Abstract
We describe a full structural model for amyloid fibrils formed by the 40-residue β-amyloid peptide associated with Alzheimer's disease (Aβ1–40), based on numerous constraints from solid state NMR and electron microscopy. This model applies specifically to fibrils with a periodically twisted morphology, with twist period equal to 120 ± 20 nm (defined as the distance between apparent minima in fibril width in negatively stained transmission electron microscope images). The structure has threefold symmetry about the fibril growth axis, implied by mass-per-length data and the observation of a single set of 13C NMR signals. Comparison with a previously reported model for Aβ1–40 fibrils with a qualitatively different, striated ribbon morphology reveals the molecular basis for polymorphism. At the molecular level, the 2 Aβ1–40 fibril morphologies differ in overall symmetry (twofold vs. threefold), the conformation of non-β-strand segments, and certain quaternary contacts. Both morphologies contain in-register parallel β-sheets, constructed from nearly the same β-strand segments. Because twisted and striated ribbon morphologies are also observed for amyloid fibrils formed by other polypeptides, such as the amylin peptide associated with type 2 diabetes, these structural variations may have general implications.
Keywords: Alzheimer's disease, electron microscopy, solid state NMR, amyloid structure, protein misfolding
Amyloid fibrils are filamentous aggregates that result from the spontaneous self-assembly of a large and diverse class of polypeptides. Amyloid formation and deposition are associated with numerous human diseases, notably Alzheimer's disease, as well as certain biological functions (1, 2). Although all amyloid fibrils contain β-sheets, formed by β-strand segments that run approximately perpendicular to the fibril growth axis in a “cross-β” motif (3), their detailed molecular structures are not determined uniquely by amino acid sequence (4, 5). Self-propagating variations in the molecular structures of amyloid fibrils or amyloid-like aggregates formed by a single protein are believed to be responsible for the existence of multiple strains of mammalian prion diseases (6, 7) and yeast prion phenotypes (8–11), and may produce variations in the toxicity (4) or patterns of deposition (12) of amyloid fibrils in Alzheimer's and other amyloid diseases. Here, we describe an experimentally-based structural model for fibrils formed by the 40-residue Alzheimer's β-amyloid peptide (Aβ1–40) that applies specifically to fibrils with a characteristic periodically-twisted morphology in transmission electron microscopy (TEM) images, sometimes described as a “twisted pair” morphology (13, 14). A combination of data from solid state NMR, TEM, and scanning transmission electron microscopy (STEM) indicates that these fibrils in fact possess threefold symmetry about the fibril growth axis. Comparison with a previously developed twofold-symmetric model for Aβ1–40 fibrils with untwisted, “striated ribbon” morphologies (15) reveals that Aβ1–40 fibrils with distinct morphologies share common secondary and tertiary structures, but differ primarily in overall symmetry, quaternary structure (as defined below), and the conformations of non-β-strand segments.
Results
Seeded Growth Produces Morphologically Homogeneous, Twisted Fibrils Comprised of 3 Cross-β Units.
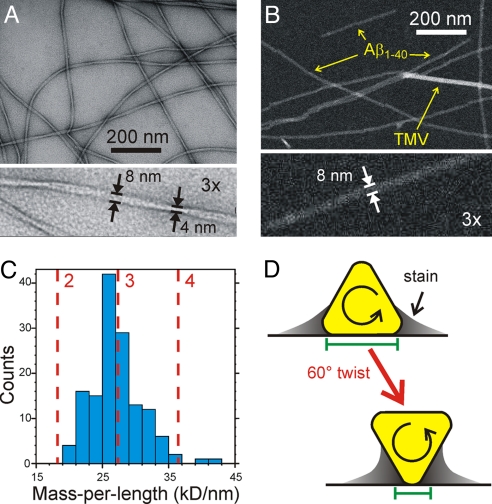
Fibrils with a homogeneous twisted morphology were prepared from synthetic Aβ1–40 by a protocol similar to our reported quiescent growth protocol (4), but with intermittent sonication during growth, and with 12 rounds of seeding (see Materials and Methods and supporting information (SI) Fig. S1). Although fibrils from the early rounds exhibited various morphologies in TEM images, morphological homogeneity increased markedly in the later rounds, indicating that fibrils with an apparent twist period of 120 ± 20 nm (spacing between apparent minima in fibril width) propagate most efficiently under our protocol. Importantly, although the width of these fibrils seems to vary periodically between 4 ± 1 nm and 8 ± 1 nm in TEM images obtained with conventional negative staining (Fig. 1A), no width modulation is observed in unstained STEM images (Fig. 1B), where the apparent width is 8 ± 1 nm. Large width modulations are also absent in positively stained TEM images, unstained TEM images, and atomic force microscope images (Fig. S2). Quantitative analysis of STEM images, in which image intensity is directly proportional to mass density (4, 13, 16–18), generated a histogram of mass-per-length (MPL) values (Fig. 1C) with a maximum at 26 ± 2 kDa/nm and no counts <19 kDa/nm. Given the 9.1 kDa/nm MPL expected for 1 layer of Aβ1–40 molecules in a cross-β motif (4.3-kDa molecular mass divided by 0.47-nm spacing in a β-sheet), the observed MPL maximum indicates that fibrils with the morphology in Fig. 1 are constructed from 3 cross-β units, i.e., 3 layers of Aβ1–40 molecules. Thus, although the appearance of these fibrils in negatively stained TEM images suggests that they are twisted pairs of filamentous subunits, this possibility is ruled out by the STEM data. Earlier STEM data for Aβ1–40 have also suggested the presence of fibrils constructed from 3 cross-β units, but in fibril preparations that were less morphologically homogeneous (4, 17).
