Abstract
The eukaryotic initiation factor 3 (eIF3) plays an important role in translation initiation, acting as a docking site for several eIFs that assemble on the 40S ribosomal subunit. Here, we use mass spectrometry to probe the subunit interactions within the human eIF3 complex. Our results show that the 13-subunit complex can be maintained intact in the gas phase, enabling us to establish unambiguously its stoichiometry and its overall subunit architecture via tandem mass spectrometry and solution disruption experiments. Dissociation takes place as a function of ionic strength to form three stable modules eIF3(c:d:e:l:k), eIF3(f:h:m), and eIF3(a:b:i:g). These modules are linked by interactions between subunits eIF3b:c and eIF3c:h. We confirmed our interaction map with the homologous yeast eIF3 complex that contains the five core subunits found in the human eIF3 and supplemented our data with results from immunoprecipitation. These results, together with the 27 subcomplexes identified with increasing ionic strength, enable us to define a comprehensive interaction map for this 800-kDa species. Our interaction map allows comparison of free eIF3 with that bound to the hepatitis C virus internal ribosome entry site (HCV-IRES) RNA. We also compare our eIF3 interaction map with related complexes, containing evolutionarily conserved protein domains, and reveal the location of subunits containing RNA recognition motifs proximal to the decoding center of the 40S subunit of the ribosome.
Keywords: hepatitis C virus internal ribosome entry site, subunit organization model3, top-down analysis of macromolecular complexes, translation regulation, in-solution disruption
Since its identification in the 1970s (1–3), the translation initiation factor eIF3 has been subjected to intense scrutiny. Despite considerable interest, knowledge of many aspects of its structure and function remain elusive because of its overall structural complexity and the lack of facile genetic approaches. It is established, however, that eIF3 is involved in both ribosome biogenesis and protein synthesis in eukaryotes (4). Concerted binding of initiation factors is required to initiate protein synthesis and recruit transfer and messenger RNAs to the 40S subunit before assembly of active ribosomes (5). eIF3 binding may take place initially during this process, together with eIF1 and eIF1A to the 40S, followed by binding of the Met-tRNAi-eIF2-GTP complex. Then mRNA binding, scanning, and AUG recognition occur, enabling the 60S subunit to join to form elongation-competent 80S ribosomes (6). An alternative pathway of initiating protein synthesis, often used by viruses, involves a structured sequence in the 5′ untranslated region of mRNA known as the internal ribosome entry site (IRES). These sequences promote translation initiation without requiring the full complement of eukaryotic initiation factors (5–7). The hepatitis C virus (HCV) IRES is recognized specifically by the small ribosomal subunit and eIF3 before viral translation initiation, forming stable complexes amenable to numerous structural studies (7, 8).
Much of the structural information on eIF3 is derived from electron microscopy images, where eIF3 is revealed as a five-lobed particle that binds to the 40S subunit of the ribosome on the side opposite the 60S subunit. HCV IRES RNA was found to bind diagonally across eIF3, and it also binds to the solvent side of the 40S subunit, inducing conformational changes in the 40S upon binding (7, 8). eIF4G binds to the same eIF3 “arm” as that occupied by part of the IRES, consistent with its role in mRNA recruitment to the 43S preinitiation complex. It is also established, from directed hydroxyl radical probing, that subunit eIF3j binds to the decoding center of the human 40S subunit (9).
Human eIF3 shares an intriguing degree of homology with two other complexes whose functions appear unrelated: the COP9 signalosome and the 19S proteasome lid. All three complexes consist of subunits with either PCI (Proteasome, COP9, eIF3) or MPN (Mpr1–Pad1 N-terminal) signature domains (10), and share a common 6PCI + 2MPN domain stoichiometry. The mammalian eIF3 also has an additional five non-PCI/MPN subunits. Two of these additional subunits (b and d) have been shown to cross-link with mRNA within both the 48S preinitiation complex (11) and the IRES–eIF3 binary complex. eIF3b, together with eIF3g that is also a non-PCI/MPN subunit, contain RNA recognition motifs (RRM), indicating their potential role in RNA binding. The roles of the MPN and PCI domains are not well understood, but current opinion is that these domains serve as a structural scaffold, interacting with each other and to other binding partners. It has also been proposed that MPN subunits have a regulatory role in coordinating overall activity (12).
Many of the interactions established in eIF3 are thought to be conserved between human and yeast (6) suggesting that they share a common core, although yeast eIF3 contains only a subset of the subunits identified in human. A comprehensive subunit interaction map for human eIF3 has not yet been reported, although models have been proposed for the yeast complex (6). We have shown that mass spectrometry (MS) can be used to uncover complete interaction maps for large multiprotein complexes, including the yeast exosome (13) and 19S proteasome lid (14). These interaction maps were achieved by generating large numbers of subcomplexes as “building blocks” of the entire complex.
