 are near null for sex determination. groE48 and run3 are null alleles. Embryos homozygous or hemizygous for scsisB3 and sisA1 were generated using the constitutive SxlM4 allele to bypass female-lethal effects (19). Null allele Sxlf1 suppressed the male lethality of the 2X P(miniw+, sisA+) and 2X P(miniw+, scsisB+) chromosome (19). Nascent transcripts from Sxlf1 and SxlM4 are not detectably different than those from wild type. Germline clones (39) were generated in larvae of genotype P{hsFLP}1, y1 w1118/w1118; P{neoFRT}82B ry506 groE48/P{neoFRT}82B P{ovoD1–18}3R; and P{hsFLP}1, y1. Females with recombinant germlines were crossed with w1118/Y males. Embryos were collected at 25 °C. Other mutations and chromosomes are described at http://flybase.bio.indiana.edu. The scsisB3 allele and transgenic XSE duplications were provided by T. Cline (University of California Berkeley). FRT82B groE48 was provided by P. Simpson (University of Cambridge). D. virilis was provided by S. Johnson (Texas A&M University). D. simulans, D. yakuba, and D. simulans were provided by D. Barbash (Cornell University). Fly stocks for FLP/FRT recombination were obtained from the Bloomington Drosophila stock center.
Supplementary Material
Acknowledgments.
We thank Tom Cline, Pat Simpson, Spencer Johnson, Dan Barbash and the Bloomington Drosophila stock center for generously providing fly stocks, and Keith Maggert, Deborah Siegele, and Teresa Lamb for their helpful criticisms of the manuscript. This work was supported by National Institutes of Health Grant GM063606.
Footnotes
The authors declare no conflicts of interest.
This article is a PNAS Direct Submission.
1Deceased December 1, 2002.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805993105/DCSupplemental.
References
- 1.Ashburner M, Golic K, Hawley R. Drosophila: A Laboratory Handbook. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2005. [Google Scholar]
- 2.Cline TW, Meyer BJ. Vive la difference: Males vs females in flies vs worms. Annu Rev Genet. 1996;30:637–702. doi: 10.1146/annurev.genet.30.1.637. [DOI] [PubMed] [Google Scholar]
- 3.Penalva LO, Sanchez L. RNA binding protein Sex-lethal (Sxl) and control of Drosophila sex determination and dosage compensation. Microbiol Mol Biol Rev. 2003;67:343–359. doi: 10.1128/MMBR.67.3.343-359.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bernstein M, Lersch RA, Subrahmanyan L, Cline TW. Transposon insertions causing constitutive Sex-lethal activity in Drosophila melanogaster affect Sxl sex-specific transcript splicing. Genetics. 1995;139:631–648. doi: 10.1093/genetics/139.2.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell LR, Horabin JI, Schedl P, Cline TW. Positive autoregulation of Sex-lethal by alternative splicing maintains the female-determined state in Drosophila. Cell. 1991;65:229–239. doi: 10.1016/0092-8674(91)90157-t. [DOI] [PubMed] [Google Scholar]
- 6.Horabin JI, Schedl P. Sex-lethal autoregulation requires multiple cis-acting elements upstream and downstream of the male exon and appears to depend largely on controlling the use of the male exon 5′ splice site. Mol Cell Biol. 1993;13:7734–7746. doi: 10.1128/mcb.13.12.7734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keyes LN, Cline TW, Schedl P. The primary sex determination signal of Drosophila acts at the level of transcription. Cell. 1992;68:993–943. doi: 10.1016/0092-8674(92)90036-c. [DOI] [PubMed] [Google Scholar]
- 8.Cline TW. The Drosophila sex determination signal: How do flies count to two? Trends Genet. 1993;9:385–390. doi: 10.1016/0168-9525(93)90138-8. [DOI] [PubMed] [Google Scholar]