Fig. 1.
Electron microscopy. (A) Negatively stained TEM image of Aβ1–40 fibrils, showing a periodically twisted morphology with apparent width modulation. (B) STEM image of the same fibrils, with tobacco mosaic virus (TMV) included for mass density calibration. (C) Mass-per-length histogram, extracted from multiple STEM images, indicating a structure comprised of 3 cross-β units. (D) Schematic explanation of the enhanced width modulation in negatively stained TEM images of fibrils with approximately triangular cross-sections. Green bars represent the apparent widths at 2 positions of twist, as measured from the approximate inner edges of the stain.
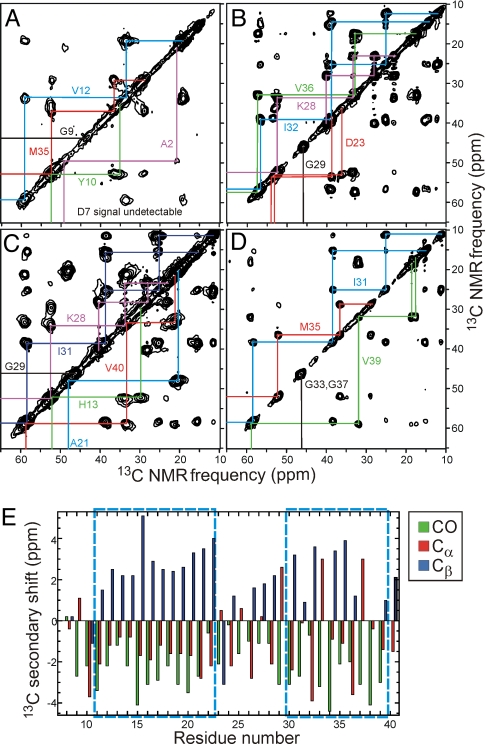
Solid State NMR Data Indicate Threefold Symmetry and Provide Constraints on Secondary, Tertiary, and Quaternary Structure.
Molecular structural constraints were derived from a variety of solid state NMR measurements on Aβ1–40 fibrils, prepared with uniformly 15N,13C-labeled residues or individual 13C-labeled and 15N-labeled sites (Table 1). Two-dimensional (2D) solid state 13C NMR spectra of hydrated fibrils that contain uniformly-labeled residues (Fig. 2 A–D and Fig. S3) show a single set of 13C chemical shifts and 1.0–1.5 ppm linewidths (full width at half maximum) for nearly all sites in residues 10–40, indicating that all Aβ1–40 molecules in these fibrils have nearly equivalent conformations and structural environments. Together with the MPL data in Fig. 1C, this NMR result implies an approximate threefold symmetry about the fibril growth axis. Absence of threefold symmetry would produce splitting or broadening of many 13C NMR lines, because of variations in the conformations or structural environments of the 3 cross-β units, contrary to our experimental observations. (Given the 0.47-nm intermolecular spacing along the fibril axis in a cross-β structure, twofold rotational symmetry would produce MPL ≈ 18 kDa/nm, as would a twofold screw axis. Twofold symmetries would require that the intermolecular spacing in the β-sheets be ≈0.33 nm, at variance with known properties of β-sheets (3).
Table 1.