Our previous MS studies of human eIF3 have shown that we can determine close to 30 phosphorylation sites, primarily on subunits eIF3a, b, and c, and through tandem MS of the intact particle, we were able to define its overall subunit stoichiometry (15). Here, we extend these studies and compare both human and yeast eIF3 complexes. The yeast complex appears less stable than its human counterpart and readily dissociates to yield subcomplexes. By contrast, human eIF3 reveals remarkably well resolved charge states for the intact complex and demonstrates a highly stable complex in solutions of low ionic strength. By increasing the ionic strength, we generate three relatively stable modules, the interactions within and between each module being defined by 27 subcomplexes. This information, together with results from immunoprecipitation (IP) and our analysis of eIF3 from yeast, enables us to produce a comprehensive interaction map for the 800-kDa human eIF3 particle [see supporting information (SI) Fig. S1].
Results
Intact eIF3 Contains 13 Subunits at Unit Stoichiometry.
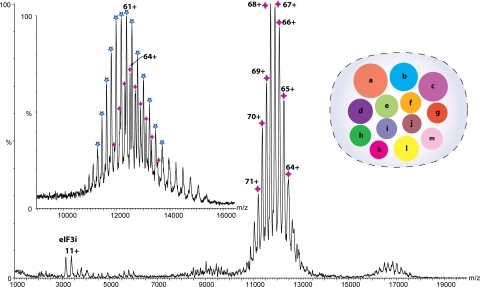
The mass spectrum of eIF3, isolated from HeLa cells as described (15), recorded in a 100 mM ammonium acetate (AmAc) solution, is shown in Fig. 1. Despite the heterogeneity of this complex observed in previous preparations (15), it was possible to obtain remarkably well resolved spectra. The principal series of charge state peaks illustrate that the complex exists as a homogeneous population with a measured mass of 797,999 ± 180 Da. Comparing this value with that calculated by summing the masses determined for each of the subunits (794,310 Da) (Table S1) demonstrates that all 13 subunits are present at unit stoichiometry. The increase of observed versus calculated mass is likely due not only to incomplete PTMs, particularly multiple substoichiometric phosphorylations (15), but also to retention of water and buffer ions under the “soft” ionization conditions used to maintain subunit interactions (16). A minor series corresponding to monomers of subunit eIF3i is also apparent, indicating the relatively labile association of this subunit with the intact complex.
Fig. 1.
MS of the intact eIF3 revealing a series of well resolved charge states consistent with the predominant species being the intact 13 subunit complex. Masses of the 13 subunits were confirmed in a separate proteomics analysis. The y axis is the relative intensity of the peaks. The inventory of subunits is shown with the radius of each subunit scaled according to mass. (Inset) High-energy MS spectrum of eIF3 from 150 mM AmAc solution, showing a second charge state series (blue star) resulting from the dissociation of eIF3j from the intact complex (purple star).
To investigate the existence of other labile subunits, we subjected the complex to increasing acceleration within the mass spectrometer. Two additional subunits, eIF3k and eIF3m, were found to dissociate, consistent with our previous study (16). A spectrum recorded in 150 mM AmAc, under high activation energy, reveals the emergence of a second charge state series close to that of the intact complex, but in this case, subunit j has dissociated (Fig. 1 Inset). Together, these results suggest that subunits eIF3i, j, k, and m locate on the periphery of the complex (14).
eIF3 Dissociates to Form Three Structural Modules.
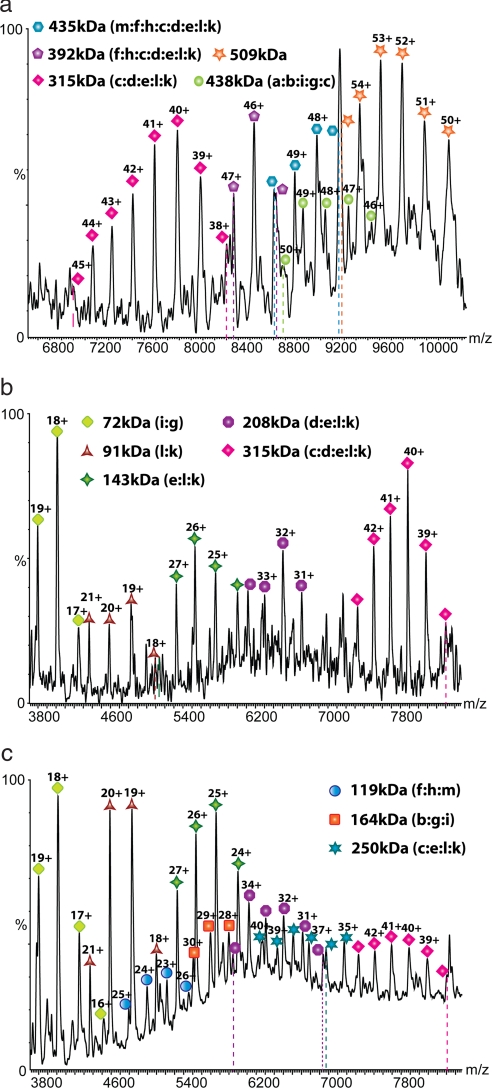
To probe the subunit organization of this 13-component complex, we increased the ionic strength of the complex-containing solution, reasoning that polar and ionic interactions of subunit interfaces would be perturbed (17). At a concentration of 250 mM AmAc, we observed several series of peaks at a lower m/z region than that of intact eIF3, characteristic of subcomplexes generated in solution rather than in the gas phase (18). Six major charge state series were identified, ranging from 315 to 509 kDa [Fig. 2a and Table 1 (2 to 7)]. As we continued to raise the ionic strength of the solution to 350 and 500 mM AmAc (Fig. 2 b and c), a number of overlapping charge state series were observed, consistent with subcomplexes each comprising a small number of subunits (≤5) (Table 1, 14–27). These subcomplexes are likely to be the most informative in defining subunit–subunit contacts (19).
