Abstract
A number of genes have been implicated in regeneration, but the regulation of these genes, particularly pertaining to regeneration in higher vertebrates, remains an interesting and mostly open question. We have studied microRNA (miRNA) regulation of regeneration and found that an intact miRNA pathway is essential for caudal fin regeneration in zebrafish. We also showed that miR-203 directly targets the Wnt signaling transcription factor Lef1 during this process. Repression of Lef1 by miR-203 blocks regeneration, whereas loss of miR-203 results in excess Lef1 levels and fin overgrowth. Expression of Lef1 from mRNAs lacking 3′ UTR recognition elements can rescue the effects of excess miR-203, demonstrating that these effects are due to specific regulation of lef1 by miR-203. Our data support a model in which regulation of Lef1 protein levels by miR-203 is a key limiting step during regeneration.
Keywords: lef1, miR-203
Most vertebrates, including humans, are unable to regenerate the majority of lost or damaged tissues. In contrast, zebrafish are able to regenerate various damaged tissues, including fins, hearts, retinas, and spinal cords (1). For fins, regeneration relies on the formation of blastema cells, stem cell-like cells that either are recruited to the damaged area or originate from the de-differentiation of cells in the area (2, 3). Zebrafish caudal fins undergo isometric growth (i.e., fin grows in proportion to body size) throughout life, and understanding the regulatory mechanisms for controlling such growth remains a key question. The fin is composed of multiple bony rays that grow autonomously and are made up of bony segments, termed lepidotrichia. Each ray is composed of two hemirays, which create a protective shell around nerves, blood vessels, and mesenchymal cells. Fins grow through the addition of bone to the distal tip of the fin. Regeneration proceeds through at least five steps: wound healing, mesenchymal disorganization or reorganization, blastema formation, outgrowth, and termination (1, 4).
miRNAs are a recently discovered class of genes that regulate gene expression at the posttranscriptional level and are required for development, stem cell maintenance, and renewal (5–18). Recently, Yin et al. (19) showed that fibroblast growth factor (Fgf) signaling alters the expression of multiple miRNAs during regeneration. One of the miRNA targets of Fgf signaling, miR-133, targets mps1, which encodes a kinase that regulates blastemal proliferation. Interestingly, these authors also found that various other markers of regeneration were indirectly activated on the reduction of miR-133 levels, suggesting that overall regulation of regeneration by miRNAs might be quite complex. Here we show that an intact miRNA pathway indeed is essential for regeneration. Furthermore, we show that in addition to regulation of Fgf signaling during regeneration, Wnt signaling also subject is to miRNA regulation through miR-203 control of Lef1.
To examine global miRNA expression patterns in regenerating fins, we first conducted microarrays. Caudal fins were amputated from adult fish, and RNA was isolated from three regenerative states: adult fins, fins undergoing active regeneration, and fins that appeared to be completely regenerated. Small RNAs from each stage were isolated, fluorescently labeled, and directly hybridized to microarrays to determine the expression patterns of 346 vertebrate miRNAs (20). To obtain sufficient RNA for three independent arrays, fins were amputated from 120 adult fish, which were then returned to the aquarium temperature of 27 °C, after which regeneration was allowed to proceed for 2 or 5 weeks at 27 °C before reamputation and another round of RNA isolation. At this temperature, and based on the position of amputation, regeneration was ≈30% complete by 2 weeks [supporting information (SI) Fig. S1] and was nearly complete by 5 weeks. Heat maps illustrating global changes in miRNA expression are given in Fig. S2; expression changes for individual miRNAs, along with corresponding fold changes and P values, are given in Fig. S3 and Table S1. Some miRNAs that change during regeneration did not appear to return to their previous expression levels after 5 weeks at 27 °C, possibly due to incomplete regeneration. We hypothesize that miRNAs exhibiting decreased expression during active regeneration enable expression of genes required for regeneration (19), whereas miRNAs that are up-regulated during regeneration repress genes that normally prevent proliferation and/or maintain terminal cell differentiation.