Isotopic labeling patterns of Aβ-1–40 samples for solid state NMR measurements
| Aβ1–40 sample | Uniformly 15N,13C-labeled residues | 13C-labeled sites | 15N-labeled residues |
|---|---|---|---|
| A | A2, D7, G9, Y10, V12, M35 | — | — |
| B | D23, K28, G29, I32, V36 | — | — |
| C | H13, A21, K28, G29, I31, V40 | ||
| D | I31, G33, M35, G37, V39 | — | — |
| E | — | V12 CO, A21 CH3 | — |
| F | — | V18 CO, M35 CH3 | L34, G37 |
| G | — | A30 CO | |
| H | K16, F19, A21, E22, I32, V36 | — | — |
| I | F19, V24, G25, A30, I31, L34, M35 | — | — |
| J | F20, D23, V24, K28, G29, A30, I31 | — | — |
| K | S8, H13, L17, V18, A30, I32, G33 | — | — |
| L | F20, S26, N27, G33, G38, V39 | — | — |
| M | H14, Q15, A21, V36, G37 | — | — |
| N | F20, E22, K28, I32, M35, V36 | — | — |
| O | E11, H13, N27, K28, G29, A30 | — | — |
Fig. 2.
Solid state NMR. (A–D) 2D 13C NMR spectra of Aβ1–40 fibril samples A–D, respectively, with chemical shift assignment paths for the indicated residues. Strong, sharp cross-peaks are observed for residues 10–40, indicating a rigid, ordered structure, whereas cross-peaks for A2, D7, and G9 are weak or absent because of dynamic disorder of the N-terminal segment. The absence of splittings of the NMR lines indicates that all Aβ1–40 molecules have equivalent conformations and structural environments, implying threefold symmetry. (E) Secondary 13C chemical shifts (i.e., deviations from random coil values) determined from 2D 13C NMR spectra. Dashed boxes indicate segments in which the secondary shifts indicate β-strands. Note that Cα secondary shifts for glycines (residues 25, 29, 33, 37, and 38) are not correlated with secondary structure.
13C-labeled sites in A2, S8, and G9 exhibit broader lines (≈4 ppm) in the dry lyophilized state and reduced intensities in the hydrated state (Fig. S3), indicating a disordered N-terminal segment that extends to Y10. Secondary shifts (i.e., deviations of 13C chemical shifts from random coil values) indicate β-strand secondary structure in residues 11–22 and 30–39 and non-β-strand conformations at certain positions in residues 23–29 (Fig. 2E), consistent with φ and ψ torsion angles predicted from the 13C chemical shifts by the TALOS program (19) (Table S1). Additional constraints on secondary structure for residues 23–30 were obtained from sequential backbone 15N–15N distances, determined from 15N–15N magnetic dipole–dipole couplings measured with the PITHIRDS-CT dipolar recoupling technique (20) (Fig. S4).
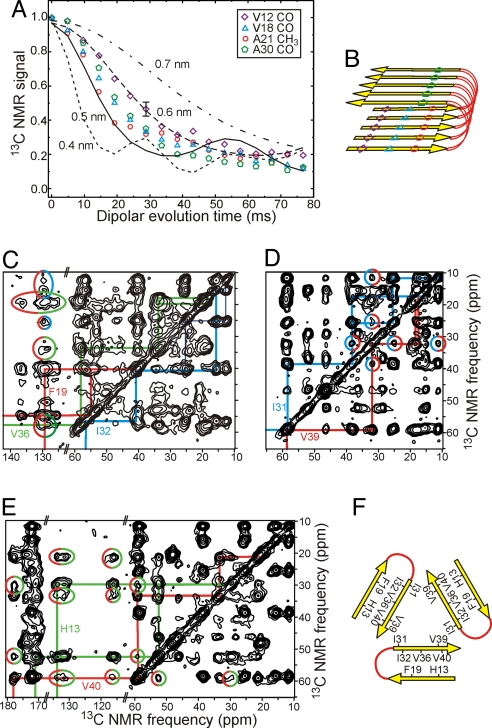
Tertiary structure (defined here and in earlier publications (3, 4, 13) as the alignment of β-strands within β-sheets) was determined with PITHIRDS-CT measurements of intermolecular 13C-13C dipole–dipole couplings (i.e., distances) in fibril samples with single-site labels. Intermolecular distances are ≈0.5 nm for backbone carbonyl sites of V12, V18, and A30, and for the methyl site of A21 (Fig. 3 A and B). These data show that the β-strands of Aβ1–40 form 2 in-register parallel β-sheets, as previously observed for other Aβ1–40 fibrils (4, 21, 22) and for amyloid fibrils formed by other polypeptides (13, 23–26).
Fig. 3.
Tertiary and quaternary constraints. (A) Measurements of intermolecular 13C-13C magnetic dipole–dipole couplings in samples E–G using the PITHIRDS-CT solid state NMR technique. Comparison with numerical simulations (lines) for linear chains of 13C nuclei with specified spacings indicates intermolecular distances of ≈0.5 nm for V12 CO, A21 CH3, and V18 CO sites, implying an in-register parallel β-sheet structure. (B) Schematic representation of the secondary and tertiary structure implied by the PITHIRDS-CT data and data in Fig. 2. (C–E) 2D 13C NMR spectra of samples H, D, and C, obtained with 0.5, 1.5, and 1.0-s RAD exchange periods, respectively. Cross-peaks enclosed in color-coded ellipses indicate F19/I32, F19/V36, I31/V39, and H13/V40 quaternary contacts. (F) Schematic representation of the quaternary structure implied by these long-range interresidue cross-peaks.