Fig. 2.
Mass spectra recorded after increasing the ionic strength from 250 mM AmAc (a) to 350 mM (b) and 500 mM (c). Series of subcomplexes, observed with decreasing m/z values as the ionic strength is increased, are assigned based on their masses, the observation of common subunit losses, and from tandem MS.
Table 1.
Calculated and observed masses for the subcomplexes generated by increasing the ionic strength from 250 mM to 500 mM AmAc
| No. | Subcomplexes | Calculated mass, Da | Observed mass, Da |
|---|---|---|---|
| 0 | (k, l, e, d, c, a, b, i, g, j, f, h, m) | 794,310 | 797,999 ± 180 |
| 1 | (k, l, e, d, c, b, i, g, f, h, m) | 598,898 | 600,501 ± 93 |
| 2 | (e, d, c, b, i, g, f, h, m) | 507,286 | 508,830 ± 23 |
| 3 | (k, l, e, d, c, h, f, m) | 434,379 | 434,885 ± 157 |
| 4 | (k, l, e, d, c, h, f) | 391,695 | 392,199 ± 36 |
| 5 | (k, l, e, d, c, h) | 354,489 | 354,978 ± 27 |
| 6 | (k, l, e, d, c) | 314,578 | 314,868 ± 93 |
| 7 | (a, b, i, g, c) | 437,811 | 438,478 ± 26 |
| 8 | (a, b, i, g, e) | 383,090 | 383,524 ± 26 |
| 9 | (k, l, e, d, c, i, g) | 386,631 | 386,950 ± 77 |
| 10 | (l, e, d, c, h, f, m) | 409,347 | 410,081 ± 72 |
| 11 | (l, e, d, c, h, f) | 366,993 | 367,285 ± 108 |
| 12 | (l, e, d, c, h) | 329,517 | 329,820 ± 90 |
| 13 | (l, e, d, c) | 289,606 | 290,003 ± 63 |
| 14 | (k, l, e, c) | 250,600 | 250,965 ± 55 |
| 15 | (k, l, e, d) | 207,791 | 207,872 ± 17 |
| 16 | (l, e, d) | 182,820 | 182,861 ± 49 |
| 17 | (e, d) | 116,180 | 116,199 ± 36 |
| 18 | (b, i, g) | 164,518 | 164,680 ± 9 |
| 19 | (i, g) | 71,985 | 72,108 ± 2 |
| 20 | (b, g) | 128,015 | 128,102 ± 21 |
| 21 | (f, h, m) | 119,740 | 119,742 ± 5 |
| 22 | (f, h) | 77,321 | 77,322 ± 5 |
| 23 | (h, m) | 82,264 | 82,268 ± 2 |
| 24 | (f, m) | 79,890 | 79,931 ± 23 |
| 25 | (k, l, e) | 143,749 | 143,819 ± 49 |
| 26 | (k, l) | 91,616 | 91,636 ± 7 |
| 27 | (l, e) | 118,777 | 118,786 ± 37 |
Calculated masses are the sum of the measured masses determined for the individual subunits by using HPLC/MS with the exception of eIF3a, which could not be measured with sufficient accuracy. The sequence mass of eIF3a was used in this case. Assignment based on mass alone was rarely possible. Tandem MS and observation of series of subcomplexes were used to limit our search and enable a unique assignment.
To assign these subcomplexes, we submitted the measured masses of all of the subunits and subcomplexes to an iterative search algorithm (SUMMIT) to explore all possible combinations that could give rise to the measured masses within a given error limit (19). Where more than one composition was possible, tandem MS experiments were performed to release component subunits and differentiate the various possibilities (Fig. S2). Three small subcomplexes with masses of 72, 143, and 119 kDa were frequently observed, implying that they dissociate readily and are located on the periphery of the complex. Tandem MS demonstrates that one of these subcomplexes is dimeric eIF3i:g, and two are trimers eIF3(e:l:k) and eIF3(f:h:m). Furthermore, our data show that the trimer eIF3(f:h:m) can form three pairwise dimers, consistent with a compact arrangement with each subunit in contact with two neighboring subunits. By contrast, both eIF3e and k can dissociate independently from the subcomplex eIF3(e:l:k) with subunit l contacting both e and k. The fact that e:k interaction was not maintained either in solution or in the gas phase strongly implicates an open conformation for the trimer.