To validate our approach, we chose to first focus on those miRNAs whose expression was altered most dramatically (either up or down) and that are predicted to target genes implicated in regeneration and/or genes whose expression changes during active regeneration (Table 1). For example, the arrays showed that expression of miR-200b increased during regeneration and that one of its predicted targets is bmp3, which has been shown to be correspondingly down-regulated (21). Similarly, miR-203 is down-regulated during regeneration and is predicted to target lef1. Regeneration requires up-regulation of lef1 in newly formed regenerative epithelia and can be used as a marker for the initiation of regeneration (21–23). Thus, significant down-regulation of miR-203 during regeneration is consistent with the up-regulation of lef1 that occurs during active regeneration. Furthermore, blastema formation and maintenance of blastema cells (24) requires an active form of the heat-shock protein 60 (hsp60) and two of the down-regulated miRNAs identified in our array (miR-2 and miR-338) are predicted to target hsp60. Finally, msxb has been postulated to regulate the rate of proliferation of blastema cells during regenerative outgrowth (25, 26), and expression of miR-301, which is predicted to target msxb, is down-regulated during regeneration. These results suggest that our array strategy was able to identify differentially expressed miRNAs that target genes involved in regeneration.
Table 1.
Differential expression of miRNAs targeting genes implicated in fin regeneration
| Down-regulated | FC | P value | Predicted targets | Up-regulated | FC | P | Predicted targets |
|---|---|---|---|---|---|---|---|
| miR-338 | −92.4 | .015 | hsp60, or2.7, six4.1 | miR-80 | 172.0 | <.001 | |
| miR-144 | −90.4 | .004 | ext1, ncb5or, tnik, gpr157 | miR-69 | 169.0 | <.001 | |
| miR-301 | −70.4 | .001 | msxb, tsc1, gls, gcl | miR-66 | 154.6 | .001 | |
| miR-2a | −68.9 | .004 | hsp60 | miR-26a | 62.5 | .059 | pdcdr |
| miR-203 | −67.2 | .033 | lef1 | miR-200b | 22.3 | .076 | bmp3, pdcd4 |
Microarrays were performed to identify differentially expressed miRNAs during caudal fin regeneration. A complete list of miRNAs is given in Fig. S3 and Table S1 (raw data is provided in Table S2). A subset of differentially expressed miRNAs, fold changes (FCs), P values, and possible mRNA targets are as indicated. Predicted targets were found using miRanda, miRTar (http://mirtar.mbc.nctu.edu.tw/index.html), and miRBase (41, 42).
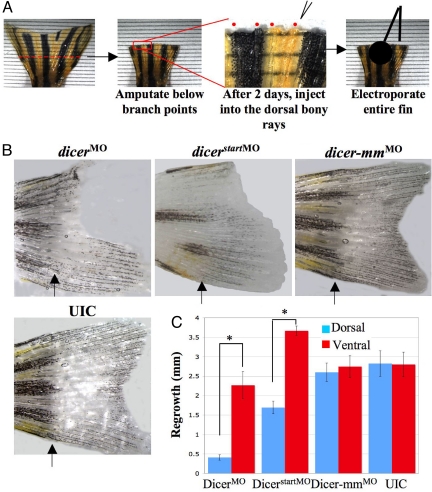
We next sought to test whether miRNAs directly regulate regeneration. First, we decided to block overall miRNA production in regenerating fins by introducing two different antisense morpholino oligonucleotides against Dicer, an enzyme required for cytoplasmic processing of miRNA precursors (27). The loss of Dicer is embryonically lethal in both fish and mice (15, 28, 29), and stem cell maintenance requires Dicer (16, 17, 30). To silence Dicer, we used a previously described antisense morpholino oligonucleotide complementary to the 5′ UTR of dicer mRNA (28) (Fig. 1), as well as a second dicer morpholino against the translational start site. For these experiments, fish were placed in tanks at 33 °C after amputation, injection, and electroporation, with regeneration then monitored over time. Regeneration proceeds normally but at about twice the rate at 33 °C compared with that at 27 °C (2) (Fig. S1). The increased rate of regeneration at this temperature allows disruption of gene function by morpholinos, which can persist for up to 72 h (31). Because the array data were obtained from fish maintained at 27 °C, we first verified, by northern blot analysis, that the patterns of expression for differentially expressed miRNAs, as determined using arrays, were similarly altered when regeneration proceeded at 33 °C (Fig. S4 a and b and data not shown). Northern blot analyses of miR-203 levels showed a dramatic decrease in expression when regeneration proceeded at 33 °C, identical to that found using microarrays performed on fins isolated from fish maintained at 27 °C.