Constraints on quaternary structure (defined as the arrangement of β-sheets within and between cross-β units) (3, 4, 13) were obtained primarily from 2D 13C-13C exchange spectroscopy with radio-frequency-assisted diffusion (27) (RAD). In these measurements, cross-peaks between 13C NMR lines of nonsequential uniformly-labeled residues arise from spatial proximity of their sidechains, with sidechain-sidechain distances up to ≈0.6 nm (15). Nonsequential cross-peaks are observed for F19/I32, F19/L34, F19/V36, Q15/V36, H13/V40, and I31/V39 pairs (Fig. 3 C–E and Fig. S5). With the exception of the I31/V39 cross-peaks, these cross-peaks are explained by “internal” quaternary contacts, i.e., contacts between the 2 β-sheets formed by residues 11–22 and 30–39 within a single cross-β unit. Because I31 and V39 are in the same β-strand, I31/V39 cross-peaks are explained by “external” quaternary contacts, i.e., contacts between different cross-β units that are related by the threefold symmetry (Fig. 3F). 2D RAD spectra of isotopically diluted samples (Fig. S5) suggest that contacts involving F19 are intermolecular, consistent with staggering of N-terminal and C-terminal β-sheets (15).
Note that we use the term “quaternary structure” to describe essential aspects of the molecular structure within a single Aβ1–40 protofilament, where the protofilament is the minimal fibrillar entity observed in experiments. Here, the Aβ1–40 protofilament is threefold symmetric, with MPL ≈ 27 kDa/nm. Other authors may use the term “quaternary structure” to describe lateral association of protofilaments to form various bundles with higher total MPL values.
Additional experimental constraints (Fig. S4 and Fig. S5) include: (i) PITHIRDS-CT data for 13C-labeled M35 methyl carbons that indicate approximate 0.7-nm distances between rows of methyl groups in the fibril core; (ii) frequency-selective rotational echo double resonance (fsREDOR) data (28) that imply an extended conformation for the M35 sidechain, with approximate 0.6-nm distances from M35 methyl carbons to backbone nitrogen sites of G37 and L34; (iii) fsREDOR measurements of 15N–13C dipole–dipole couplings between carboxylate groups of E11, E22, D23, and V40 and amino groups of K28 and K16 that indicate an approximate 0.5-nm D23/K28 distance, but distances >0.5 nm for other oppositely charged pairs; (iv) 2D TEDOR spectra (29) that confirm proximity of the V39 backbone nitrogen to I31 methyl carbons.
Alternative assignments of the observed quaternary contacts to the “internal” and “external” categories lead to impossible structures. For example, if the F19/I32, F19/L34, and F19/V36 contacts were external, then the Q15/V36 and H13/V40 contacts would also have to be external, given that residues 13–19 are in 1 β-sheet and residues 32–40 are in another β-sheet. The 2 interacting β-sheets would necessarily stack in an antiparallel manner. But then, in a threefold symmetric structure, connecting the N- and C-terminal β-strands of a given Aβ1–40 molecule would require the intervening non-β-strand segment to be much longer than the 7 residues indicated by the data above.
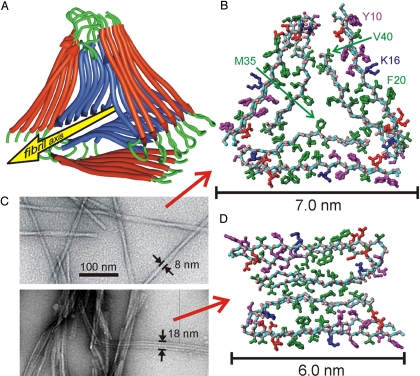
Structural Model Determined from Experimental Constraints.
We generated full molecular models for fibrils with the twisted morphology in Fig. 1, using a computational protocol that began with fully extended Aβ9–40 chains and applied experimentally based constraints (Table S2) on backbone conformation, sidechain conformation, and intermolecular distances in 2 stages of simulated annealing (see Materials and Methods). The resulting models (Fig. 4 A and B and Fig. S6) are consistent with existing experimental data, and we are unable to find qualitatively different models that are also consistent with the data. In these models, the majority of hydrophobic amino acid sidechains reside in the interior of the fibril, whereas the majority of charged and polar sidechains reside on the exterior, consistent with hydrophobic stabilization of the fibril structure (30). In addition to creating a hydrophobic core, the parallel nature of the β-sheets may permit “polar zipper” interactions among Q15 sidechains (31), which fit into a cavity created by G37 and G38 in a manner reminiscent of the “steric zippers” that have been observed in amyloid-like peptide crystal structures (32, 33) and other amyloid models (34).