Assuming a peripheral location of these subcomplexes, we looked for complexes that differ by additional subunits attached to these species. Interestingly, we noticed that at intermediate ionic strength, four of the large subcomplexes differ by the masses of f, h, and m (Table 1, 3–6). Additionally, MS/MS experiments reveal that they all contain subunit k (Fig. S2 f and g). Assuming that the four subcomplexes result from successive loss of the subunits f, h, and m, we were able to assign the series to subcomplexes containing c, d, e, l, and k with successive addition of h, f, and m. The fact that f and m were only able to bind to (c:d:e:l:k) in the presence of h strongly suggests that these two proteins bind to the subcomplex indirectly via subunit h.
In determining interactions within the group (c:d:e:l:k), six subcomplexes were identified, comprising three dimers l:k, e:l, and e:d; one trimer, e:l:k; and two tetramers, d:e:l:k and c:e:l:k (Table 1). These subcomplexes are consistent with three possible arrangements, with subunit c contacting e, l, or k. The intense peaks observed for the l:k dimer, however, are indicative of a relatively weak association of the dimer to the complex. Therefore, the most likely scenario is that eIF3c contacts eIF3e. Next, we consider the assignment of the species with masses 438 and 383 kDa. We looked for evidence of similar dissociation patterns as above but were unable to identify any series corresponding to loss of eIF3f, h, or m from these subcomplexes, implying that these subunits are not present. By excluding these three subunits from our search, we were able to assign two pentamers eIF3(a:b:i:g:c) and eIF3(a:b:i:g:e) sharing a common core (a:b:i:g) with either subunit c or e attached (Table 1, 7 and 8). For the group (a:b:i:g), we were able to observe three subcomplexes: two dimers b:g and i:g together with the trimer (b:g:i) allowing us to define the arrangement for the trimer b:g:i with g contacting both b and i. A number of these subcomplexes have also been generated in baculovirus (20) including (a:b:i:g:c), which comprises the five human subunits that have homologs in Saccharomyces cerevisiae (6). Identification of this subcomplex for human eIF3 in our spectra suggests its resemblance to the yeast eIF3 core complex.
MS of Yeast eIF3 and IP Confirm Our Interaction Network.
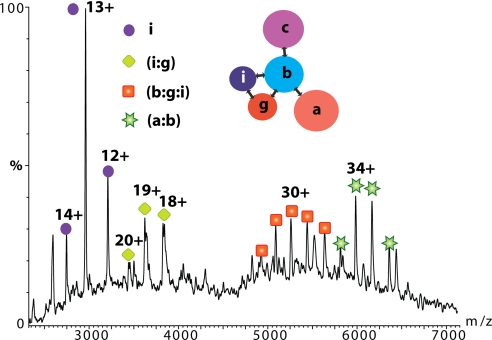
To provide the missing links within the subcomplex (a:b:c:i:g), we isolated eIF3 from yeast using a tandem affinity protocol (21). We were unable to record a spectrum of the intact complex despite confirmation from proteomics experiments that subunits a, b, c, g, and i, but not j, together with eIF5 were present in the complex (SI Text and Fig. S5). This implies that the complex is less stable under our conditions than that of its human counterpart. However, we were able to detect pairwise dimers eIF3g:i and eIF3a:b and the trimer eIF3b:g:i when the affinity tag is placed on subunit b (Fig. 3). By tagging subunit c, we observed additional subcomplexes assigned as eIF3(c:b:i), b:i, b:g, and (c:b:g) (data not shown). Together, MS data from yeast establish seven subcomplexes, enabling us to derive an interaction network for eIF3(a:b:c:i:g) with eIF3b serving as the scaffolding protein connecting the remaining subunits a,c,i and g (Fig. 3).
Fig. 3.
MS spectrum of the yeast eIF3 isolated by tagging subunit eIF3b. Charge state series are assigned on the basis of masses to subcomplexes eIF3i:g, eIF3b:g:i, and eIF3a:b. eIF3i is observed dissociating from the yeast complex at ≈m/z 3,000. (Inset) The interaction network for yeast eIF3 derived from seven subcomplexes observed by MS.
The results from our investigation of yeast eIF3 allow us to define analogous interactions within the human complex, which resembles closely one of the three relatively tightly associated modules generated by manipulating the ionic strength. These modules are denoted A(eIF3a:b:i:g), B(eIF3f:h:m), and C(eIF3k:l:e:d:c). The remaining subunit eIF3j, a labile subunit, attaches to the complex via eIF3b as previously defined (9, 22). The fact that all of the subcomplexes with masses <500 kDa are composed of members from only one or at most two of these modules supports our proposal of three tightly associated modules with weaker interactions occurring between them.