Fig. 1.
An intact miRNA pathway is required for fin regeneration. (A) Schematic of the injection/electroporation strategy. (B) Caudal fins were amputated at the indicated line, and dorsal halves were injected at 2 dpa with either antisense morpholino oligonucleotides against dicer (dicerMO), a morpholino against a region including the start codon of dicer (dicerstartMO), a mismatch morpholino oligonucleotide (dicer-mmMO), or UICs. Fins are shown at 6 dpa. (C) Quantitative analysis of regeneration. Fin growth was measured, and the difference between dorsal and ventral regeneration was plotted (n = 12 fins at 6 dpa, P = .0001, except for dicerstartMO, n = 6, P ≪ .001).
The morpholinos against dicer were injected into the lepidotrichia (bony rays) on the dorsal half of amputated fins, followed by electroporation to facilitate cellular uptake (25, 32) (Fig. 1A). Because newly amputated fins cannot be injected immediately, we waited 2 days after amputation, to allow wound healing, before injectiing morpholinos or RNAs. The entire fin was subjected to electroporation, but only the dorsal half was injected with morpholinos or RNAs, such that the ventral half served as an internal control. After injection of the two different morpholinos against dicer and a mismatched control morpholino, we measured the microns of regrowth on the dorsal halves versus the ventral halves of electroporated fins. As shown in Fig. 1 B and C, regeneration was significantly inhibited in the dorsal halves of fins injected with the dicer morpholinos compared with those injected with either the mismatch morpholino or uninjected controls (UICs). The effect was pronounced at 6 days postamputation (dpa), after which the fins began to regenerate over time (data not shown), which is consistent with the half-life of the morpholinos but also indicates that the injections did not irreversibly block regeneration. These results demonstrate that an intact miRNA pathway is essential for regeneration and are consistent with the hypothesis that induction of specific miRNAs is needed for proper regeneration (19).
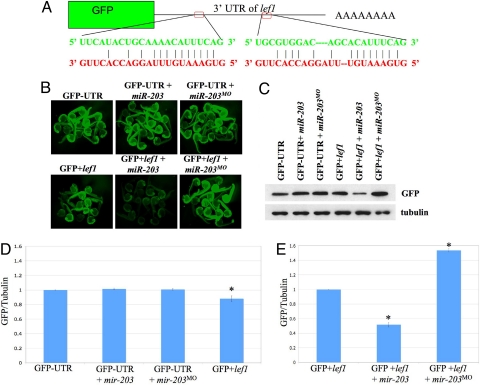
To investigate the function of miRNAs that are down-regulated during regeneration and to test the role of individual miRNAs, we focused on miR-203. miR-203 was found to be significantly down-regulated during regeneration and is predicted to target lef1 (Table 1), a Wnt signaling-regulated transcription factor, the transcription of which is induced during regeneration and serves as a marker of the basal epidermal layer during blastema formation (22). The 3′ UTR of lef1 contains two potential miRNA recognition elements (MREs) for miR-203 (Fig. 2A). As a first test of the hypothesis that miR-203 targets lef1, we created a construct that fused the 3′ UTR of lef1 to the coding region of GFP (Fig. 2A). We then injected in vitro transcribed GFP fusion transcripts into single-cell zebrafish embryos in the presence or absence of exogenous miR-203 and examined fluorescence levels in living embryos at 1 day postfertilization (dpf) (Fig. 2B). We also co-injected antisense morpholinos complementary to miR-203 (i.e., miR-203MO) that function to block the activity of endogenous miR-203 and thus should lead to increased fluorescence (31, 33). As a control, GFP lacking the lef1 3′ UTR also was injected into single-cell zebrafish embryos with and without exogenous miR-203 and miR-203MO (Fig. 2B). As shown in Fig. 2B, co-injection of miR-203 and GFP+lef1 clearly reduced GFP levels in embryos, whereas co-injection of miR-203MO and GFP+lef1 resulted in increased fluorescence. In parallel assays, western blot analyses of pooled embryo lysates were performed using antibodies against GFP and α-tubulin (Fig. 2 C and D). Consistent with the fluorescence experiments, GFP levels were decreased significantly on co-injection of miR-203, and knockdown of endogenous miR-203 by injection of miR-203MO led to increased GFP levels (Fig. 2 D and E). GFP levels were significantly lower in the GFP+lef1 injected embryos compared with the GFP-UTR control. This lower fluorescence can be attributed to basal miR-203 expression in 1 dpf embryos (Fig. S4a). Together, the GFP reporter assays suggest that miR-203 specifically targets lef1.