- 9.Erickson JW, Quintero JJ. Indirect effects of ploidy suggest X chromosome dose, not the X:A ratio, signals sex in Drosophila. PLoS Biol. 2007;5:e332. doi: 10.1371/journal.pbio.0050332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu C, Urano J, Bell LR. The Sex-lethal early splicing pattern uses a default mechanism dependent on the alternative 5′ splice sites. Mol Cell Biol. 1997;17:1674–1681. doi: 10.1128/mcb.17.3.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Horabin JI, Schedl P. Splicing of the Drosophila Sex-lethal early transcripts involves exon skipping that is independent of Sex-lethal protein. RNA. 1996;2:1–10. [PMC free article] [PubMed] [Google Scholar]
- 12.Salz HK, et al. The Drosophila female-specific sex-determination gene, Sex-lethal, has stage-, tissue-, and sex-specific RNAs suggesting multiple modes of regulation. Genes Dev. 1989;3:708–719. doi: 10.1101/gad.3.5.708. [DOI] [PubMed] [Google Scholar]
- 13.Samuels ME, Schedl P, Cline TW. The complex set of late transcripts from the Drosophila sex determination gene Sex-lethal encodes multiple related polypeptides. Mol Cell Biol. 1991;11:3584–3602. doi: 10.1128/mcb.11.7.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Louis M, Holm L, Sanchez L, Kaufman M. A theoretical model for the regulation of Sex-lethal, a gene that controls sex determination and dosage compensation in Drosophila melanogaster. Genetics. 2003;165:1355–1384. doi: 10.1093/genetics/165.3.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barbash DA, Cline TW. Genetic and molecular analysis of the autosomal component of the primary sex determination signal of Drosophila melanogaster. Genetics. 1995;141:1451–1471. doi: 10.1093/genetics/141.4.1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keyes LN. Princeton, NJ: Princeton Univ; 1995. Sex-specific activation of the Sex-lethal gene in Drosophila melanogaster. PhD thesis. [Google Scholar]
- 17.Lu H, et al. Maternal Groucho and bHLH repressors amplify the dose-sensitive X chromosome signal in Drosophila sex determination. Dev Biol. 2008 doi: 10.1016/j.ydbio.2008.08.012. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avila FW, Erickson JW. Drosophila JAK/STAT pathway reveals distinct initiation and reinforcement steps in early transcription of Sxl. Curr Biol. 2007;17:643–648. doi: 10.1016/j.cub.2007.02.038. [DOI] [PubMed] [Google Scholar]
- 19.Cline TW. Evidence that sisterless-a and sisterless-b are two of several discrete “numerator elements” of the X:A sex determination signal in Drosophila that switch Sex-lethal between two alternative stable expression states. Genetics. 1988;119:829–862. doi: 10.1093/genetics/119.4.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grosshans J, Muller HA, Wieschaus E. Control of cleavage cycles in Drosophila embryos by fruhstart. Dev Cell. 2003;5:285–294. doi: 10.1016/s1534-5807(03)00208-9. [DOI] [PubMed] [Google Scholar]
- 21.Foe VA, Odell GM, Edgar BA. Mitosis and morphogenesis in the Drosophila embryo: Point and counterpoint. In: Bate M, Martinez-Arias A, editors. The Development of Drosophila Melanogaster. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 1993. pp. 149–300. [Google Scholar]
- 22.Erickson JW, Cline TW. Key aspects of the primary sex determination mechanism are conserved across the genus Drosophila. Development. 1998;125:3259–3268. doi: 10.1242/dev.125.16.3259. [DOI] [PubMed] [Google Scholar]
- 23.Yang D, et al. Interpretation of X chromosome dose at Sex-lethal requires non–E-box sites for the basic helix-loop-helix proteins SISB and daughterless. Mol Cell Biol. 2001;21:1581–1592. doi: 10.1128/MCB.21.5.1581-1592.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kramer SG, Jinks TM, Schedl P, Gergen JP. Direct activation of Sex-lethal transcription by the Drosophila runt protein. Development. 1999;126:191–200. doi: 10.1242/dev.126.1.191. [DOI] [PubMed] [Google Scholar]