Having established interaction networks within these three modules, we then looked for connections between them. We noted that the effect of increasing ionic strength on the subcomplex (eIF3k:l:e:d:c:f:h:m) was formation of smaller subcomplexes l:k, e:l:k, and d:e:l:k while the intact (k:l:e:d:c:f:h:m) subcomplex persists, implying that it is not likely that the four proteins k, l, e, d interact strongly with the trimer f:h:m. Together with our earlier conclusion that the presence of subunit h is essential for binding the trimer f:h:m, it is reasonable to propose that c and h provide one of the electrostatic interactions between modules B and C. Similarly, establishment of the eIF3b:c interaction in yeast and the observation of subcomplex (a:b:c:i:g) strongly suggest the role of eIF3c in recruiting module C to A. It is, however, difficult to define unambiguously contacts between the module B and A based on MS data alone. We therefore supplemented our data with results from IP. A number of binary interactions were found that support our subunit contacts for modules B and C (Fig. S3). Moreover, those between eIF3b:f and b:h are important for determining interactions between the modules A and B.
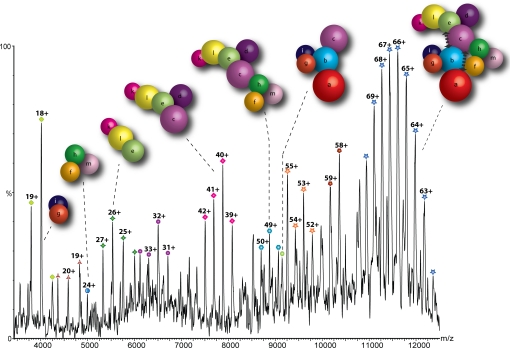
Taking this data together, we are able to propose a detailed interaction network for the stable interactions within the entire human eIF3 (Fig. 4). In this model, eIF3 subunits are represented as spheres scaled according to their mass. It was not possible to take into account subunits that may deviate from globular conformations because a complete atomic structure of only one subunit (eIF3k) has been reported to date (23). This simplistic interaction model is, however, consistent with the IP data, the interactions within yeast eIF3, and the 27 subcomplexes assigned for human eIF3 as a function of ionic strength.
Fig. 4.
Model of the human eIF3 derived from 27 subcomplexes, IP data, and interactions identified in the yeast complex. The complex dissociates into distinct modules in response to changes in ionic strength; in this case, the spectrum shown was recorded at intermediate ionic strength (350 mM AmAc). Arrows denote additional interactions not readily represented in this model.
Comparison of eIF3 with eIF3:IRES.
If we now compare the mass spectra of free human eIF3 with that bound to HCV IRES, we observe an increase in m/z value consistent with the association of a 332-nucleotide RNA of 107 kDa with eIF3 (Fig. S4). MS/MS of the IRES:eIF3 peak isolated at m/z 13,800 yielded subunit k (Fig. S4c), indicating that gas-phase dissociation of eIF3k is not inhibited by IRES binding. Moreover, these high energy conditions give rise to marginally resolved charge states enabling the mass of the IRES:eIF3 complex to be measured as 907 kDa, confirming binary complex formation. If we compare the activated spectra of the intact eIF3 complex with those recorded for the complex bound to IRES, we note several differences: first, the disappearance of peaks corresponding to relatively large subcomplexes in the IRES:eIF3 spectrum, indicating that when the IRES is bound, subcomplex formation is diminished, implying a global stabilization. Particularly interesting is the observation that eIF3i and the l:k dimer can still dissociate in solution from the binary complex, suggesting that these three subunits are not in direct contact with the IRES. By contrast, two dimers, i:g and f:h, are not present in the IRES:eIF3 spectrum, suggesting that the subunits g, f, and h play a role in IRES binding (Fig. 5a).
Fig. 5.
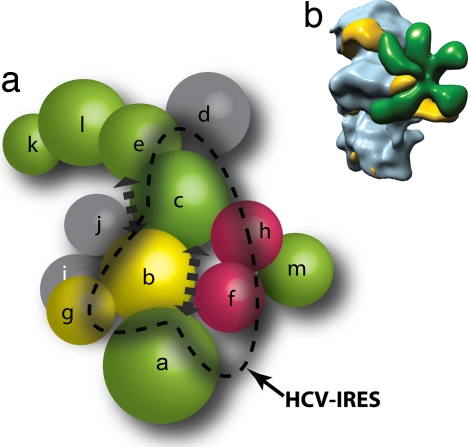
Proposed model for the eIF3:HCV IRES interaction. (a) Subunit organization colored according to signature domains contained within the various subunits. PCI-containing domains (green), MPN domains (red), and RNA recognition motifs (yellow) show direct interactions with the exception of eIF3m and eIF3a. Subunits with no common signature domains are shown in gray. Subunits that are affected by binding of HCV IRES are within the dashed line. The location of subunits satisfies the interaction network and is not indicative of their location within the EM density. (b) EM model of eIF3-IRES-40S complex showing the binding of IRES (yellow) to both eIF3 (green) and the 40S (blue).