Fig. 2.
miR-203 Targets lef1. (A) The 3′ UTR of lef1 contains two recognition sites (MREs) for miR-203. The predicted pairing is shown with miR-203 sequences in red and lef1 mRNA in green. (B) Single-cell zebrafish embryos were injected with RNA encoding GFP lacking a 3′ UTR or the GFP+lef1 fusion construct above in the presence or absence of exogenous miR-203 or miR-203MO. Fluorescent images of representative embryos were obtained at 1 day post fertilization. (C) Lysates were prepared from embryos injected as above, and western blot analyses were performed using antibodies against GFP or α-tubulin as a loading control. Each lane contains lysates from ≈5 embryos. (D and E) Quantitation of GFP/tubulin ratios from multiple western blot analyses (n = 6) as in (C). Asterisks indicate P < .01. In (D), all lanes are normalized to GFP-UTR; in (E), all lanes are normalized to GFP+lef1.
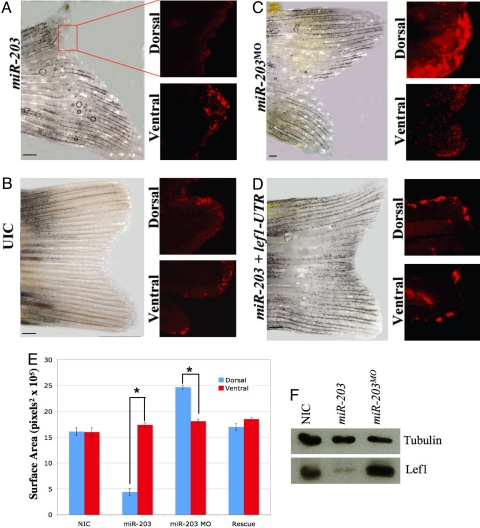
To test whether miR-203 regulates lef1 during actual regeneration, we amputated caudal fins and injected excess miR-203, then performed electroporation. Evaluation of fin regeneration under these conditions revealed a definite loss of regenerative outgrowth in the dorsal halves of fins injected with miR-203 compared with the control ventral halves (UICs) (Fig. 3 A and B) or with injection of miR-15b, a miRNA whose expression does not change during regeneration (Fig. S5a). To directly examine Lef1 levels under these conditions, we immunostained whole regenerating fins with antibodies against Lef1. Lef1 levels were dramatically decreased in the outermost epithelial layer on injection of miR-203 in the dorsal halves, but not in the ventral halves, of regenerating fins (Fig. 3A). Similar Lef1 levels were detected in both the dorsal and ventral halves of regenerating UIC fins (Fig. 3B). As an additional control, we injected morpholinos against a region including the translation start site of lef1 mRNA to determine whether a similar phenotype to miR-203 overexpression could be found. Consistent with the regulation of lef1 by miR-203, knockdown of Lef1 inhibited regeneration in the dorsal halves of injected fins (Fig. S5b). These results are entirely consistent with the hypothesis that miR-203 regulates Lef1 levels during regeneration.