- 25.Erickson JW, Cline TW. Molecular nature of the Drosophila sex determination signal and its link to neurogenesis. Science. 1991;251:1071–1074. doi: 10.1126/science.1900130. [DOI] [PubMed] [Google Scholar]
- 26.Erickson JW, Cline TW. A bZIP protein, SISTERLESS-A, collaborates with bHLH transcription factors early in Drosophila development to determine sex. Genes Dev. 1993;7:1688–1702. doi: 10.1101/gad.7.9.1688. [DOI] [PubMed] [Google Scholar]
- 27.Walker JJ, Lee KK, Desai RN, Erickson JW. The Drosophila melanogaster sex determination gene sisA is required in yolk nuclei for midgut formation. Genetics. 2000;155:191–202. doi: 10.1093/genetics/155.1.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Estes PA, Keyes LN, Schedl P. Multiple response elements in the Sex-lethal early promoter ensure its female-specific expression pattern. Mol Cell Biol. 1995;15:904–917. doi: 10.1128/mcb.15.2.904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Davuluri RV, Suzuki Y, Sugano S, Plass C, Huang TH. The functional consequences of alternative promoter use in mammalian genomes. Trends Genet. 2008;24:167–177. doi: 10.1016/j.tig.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 30.Graveley BR. Sex, AGility, and the regulation of alternative splicing. Cell. 2002;109:409–412. doi: 10.1016/s0092-8674(02)00750-x. [DOI] [PubMed] [Google Scholar]
- 31.Lallena MJ, Chalmers KJ, Llamazares S, Lamond AI, Valcarcel J. Splicing regulation at the second catalytic step by Sex-lethal involves 3′ splice site recognition by SPF45. Cell. 2002;109:285–296. doi: 10.1016/s0092-8674(02)00730-4. [DOI] [PubMed] [Google Scholar]
- 32.Gladden JM, Farboud B, Meyer BJ. Revisiting the X:A signal that specifies Caenorhabditis elegans sexual fate. Genetics. 2007;177:1639–1654. doi: 10.1534/genetics.107.078071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gladden JM, Meyer BJ. A ONECUT homeodomain protein communicates X chromosome dose to specify Caenorhabditis elegans sexual fate by repressing a sex switch gene. Genetics. 2007;177:1621–1637. doi: 10.1534/genetics.106.061812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ohtsuki S, Levine M, Cai HN. Different core promoters possess distinct regulatory activities in the Drosophila embryo. Genes Dev. 1998;12:547–556. doi: 10.1101/gad.12.4.547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arnosti DN. Design and function of transcriptional switches in Drosophila. Insect Biochem Mol Biol. 2002;32:1257–1273. doi: 10.1016/s0965-1748(02)00089-9. [DOI] [PubMed] [Google Scholar]
- 36.Foley KP, Engel JD. Individual stage selector element mutations lead to reciprocal changes in beta- vs. epsilon-globin gene transcription: Genetic confirmation of promoter competition during globin gene switching. Genes Dev. 1992;6:730–744. doi: 10.1101/gad.6.5.730. [DOI] [PubMed] [Google Scholar]
- 37.Tadros W, Westwood JT, Lipshitz HD. The mother-to-child transition. Dev Cell. 2007;12:847–849. doi: 10.1016/j.devcel.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 38.Lehmann R, Tautz D. In situ hybridization to RNA. In: Goldstein LSB, Fyrberg EA, editors. Drosophila Melanogaster: Practical Uses in Cell and Molecular Biology. San Diego, CA: Academic; 1994. pp. 576–597. [Google Scholar]
- 39.Chou TB, Perrimon N. The autosomal FLP-DFS technique for generating germline mosaics in Drosophila melanogaster. Genetics. 1996;144:1673–1679. doi: 10.1093/genetics/144.4.1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.