Discussion
Here, we report a model of the subunit organization of human eIF3, the largest and most complex of the eukaryotic initiation factors. Our model, is based primarily on data from MS of intact subcomplexes generated in solution and is supported by a subset of interactions generated independently by IP and our analysis of the homologous yeast complex. Our results reveal facile dissociation of subunits eIF3i, j, k, and m as well as three well defined interaction modules generated by manipulating the ionic strength of the complex-containing solution. Furthermore, results from binding of human eIF3 to the IRES shows that although dissociation of larger subcomplexes is prevented, eIF3i and the eIF3l:k dimer are still able to dissociate, suggesting that these subunits are not directly involved in the interactions with the IRES.
If we compare our model with data for other protein complexes containing both MPN and PCI domains (10), we note that the two MPN domain-containing subunits, eIF3f and h, interact directly in our subunit organization map (Fig. 5a), a phenomenon also observed with another PCI-containing complex, the proteasome lid (14). Moreover, several directly interacting PCI domains are found in both complexes, supporting their role in providing a scaffold for active subunits. Our interaction map for human eIF3 also establishes direct interactions among eIF3b, g, i, and j, four of the five non-PCI/MPN subunits absent in the other two PCI complexes.
The high sensitivity of eIF3 to dissociation into subcomplexes as the ionic strength is raised suggests that electrostatic interactions may play a central role in maintaining the integrity of the complex and in regulating its function. The three largest subunits eIF3a, b, and c are extensively phosphorylated, occupy a central position in our model, and are conserved between human and yeast (16). Increasing ionic strength leads primarily to dissociation of interactions between eIF3b:c and eIF3c:h, suggesting that salt bridges between charged groups, such as phosphorylated side-chains, are directly involved in the interfaces between these subunits. It is therefore tempting to speculate that phosphorylation of eIF3 is involved in regulating labile interactions, both between eIF3 modules and subcomplexes and maybe also with other parts of the translational apparatus. It is also conceivable that the three modules, observed in nonphysiological high ionic-strength buffers, may mimic dephosphorylated states of the complex in vivo.
It is interesting to compare our results with a recent report of coexpression of subunits and formation of a reconstituted functional “core” for the mammalian eIF3 comprising subunits eIF3a, b, c, e, f, and h (20). Interestingly, all of the “functionally dispensable” subunits according to this study are located on the periphery of the complex in our model and are not involved in interactions between our three modules, consistent with our overall view of this assembly.
Our results also show that one of the core subunits eIF3c is observed in a series of subcomplexes containing up to seven noncore mammalian subunits not present in the yeast complex (f, h, m, d, e, l, and k). The role of eIF3c may therefore involve facilitating the recruitment and assembly of these noncore subunits into the intact eIF3 complex. This proposal is supported by deletion of eIF3c from a functional subcomplex that resulted in loss of its ability to promote binding of mRNA to the 40S ribosomal subunit (20). eIF3b also has an established scaffolding role (24) and is found involved in mRNA binding (11, 25). In our network, it interacts directly with the other RNA recognition motif (RRM)-containing subunit eIF3g. Both are located in close proximity to eIF3j, known to be located at the decoding centre of the 40S subunit (9) (Fig. 5b). This is an intriguing observation because it implies a role of RNA recognition for these two subunits (eIF3b and g) in binding and presenting mRNA to the decoding centre of the ribosome.
The labile binding of eIF3j, as shown by its absence from all of our subcomplexes, implies its ready dissociation from the complex. Interestingly, eIF3j has been shown to dissociate from eIF3 in quiescent T lymphocytes, but upon activation, eIF3j binds eIF3 and 40S ribosomal subunits, implying that the interaction of subunit j with eIF3 may well be regulated (26). Moreover, eIF3j has been shown to have more than one role, not only in the decoding center (9), but also taking part in 20S pre-rRNA processing (4). The fact that this critical subunit is only loosely associated and is located close to eIF3b:g:i and the observation that subunit eIF3i is also labile implies that dissociation of eIF3j, with or without its neighboring subunits, may well be associated with its regulatory role in translation initiation and in ribosome biogenesis (4).
Overall, in this study, we have derived a subunit interaction map for a 13-subunit complex using an emerging MS approach (13, 14, 27). If we consider our interaction map with the five-lobed architecture revealed by cryo-EM (8), it is tempting to speculate that the subcomplexes frequently observed in our MS experiments may correspond to the lobes identified within the EM structure. The assignment of these subcomplexes to the different domains of the 3D EM structure remains to be completed. Overall, the subunit architecture presented here, together with knowledge of the ease of dissociation and interactions that link the three stable modules provide an intriguing insight into the relationships between subunits and their possible functional significance in this complex initiation factor.
Experimental Methods
The eIF3 sample was purified from HeLa cell cytosol as described (8, 15).
LC-MS Analysis of eIF3 Subunits.
Capillary HPLC-MS analysis of individual eIF3 subunits was carried out by using a LC-Packings Ultimate System (Dionex) equipped with a capillary UV detector set at 214 and 280 nm. eIF3 was prepared in a 1:1 (vol/vol) of 0.1% TFA and 1 μl of sample applied to a capillary PS-DVB reverse-phase monolithic column (200 μm i.d.× 5 cm; Dionex) equilibrated at 90% solvent A (0.05% TFA) and 10% solvent B (0.04% TFA, 90% ACN). A linear gradient was used of 10–70% solvent B in 25 min at a flow rate of 3 μl/min. The column effluent was analyzed by both electrospray and MALDI MS (see SI Text).