Fig. 4.
Experimentally based structural models. (A) Ribbon representation of the lowest-energy model for fibrils with the twisted morphology in Fig. 1. Modeling calculations assume threefold symmetry, consistent with STEM data and solid state NMR spectra, and are constrained by specific secondary, tertiary, and quaternary structural data from solid state NMR. (B) Atomic representation, viewed down the fibril axis. Hydrophobic, polar, negatively-charged, and positively charged amino acid sidechains are green, magenta, red, and blue, respectively. Backbone nitrogen and carbonyl oxygen atoms are cyan and pink. Unstructured N-terminal residues 1–8 are omitted. (C) Comparison of twisted (Upper) and striated ribbon (Lower) fibril morphologies in negatively stained TEM images. (D) Atomic representation of a model for striated ribbon fibrils developed previously by Petkova et al. (4, 15).
The model in Fig. 4B contains interior channels adjacent to the M35 sidechains in the center of the fibrils. These channels are created by glycine residues at positions 33 and 37, and may contain rows of water in the hydrated state. As a further test of the model, we examined whether G33N and G37N mutants, in which asparagine sidechains are predicted to fill the interior channels, could be seeded by wild-type Aβ1–40 fibrils with the twisted morphology. Seeding experiments demonstrate that both mutants are indeed compatible with the twisted Aβ1–40 fibril morphology (Fig. S7). Measurements of paramagnetic enhancement of 13C NMR spin-lattice relaxation rates provide additional support (Fig. S3).
Assuming a gradual twist about the fibril growth axis (18), the roughly triangular cross-section in Fig. 4 A and B implies a modulation of fibril width between 6 nm and 7 nm (ignoring the disordered N-terminal segment) when projected onto a plane parallel to the growth axis. This relatively small modulation is consistent with STEM images (Fig. 1B) and other types of images (Fig. S2), but is at first glance inconsistent with negatively stained TEM images. However, it is important to realize that contrast in the negatively stained images arises primarily from the uranyl acetate stain, not from the fibrils themselves. Accumulation of stain around areas of contact between fibrils and the carbon film on the TEM grid produces an amplification of the apparent width modulation, as shown schematically in Fig. 1D.
Discussion
Fig. 4C compares the morphologies of twisted and striated ribbon Aβ1–40 fibrils in TEM images. Solid state NMR data for striated ribbons, reported previously (4, 15), are significantly different from the corresponding data for twisted fibrils. Differences include 13C chemical shifts (differences >1 ppm for certain sites in residues 10, 13–15, 18, 22–25, 28, 29, 32, 33, 36, and 37), 2D RAD cross-peaks (I31/V39 and H13/V40 cross-peaks for twisted fibrils only; I31/G37 and I31/M35 cross-peaks for striated ribbons only), salt bridge distances indicated by D23/K28 fsREDOR data (0.37 nm for striated ribbons, 0.5 nm for twisted fibrils), and lateral distances among M35 methyl carbons (Fig. S4). The protofilament in striated ribbon fibrils has an MPL value of ≈20 kDa/nm (4), which, together with the solid state NMR data, supports the twofold-symmetric model for the striated ribbon protofilament in Fig. 4D. Our data show that both Aβ1–40 fibril morphologies contain in-register parallel β-sheets, nearly the same locations of β-strand, non-β-strand, and disordered segments, and similar internal quaternary contacts. The principal structural differences are in the conformations of the non-β-strand segments, the external quaternary contacts, and the overall symmetry.
The interior channels adjacent to M35 sidechains discussed above are not present in Fig. 4D. The twofold-symmetric structure is apparently disfavored by G33N and G37N mutations, as the striated ribbon morphology does not propagate faithfully when wild-type Aβ1–40 fibrils with this morphology are used to seed fibril growth by G33N and G37N peptides (Fig. S7).
Given the similarity of secondary and tertiary structures within twisted and striated ribbon Aβ1–40 fibrils, it is remarkable that both morphologies are observed and can be prepared in pure form (>90% morphological homogeneity by TEM and NMR) under identical conditions of peptide concentration, pH, ionic strength, buffer composition, and temperature. As previously shown (4), agitation of an unseeded, initially monomeric Aβ1–40 solution during fibril growth produces the striated ribbon morphology, whereas quiescent conditions produce twisted morphologies. The predominant morphology may be influenced by a variety of competing factors, including rates of spontaneous nucleation, fibril extension, and fibril fragmentation (35, 36). Agitation may influence both nucleation and fragmentation rates. However, once a fibril forms with a particular morphology, either by spontaneous nucleation or by seeded growth, the kinetic barrier to conversion to a different morphology is apparently high enough to prevent conversion on the time scale of our experiments (i.e., at least several months). Quantitative characterization of thermodynamic properties and kinetic factors, and of the effects of solvent conditions on these quantities, will be a subject of future studies.