Fig. 3.
miR-203 regulates lef1 during fin regeneration. (A–C) Caudal fins were amputated and then injected at 2 dpa with miR-203, antisense morpholinos against miR-203 (miR-203MO), or UICs. Whole fins were allowed to regenerate for 6 days, after which they were photographed as shown in Fig. 1 with dorsal up and ventral down. Subsequently, the same fins were fixed and immunostained with anti-lef1 antibodies. Shown are regions corresponding to single bony rays from either the dorsal or ventral halves. Scale bars indicate 500 μ. (D) Inhibition of regeneration caused by overexpression of miR-203 was rescued by co-injection of lef1 mRNA lacking its normal 3′ UTR. (E) Quantitative analysis of regeneration. Surface area was calculated using Image J (n = 9; P ≪ .001 for miR-203, P < .001 for miR-203MO). (F) Total proteins were isolated from the distal tips (500 μ) of the dorsal halves of six fins at 6 dpa, and western blot analyses were performed using antibodies against α-tubulin or lef1.
We next injected miR-203MO into regenerating fins to determine the effect of loss of miR-203 on regeneration. Two different morpholinos were used, the same miR-203MO as before and a second morpholino against the pre-miR-203 loop region. In both cases, we observed a dramatic and remarkable increase in fin growth (Fig. 3C and Fig. S5 c and d). Some of the morpholino experiments resulted in increased fin length, whereas others demonstrated widening of the regenerating dorsal side, such that we quantitated the increase in regeneration for these experiments by measuring the increase in total surface area between the dorsal and ventral halves (Fig. 3E). Consistent with the increase in size being due to knockdown of miR-203, immunostaining and western blot analyses of Lef1 protein levels in the dorsal halves of injected fins demonstrated significantly increased expression of Lef1 (Fig. 3 C and F). Thus, regulation of Lef1 by miR-203 appears to be a critical limiting step in regeneration. The data also suggest that reestablishment of miR-203 levels is necessary for proper termination of regeneration.
To further test the hypothesis that miR-203 regulates lef1, we performed in situ hybridization experiments to localize miR-203 and lef1 RNAs on sections from a single bony ray flanked by adjacent mesenchymal tissue. We found a perfect correlation between the presence of miR-203 and the loss of lef1 (Fig. S4c). Normally, miRNAs repress translation, but continued repression can lead to mRNA decay (34, 35). We also sectioned fins and conducted immunostaining with antibodies against Lef1 or, as a control, β-catenin. As before, Lef1 levels increased during normal regeneration, decreased on co-injection of miR-203, and were greatly elevated by co-injection of miR-203MO, whereas the β-catenin levels did not change under the different conditions (data not shown).
To ensure that the effect of miR-203 is direct, we evaluated whether the expression of Lef1 from mRNAs lacking a normal 3′ UTR would be able to rescue the repression observed in the presence of miR-203. Co-injection of miR-203 with lef1 mRNAs lacking MREs led to a substantial rescue of both fin regeneration and Lef1 protein levels (Fig. 3 D and E). The overall rescue was dose-dependent (Fig. S6). Finally, we injected morpholinos against lef1 in the presence and absence of co-injected miR-203MO (Fig. S5b, e, and f). The loss of lef1 led to inhibition of regeneration, and the overgrowth normally observed on injection of miR-203MO was blocked by co-injection of lef1MO, consistent with the hypothesis that miR-203 directly regulates lef1 expression.
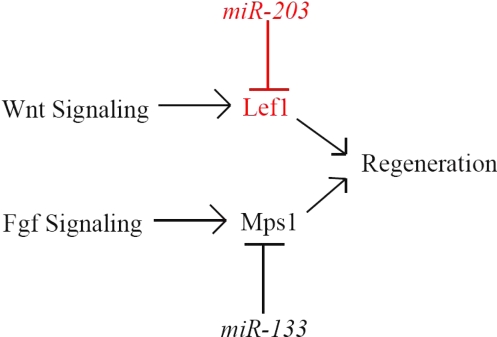
Our gain-of-function and loss-of-function regeneration experiments validate our array data and also suggest that a possible mechanism for termination of regeneration may be the increased expression of specific miRNAs that control the genes essential for regeneration. Consistent with this, the microarray and northern blot analysis results (Fig. S4b) demonstrate that miR-203 is readily detectable in adult fins and postregeneration fins but mostly undetectable in regenerating fins, suggesting transcriptional regulation of miR-203. Our results point to a key limiting role for lef1 in regeneration, but the possibility remains that miR-203 regulates other genes involved in regeneration as well. Considering the Dicer experiments shown in Fig. 1, along with data indicating a role for miR-133 in regeneration (19), it seems clear that the overall regeneration process is subject to extensive control by miRNAs (Fig. 4).