MS of Intact eIF3 and Subcomplexes.
For MS of the intact complex, 20 μl of purified eIF3 (1 μg/μl) was buffer exchanged into AmAc buffer (pH 7.5) containing 1 mM DTT by using micro biospin 6 columns (Bio-Rad). 2-μl aliquots were electrosprayed from gold-coated borosilicate capillaries prepared in house. Spectra were recorded on a QSTAR XL (Applied Biosystems) modified for high mass detection (28) and adjusted to preserve noncovalent interactions (29). MS experiments were performed at a capillary voltage of 1,200 V and declustering potentials of 40 V and 15 V. In tandem MS experiments, ions were isolated in the quadrupole and subjected to collision-induced dissociation (acceleration energy up to 170 V). To generate subcomplexes, a 7.5 M AmAc solution (Sigma) was diluted stepwise and added to the eIF3 solution at a ratio of 1:10 (vol/vol) to give the required AmAc concentration and incubated on ice for 10 min before analysis.
HCV-IRES Binding to eIF3.
Wild-type HCV-IRES was transcribed in vitro and gel-purified (30). eIF3 was incubated with HCV-IRES at a molar ratio of 1:1 for 3 min at 37°C in 20 mM Tris (pH 7.5), 100 mM KCl, 3 mM Mg(OAc)2, and 1 mM DTT. A control solution of eIF3 alone in the same buffer was prepared. Both the control and the experiment were then buffer-exchanged into 150 mM AmAc (pH 7.5) containing 3 mM Mg2+ acetate and 1 mM DTT.
Expression, Purification, and MS of Yeast eIF3.
S. cerevisiae eIF3 was purified from TAP-tag yeast strains (Open Biosystems), modified to express endogenous proteins fused with a C-terminal tandem affinity tag on eIF3b or c (21). See SI Text and Fig. S5. Spectra were recorded on the QSTAR XL at a capillary voltage of 1,200 V and declustering potentials of 150 V and 15 V.
Acknowledgments.
We acknowledge with thanks funding from EU 3D Repertoire LSHG-CT-2005-512028, the Waters Kundert Trust, the Biotechnology and Biological Sciences Research Council, and the Royal Society (to C.V.R.), sabbatical funding from Lawrence Livermore National Laboratory (to D.B.), and a National Institutes of Health program grant (to J.A.D., J.B.H., and J.A.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801313105/DCSupplemental.
References
- 1.Prichard PM, Gilbert JM, Shafritz DA, Anderson WF. Factors for the initiation of haemoglobin synthesis by rabbit reticulocyte ribosomes. Nature. 1970;226:511–514. doi: 10.1038/226511a0. [DOI] [PubMed] [Google Scholar]
- 2.Safer B, et al. Purification and characterization of two initiation factors required for maximal activity of a highly fractionated globin mRNA translation system. Proc Natl Acad Sci USA. 1976;73:2584–2588. doi: 10.1073/pnas.73.8.2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benne R, Hershey JW. Purification and characterization of initiation factor IF-E3 from rabbit reticulocytes. Proc Natl Acad Sci USA. 1976;73:3005–3009. doi: 10.1073/pnas.73.9.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Valasek L, Hasek J, Nielsen KH, Hinnebusch AG. Dual function of eIF3j/Hcr1p in processing 20 S pre-rRNA and translation initiation. J Biol Chem. 2001;276:43351–43360. doi: 10.1074/jbc.M106887200. [DOI] [PubMed] [Google Scholar]
- 5.Pestova TV, Lorsch JR, Hellen CUT. The Mechanism of Translation Initiation in Eukaryotes. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2007. [Google Scholar]
- 6.Hinnebusch AG. eIF3: A versatile scaffold for translation initiation complexes. Trends Biochem Sci. 2006;31:553–562. doi: 10.1016/j.tibs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 7.Spahn CM, et al. Hepatitis C virus IRES RNA-induced changes in the conformation of the 40s ribosomal subunit. Science. 2001;291:1959–1962. doi: 10.1126/science.1058409. [DOI] [PubMed] [Google Scholar]
- 8.Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- 9.Fraser CS, Berry KE, Hershey JW, Doudna JA. eIF3j is located in the decoding center of the human 40S ribosomal subunit. Mol Cell. 2007;26:811–819. doi: 10.1016/j.molcel.2007.05.019. [DOI] [PubMed] [Google Scholar]
- 10.Hofmann K, Bucher P. The PCI domain: A common theme in three multiprotein complexes. Trends Biochem Sci. 1998;23:204–205. doi: 10.1016/s0968-0004(98)01217-1. [DOI] [PubMed] [Google Scholar]