Amyloid fibrils with twisted morphologies in negatively stained TEM images (similar to Fig. 1A) are formed by many other polypeptides, including amylin (18), insulin (14), and mammalian prion proteins (37). Interestingly, Perutz et al. interpreted MPL data as evidence for threefold symmetry in twisted fibrils formed by a 22-residue Q,N-rich peptide derived from the Sup35 yeast prion protein (38). The model in Fig. 4 A and B may represent a general structural motif for a class of fibrils with similar morphologies.
Wasmer et al. have recently reported a full structural model for amyloid fibrils formed by residues 218–289 of the Podospora anserina HET-s protein (39). Although the HET-s fibrils have a roughly triangular cross-section that is superficially similar to our threefold-symmetric Aβ1–40 fibril model, the details of the 2 models are quite different. Specifically, (i) β-sheets in HET-s fibrils contain both intramolecular and intermolecular backbone hydrogen bonds, whereas β-sheets in Aβ1–40 fibrils contain only intermolecular hydrogen bonds; (ii) each side of the triangular cross-section in HET-s fibrils is formed by a single β-sheet layer, whereas each side of the triangular cross-section in threefold-symmetric Aβ1–40 fibrils is formed by 2 β-sheet layers; (iii) the MPL of HET-s fibrils (40) is ≈0.5 × MW/(0.47 nm), where MW is the molecular weight, whereas the MPL of threefold-symmetric Aβ1–40 fibrils is 3 × MW/(0.47 nm); (iv) HET-s fibrils have no symmetry about their long axes.
Finally, the structural models in Fig. 4 raise the possibility that small molecules may be designed to inhibit, stabilize, or label particular fibril morphologies, perhaps by interacting specifically with the regions of Aβ1–40 that exhibit the greatest structural variations in different morphologies. Morphology-specific labeling may permit direct identification of specific amyloid structures in the senile plaques that develop in Alzheimer's disease, and could facilitate investigations of correlations between amyloid structure and cognitive impairment or between amyloid structure and plaque location within the brain.
Materials and Methods
Aβ1–40 (sequence DAEFRHDSGYEVHHQKLVFFAEDVGSNKGAIIGLMVGGVV) was synthesized and purified as described (4, 15). Fibrils were grown initially at 210 μM peptide concentration in 10 mM phosphate buffer, pH 7.4, without continual agitation but with brief sonication once per hour for 9 days. The most efficiently propagating fibril morphology was isolated from this initial polymorphic sample by 12 rounds of seeded growth from fresh Aβ1–40 solutions (Fig. S1). The resulting sample, which exhibited uniform fibril morphologies, was used as the source of seeds for subsequent preparations of isotopically labeled fibrils for solid state NMR.
TEM and STEM images were obtained as described (4, 13). Solid state NMR measurements were performed in 9.4 T and 14.1 T magnets, using Varian Infinity and InfinityPlus spectrometers and Varian magic-angle spinning (MAS) probes at room temperature. Fibrils (typically 4–8 mg) were lyophilized, packed in 3.2 mm MAS rotors, and rehydrated by addition of 3–5 μL of deionized water. 2D spectra in Fig. 2 were recorded at 20 kHz MAS frequency, using finite-pulse radiofrequency-driven recoupling (41) during 2.4-ms mixing periods between t1 and t2 periods. 2D spectra in Fig. 3 were recorded at 18.3 kHz MAS frequency, using RAD mixing periods (27) between 500 and 1500 ms. PITHIRDS-CT measurements and simulations in Fig. 3 were performed as described in ref. 20.
Molecular modeling was performed with AMBER. Initially, Aβ9–40 hexamers with a double-layered cross-β structure were created from fully extended, well-separated chains by applying experimentally-based restraints on backbone conformation, β-sheet alignment, and internal quaternary contacts during simulated annealing with Langevin dynamics. Three copies of a hexamer were then assembled into a final model by adding restraints that represent external quaternary contacts and carrying out additional annealing.
Additional details of sample preparation, microscopy, solid state NMR measurements, and molecular modeling are given in the SI Text.
Supplementary Material
Acknowledgments.
We thank Dr. Sorin Luca for assistance with molecular modeling. This work was supported by the Intramural Research Programs of the National Institute of Diabetes and Digestive and Kidney Diseases and the National Institute of Biomedical Imaging and Bioengineering.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806270105/DCSupplemental.