Fig. 4.
Regeneration model. Fin regeneration requires signaling cascades from both the Fgf and Wnt pathways, both of which are regulated by miRNAs.
The goal of regenerative research is to decipher the underlying mechanisms that allow organisms to recover from loss or damage caused by tissue injury, disease, and aging. Functional studies examining gain and loss of function of differentially expressed miRNAs in regenerating fins raise the possibility that identification of specific miRNA targets could lead to therapeutic targets that might ultimately allow regeneration in higher organisms. In adult humans, miR-203 is expressed in all tissues except liver, which may be one reason why regeneration is largely restricted to the liver (1, 36). Beyond regeneration, it is possible that similar mechanisms and genes may play important roles in proliferation versus terminal differentiation. This is especially intriguing for miR-203, which has been shown to promote differentiation of stratified epithelial cells (37, 38). Maintenance of the epidermis requires a balance between the proliferative capacity of the innermost basal layer and the differentiation and stratification of outer layers. This process depends on the expression of p63, a member of the p53 family of transcription factors (37). Interestingly, p63 is a target of miR-203, and p63 and Lef1 co-localize in regenerating fin sections (data not shown). The role of p63 in fin regeneration is unknown, but the possible regulation of p63 by miR-203 in regenerating fins would be consistent with epithelial differentiation models in which miR-203 has been hypothesized to block proliferation and/or “stemness.” This also is consistent with the ideas that overall regulation of regeneration requires the action of multiple miRNAs and that individual miRNAs may regulate multiple targets.
Methods
Microinjections.
Single-cell embryos were injected with 200 pg of miR-203, 5 ng of miR-203MO, and/or 50 pg of in vitro-transcribed, capped GFP reporter with or without the lef1 3′ UTR. Titrations were performed to determine the optimal amount of miR-203 and miR-203MO to use in the GFP experiments. Zebrafish lef1 3′ UTR sequences were amplified by RT-PCR and subcloned downstream of the GFP ORF inserted into pCS2+ (39).
Western Blot Analyses.
At 1 dpf, embryos were dechorionated, deyolked, and sonicated in lysis buffer as described previously (33). Then ≈30 embryos were pooled, and one-sixth of the resultant volume was loaded into each lane (Fig. 2C). For Fig. 3F, the distal tip of the dorsal half (500 μ) was amputated and pooled with six fins, and approximately one-sixth of this volume was loaded into each lane. Membranes were probed with antibodies against α-tubulin (Abcam, ab15246), GFP (Torrey Pines, TP401), or Lef1 (Abcam, ab52017). For detection, anti-rabbit HRP-conjugated secondary antibodies were used, followed by visualization with ECL.
Regeneration Experiments.
Caudal fins were amputated and allowed to regrow for 2 days at 33 °C. On the third day, each of the bony rays in the dorsal half of the fin were injected with 100 ng of dicerMO, dicer-mmMO (28), dicer-start (TCTTTCTCTTCATCTTCCTCCGATC), miR-203, miR-203MO, miR-203loopMO (TTGAGATAGAACTGTTGAACTGTTA), lef1MO (CTCCTCCACCTGACAACTGCGGCAT), or miR-15b or 30–150 ng of lef1-UTR (forward primer: CGGGATATCACTCAGCATAATG; reverse primer: TCGAGAACTTCTTTTAGGCCAG). After injection, electroporation was performed as described previously (25). The lef1 mRNA was transcribed from a construct provided by Dr. Richard Dorsky (40).
Whole-Mount Immunofluorescence.