- 11.Unbehaun A, Borukhov SI, Hellen CU, Pestova TV. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon–anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes Dev. 2004;18:3078–3093. doi: 10.1101/gad.1255704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim T, Hofmann K, von Arnim AG, Chamovitz DA. PCI complexes: Pretty complex interactions in diverse signaling pathways. Trends Plants Sci. 2001;6:379–386. doi: 10.1016/s1360-1385(01)02015-5. [DOI] [PubMed] [Google Scholar]
- 13.Hernandez H, Dziembowski A, Taverner T, Seraphin B, Robinson CV. Subunit architecture of multimeric complexes isolated directly from cells. EMBO Rep. 2006;7:605–610. doi: 10.1038/sj.embor.7400702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharon M, Taverner T, Ambroggio XI, Deshaies RJ, Robinson CV. Structural organization of the 19S proteasome lid: Insights from MS of intact complexes. PLoS Biol. 2006;4:1314–1323. doi: 10.1371/journal.pbio.0040267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Damoc E, et al. Structural characterization of the human eukaryotic initiation factor 3 protein complex by mass spectrometry. Mol Cell Proteomics. 2007;6:1135–1146. doi: 10.1074/mcp.M600399-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.McKay AR, Ruotolo BT, Ilag LL, Robinson CV. Mass measurements of increased accuracy resolve heterogeneous populations of intact ribosomes. J Am Chem Soc. 2006;128:11433–11442. doi: 10.1021/ja061468q. [DOI] [PubMed] [Google Scholar]
- 17.Levy ED, Boeri Erba E, Robinson CV, Teichmann SA. Assembly reflects evolution of protein complexes. Nature. 2008;453:1262–1265. doi: 10.1038/nature06942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Benesch JL, Aquilina JA, Ruotolo BT, Sobott F, Robinson CV. Tandem mass spectrometry reveals the quaternary organization of macromolecular assemblies. Chem Biol. 2006;13:597–605. doi: 10.1016/j.chembiol.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 19.Taverner T, et al. Subunit architecture of intact protein complexes from mass spectrometry and homology modeling. Acc Chem Res. 2008;41:617–627. doi: 10.1021/ar700218q. [DOI] [PubMed] [Google Scholar]
- 20.Masutani M, Sonenberg N, Yokoyama S, Imataka H. Reconstitution reveals the functional core of mammalian eIF3. EMBO J. 2007;26:3373–3383. doi: 10.1038/sj.emboj.7601765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rigaut G, et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat Biotechnol. 1999;17:1030–1032. doi: 10.1038/13732. [DOI] [PubMed] [Google Scholar]
- 22.ElAntak L, Tzakos AG, Locker N, Lukavsky PJ. Structure of eIF3b RNA recognition motif and its interaction with eIF3j: Structural insights into the recruitment of eIF3b to the 40 S ribosomal subunit. J Biol Chem. 2007;282:8165–8174. doi: 10.1074/jbc.M610860200. [DOI] [PubMed] [Google Scholar]
- 23.Wei Z, et al. Crystal structure of human eIF3k, the first structure of eIF3 subunits. J Biol Chem. 2004;279:34983–90. doi: 10.1074/jbc.M405158200. [DOI] [PubMed] [Google Scholar]
- 24.Fraser CS, et al. The j-subunit of human translation initiation factor eIF3 is required for the stable binding of eIF3 and its subcomplexes to 40 S ribosomal subunits in vitro. J Biol Chem. 2004;279:8946–8956. doi: 10.1074/jbc.M312745200. [DOI] [PubMed] [Google Scholar]
- 25.Sizova DV, Kolupaeva VG, Pestova TV, Shatsky IN, Hellen CU. Specific interaction of eukaryotic translation initiation factor 3 with the 5′ nontranslated regions of hepatitis C virus and classical swine fever virus RNAs. J Virol. 1998;72:4775–4782. doi: 10.1128/jvi.72.6.4775-4782.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Miyamoto S, Patel P, Hershey JW. Changes in ribosomal binding activity of eIF3 correlate with increased translation rates during activation of T lymphocytes. J Biol Chem. 2005;280:28251–28264. doi: 10.1074/jbc.M414129200. [DOI] [PubMed] [Google Scholar]
- 27.Robinson CV, Sali A, Baumeister W. Molecular sociology of the cell. Nature. 2007;450:973–982. doi: 10.1038/nature06523. [DOI] [PubMed] [Google Scholar]
- 28.Sobott F, Hernandez H, McCammon MG, Tito MA, Robinson CV. A tandem mass spectrometer for improved transmission and analysis of large macromolecular assemblies. Anal Chem. 2002;74:1402–1407. doi: 10.1021/ac0110552. [DOI] [PubMed] [Google Scholar]
- 29.Hernandez H, Robinson CV. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat Protoc. 2007;2:715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- 30.Kieft JS, Zhou K, Jubin R, Doudna JA. Mechanism of ribosome recruitment by hepatitis C IRES RNA. RNA. 2001;7:194–206. doi: 10.1017/s1355838201001790. [DOI] [PMC free article] [PubMed] [Google Scholar]