References
- 1.Fowler DM, Koulov AV, Balch WE, Kelly JW. Functional amyloid: From bacteria to humans. Trends BiochemSci. 2007;32:217–224. doi: 10.1016/j.tibs.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 2.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 3.Tycko R. Molecular structure of amyloid fibrils: Insights from solid state NMR. Q Rev Biophys. 2006;39:1–55. doi: 10.1017/S0033583506004173. [DOI] [PubMed] [Google Scholar]
- 4.Petkova AT, et al. Self-propagating, molecular-level polymorphism in Alzheimer's β-amyloid fibrils. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 5.Wetzel R, Shivaprasad S, Williams AD. Plasticity of amyloid fibrils. Biochemistry. 2007;46:1–10. doi: 10.1021/bi0620959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Telling GC, et al. Evidence for the conformation of the pathologic isoform of the prion protein enciphering and propagating prion diversity. Science. 1996;274:2079–2082. doi: 10.1126/science.274.5295.2079. [DOI] [PubMed] [Google Scholar]
- 7.Bessen RA, et al. Nongenetic propagation of strain-specific properties of scrapie prion protein. Nature. 1995;375:698–700. doi: 10.1038/375698a0. [DOI] [PubMed] [Google Scholar]
- 8.Wickner RB, Edskes HK, Shewmaker F, Nakayashiki T. Prions of fungi: Inherited structures and biological roles. Nat Rev Microbiol. 2007;5:611–618. doi: 10.1038/nrmicro1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toyama BH, Kelly MJS, Gross JD, Weissman JS. The structural basis of yeast prion strain variants. Nature. 2007;449:233-U8. doi: 10.1038/nature06108. [DOI] [PubMed] [Google Scholar]
- 10.Chien P, Weissman JS, DePace AH. Emerging principles of conformation based prion inheritance. Annu Rev Biochem. 2004;73:617–656. doi: 10.1146/annurev.biochem.72.121801.161837. [DOI] [PubMed] [Google Scholar]
- 11.Uptain SM, Sawicki GJ, Caughey B, Lindquist S. Strains of [PSI+] are distinguished by their efficiencies of prion-mediated conformational conversion. EMBO J. 2001;20:6236–6245. doi: 10.1093/emboj/20.22.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer-Luehmann M, et al. Exogenous induction of cerebral β-amyloidogenesis is governed by agent and host. Science. 2006;313:1781–1784. doi: 10.1126/science.1131864. [DOI] [PubMed] [Google Scholar]
- 13.Antzutkin ON, Leapman RD, Balbach JJ, Tycko R. Supramolecular structural constraints on Alzheimer's β-amyloid fibrils from electron microscopy and solid state nuclear magnetic resonance. Biochemistry. 2002;41:15436–15450. doi: 10.1021/bi0204185. [DOI] [PubMed] [Google Scholar]
- 14.Jimenez JL, et al. The protofilament structure of insulin amyloid fibrils. Proc Natl Acad Sci USA. 2002;99:9196–9201. doi: 10.1073/pnas.142459399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petkova AT, Yau WM, Tycko R. Experimental constraints on quaternary structure in Alzheimer's β-amyloid fibrils. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wall JS, Hainfeld JF. Mass mapping with the scanning-transmission electron-microscope. Annu Rev Biophys Biophys Chem. 1986;15:355–376. doi: 10.1146/annurev.bb.15.060186.002035. [DOI] [PubMed] [Google Scholar]
- 17.Goldsbury C, Frey P, Olivieri V, Aebi U, Muller SA. Multiple assembly pathways underlie amyloid-β fibril polymorphisms. J Mol Biol. 2005;352:282–298. doi: 10.1016/j.jmb.2005.07.029. [DOI] [PubMed] [Google Scholar]
- 18.Goldsbury C, et al. Amyloid fibril formation from full-length and fragments of amylin. J Struct Biol. 2000;130:352–362. doi: 10.1006/jsbi.2000.4268. [DOI] [PubMed] [Google Scholar]
- 19.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 20.Tycko R. Symmetry-based constant-time homonuclear dipolar recoupling in solid state NMR. J Chem Phys. 2007:126. doi: 10.1063/1.2437194. [DOI] [PubMed] [Google Scholar]
- 21.Balbach JJ, et al. Supramolecular structure in full-length Alzheimer's β-amyloid fibrils: Evidence for a parallel β-sheet organization from solid state nuclear magnetic resonance. Biophys J. 2002;83:1205–1216. doi: 10.1016/S0006-3495(02)75244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torok M, et al. Structural and dynamic features of Alzheimer's Aβ peptide in amyloid fibrils studied by site-directed spin labeling. J Biol Chem. 2002;277:40810–40815. doi: 10.1074/jbc.M205659200. [DOI] [PubMed] [Google Scholar]