Fins were fixed in 4% PFA overnight at 4 °C and washed in PBT before digestion with proteinase K for 10 min. The fins were then washed in PBT-DMSO before blocking for 1 h at room temperature (PBT-DMSO, 2% BSA, 5% goat serum). Primary Lef1 antibodies (Abcam, 1:100 dilution) were incubated for 4 h at room temperature, washed with PBT-DMSO, and then incubated with Cy5-conjugated donkey anti-goat antibodies (Jackson ImmunoResearch) for 4 h at room temperature. Before mounting and visualization, the fins were washed with PBT-DMSO.
Statistics.
Microarray data were analyzed using GeneSpring software. ANOVA using nonparametric parameters with unequal variances was used to determine P values. Paired Student t-tests were performed for the regrowth calculations (Fig. 2: n = 12, P = .0001; Fig. 3: n = 9, P ≪ .001 for miR-203, P < .001 for miR-203MO).
Supplementary Material
Acknowledgments.
This work was supported by National Institutes of Health Grants GM 075790 (to J.G.P.) and Training Fellowship GM62758 (to E.J.T.). We thank Drs. Liliana Solnica-Krezel and Bruce Appel for their advice and critical reading of the manuscript. We also thank Jenifer Ferguson and Jennell Talley for their assistance with fish maintenance and regeneration experiments and Robert Taylor for his assistance with imaging.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803713105/DCSupplemental.
References
- 1.Stoick-Cooper CL, Moon RT, Weidinger G. Advances in signaling in vertebrate regeneration as a prelude to regenerative medicine. Genes Dev. 2007;21:1292–1315. doi: 10.1101/gad.1540507. [DOI] [PubMed] [Google Scholar]
- 2.Johnson SL, Weston JA. Temperature-sensitive mutations that cause stage-specific defects in zebrafish fin regeneration. Genetics. 1995;141:1583–1595. doi: 10.1093/genetics/141.4.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Poss KD, Wilson LG, Keating MT. Heart regeneration in zebrafish. Science. 2002;298:2188–2190. doi: 10.1126/science.1077857. [DOI] [PubMed] [Google Scholar]
- 4.Akimenko MA, Mari-Beffa M, Becerra J, Geraudie J. Old questions, new tools, and some answers to the mystery of fin regeneration. Dev Dyn. 2003;226:190–201. doi: 10.1002/dvdy.10248. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP, Chen CZ. Micromanagers of gene expression: The potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004;5:396–400. doi: 10.1038/nrg1328. [DOI] [PubMed] [Google Scholar]
- 6.Carthew RW. Gene regulation by microRNAs. Curr Opin Genet Dev. 2006;16:203–208. doi: 10.1016/j.gde.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 7.Hobert O. Common logic of transcription factor and microRNA action. Trends Biochem Sci. 2004;29:462–468. doi: 10.1016/j.tibs.2004.07.001. [DOI] [PubMed] [Google Scholar]
- 8.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 9.Naguibneva I, et al. The microRNA miR-181 targets the homeobox protein Hox-A11 during mammalian myoblast differentiation. Nat Cell Biol. 2006;8:278–284. doi: 10.1038/ncb1373. [DOI] [PubMed] [Google Scholar]
- 10.O'Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT. c-Myc-regulated microRNAs modulate E2F1 expression. Nature. 2005;435:839–843. doi: 10.1038/nature03677. [DOI] [PubMed] [Google Scholar]
- 11.Pasquinelli AE, et al. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature. 2000;408:86–89. doi: 10.1038/35040556. [DOI] [PubMed] [Google Scholar]
- 12.Plasterk RH. Micro RNAs in animal development. Cell. 2006;124:877–881. doi: 10.1016/j.cell.2006.02.030. [DOI] [PubMed] [Google Scholar]
- 13.Reinhart BJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 14.Voorhoeve PM, et al. A genetic screen implicates miRNA-372 and miRNA-373 as oncogenes in testicular germ cell tumors. Cell. 2006;124:1169–1181. doi: 10.1016/j.cell.2006.02.037. [DOI] [PubMed] [Google Scholar]