- 23.Shewmaker F, Wickner RB, Tycko R. Amyloid of the prion domain of Sup35p has an in-register parallel β-sheet structure. Proc Natl Acad Sci USA. 2006;103:19754–19759. doi: 10.1073/pnas.0609638103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benzinger TLS, et al. Propagating structure of Alzheimer's β-amyloid(10–35) is parallel β-sheet with residues in exact register. Proc Natl Acad Sci USA. 1998;95:13407–13412. doi: 10.1073/pnas.95.23.13407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan JCC, Oyler NA, Yau WM, Tycko R. Parallel β-sheets and polar zippers in amyloid fibrils formed by residues 10–39 of the yeast prion protein Ure2p. Biochemistry. 2005;44:10669–10680. doi: 10.1021/bi050724t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luca S, Yau WM, Leapman R, Tycko R. Peptide conformation and supramolecular organization in amylin fibrils: Constraints from solid state NMR. Biochemistry. 2007;46:13505–13522. doi: 10.1021/bi701427q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morcombe CR, Gaponenko V, Byrd RA, Zilm KW. Diluting abundant spins by isotope edited radio frequency field assisted diffusion. J Am Chem Soc. 2004;126:7196–7197. doi: 10.1021/ja047919t. [DOI] [PubMed] [Google Scholar]
- 28.Jaroniec CP, Tounge BA, Herzfeld J, Griffin RG. Frequency selective heteronuclear dipolar recoupling in rotating solids: Accurate 13C–15N distance measurements in uniformly 13C,15N-labeled peptides. J Am Chem Soc. 2001;123:3507–3519. doi: 10.1021/ja003266e. [DOI] [PubMed] [Google Scholar]
- 29.Jaroniec CP, Filip C, Griffin RG. 3D TEDOR NMR experiments for the simultaneous measurement of multiple carbon-nitrogen distances in uniformly 13C,15N-labeled solids. J Am Chem Soc. 2002;124:10728–10742. doi: 10.1021/ja026385y. [DOI] [PubMed] [Google Scholar]
- 30.Soreghan B, Kosmoski J, Glabe C. Surfactant properties of Alzheimers Aβ peptides and the mechanism of amyloid aggregation. J Biol Chem. 1994;269:28551–28554. [PubMed] [Google Scholar]
- 31.Perutz MF, Johnson T, Suzuki M, Finch JT. Glutamine repeats as polar zippers: Their possible role in inherited neurodegenerative diseases. Proc Natl Acad Sci USA. 1994;91:5355–5358. doi: 10.1073/pnas.91.12.5355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sawaya MR, et al. Atomic structures of amyloid cross-β spines reveal varied steric zippers. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 33.Nelson R, et al. Structure of the cross-β spine of amyloid-like fibrils. Nature. 2005;435:773–778. doi: 10.1038/nature03680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferguson N, et al. General structural motifs of amyloid protofilaments. Proc Natl Acad Sci USA. 2006;103:16248–16253. doi: 10.1073/pnas.0607815103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paravastu AK, Petkova AT, Tycko R. Polymorphic fibril formation by residues 10–40 of the Alzheimer's β-amyloid peptide. Biophys J. 2006;90:4618–4629. doi: 10.1529/biophysj.105.076927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442:585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- 37.Makarava N, et al. Dichotomous versus palm-type mechanisms of lateral assembly of amyloid fibrils. Protein Sci. 2006;15:1334–1341. doi: 10.1110/ps.052013106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Perutz MF, Finch JT, Berriman J, Lesk A. Amyloid fibers are water-filled nanotubes. Proc Natl Acad Sci USA. 2002;99:5591–5595. doi: 10.1073/pnas.042681399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wasmer C, et al. Amyloid fibrils of the HET-s(218–289) prion form a β-solenoid with a triangular hydrophobic core. Science. 2008;319:1523–1526. doi: 10.1126/science.1151839. [DOI] [PubMed] [Google Scholar]
- 40.Sen A, et al. Mass analysis by scanning transmission electron microscopy and electron diffraction validate predictions of stacked β-solenoid model of HET-s prion fibrils. J Biol Chem. 2007;282:5545–5550. doi: 10.1074/jbc.M611464200. [DOI] [PubMed] [Google Scholar]
- 41.Ishii Y. 13C-13C dipolar recoupling under very fast magic angle spinning in solid state nuclear magnetic resonance: Applications to distance measurements, spectral assignments, and high-throughput secondary structure determination. J Chem Phys. 2001;114:8473–8483. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.