- 15.Bernstein E, et al. Dicer is essential for mouse development. Nat Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- 16.Hatfield SD, et al. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435:974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- 17.Kanellopoulou C, et al. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev. 2005;19:489–501. doi: 10.1101/gad.1248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Lett. 2005;579:5911–5922. doi: 10.1016/j.febslet.2005.07.070. [DOI] [PubMed] [Google Scholar]
- 19.Yin VP, et al. Fgf-dependent depletion of microRNA-133 promotes appendage regeneration in zebrafish. Genes Dev. 2008;22:728–733. doi: 10.1101/gad.1641808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thatcher EJ, Flynt AS, Li N, Patton JR, Patton JG . MiRNA expression analysis during normal zebrafish development and following inhibition of the Hedgehog and Notch signaling pathways. Dev Dyn. 2007;236:2172–2180. doi: 10.1002/dvdy.21211. [DOI] [PubMed] [Google Scholar]
- 21.Schebesta M, Lien CL, Engel FB, Keating MT. Transcriptional profiling of caudal fin regeneration in zebrafish. ScientificWorldJournal. 2006;6:38–54. doi: 10.1100/tsw.2006.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poss KD, Shen J, Keating MT. Induction of lef1 during zebrafish fin regeneration. Dev Dyn. 2000;219:282–286. doi: 10.1002/1097-0177(2000)9999:9999<::aid-dvdy1045>3.3.co;2-3. [DOI] [PubMed] [Google Scholar]
- 23.Stoick-Cooper CL, et al. Distinct Wnt signaling pathways have opposing roles in appendage regeneration. Development. 2007;134:479–489. doi: 10.1242/dev.001123. [DOI] [PubMed] [Google Scholar]
- 24.Makino S, et al. Heat-shock protein 60 is required for blastema formation and maintenance during regeneration. Proc Natl Acad Sci U S A. 2005;102:14599–14604. doi: 10.1073/pnas.0507408102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thummel R, et al. Inhibition of zebrafish fin regeneration using in vivo electroporation of morpholinos against fgfr1 and msxb. Dev Dyn. 2006;235:336–346. doi: 10.1002/dvdy.20630. [DOI] [PubMed] [Google Scholar]
- 26.Poss KD, Nechiporuk A, Hillam AM, Johnson SL, Keating MT. Mps1 defines a proximal blastemal proliferative compartment essential for zebrafish fin regeneration. Development. 2002;129:5141–5149. doi: 10.1242/dev.129.22.5141. [DOI] [PubMed] [Google Scholar]
- 27.Bernstein E, Caudy A, Hammond S, Hannon G. Role for a bidentate ribonuclease in the intiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 28.Wienholds E, Koudijs MJ, van Eeden FJ, Cuppen E, Plasterk RH. The microRNA-producing enzyme Dicer1 is essential for zebrafish development. Nat Genet. 2003;35:217–218. doi: 10.1038/ng1251. [DOI] [PubMed] [Google Scholar]
- 29.Giraldez AJ, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- 30.Park JK, Liu X, Strauss TJ, McKearin DM, Liu Q. The miRNA pathway intrinsically controls self-renewal of Drosophila germline stem cells. Curr Biol. 2007;17:533–538. doi: 10.1016/j.cub.2007.01.060. [DOI] [PubMed] [Google Scholar]
- 31.Kloosterman WP, Lagendijk AK, Ketting RF, Moulton JD, Plasterk RH. Targeted inhibition of miRNA maturation with morpholinos reveals a role for miR-375 in pancreatic islet development. PLoS Biol. 2007;5:e203. doi: 10.1371/journal.pbio.0050203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jazwinska A, Badakov R, Keating MT. Activin-betaA signaling is required for zebrafish fin regeneration. Curr Biol. 2007;17:1390–1395. doi: 10.1016/j.cub.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 33.Flynt AS, Li N, Thatcher EJ, Solnica-Krezel L, Patton JG. Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat Genet. 2007;39:259–263. doi: 10.1038/ng1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baek D, et al . The impact of microRNAs on protein output. Nature. 2008;455:64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Selbach M, et al . Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 36.Barad O, et al. MicroRNA expression detected by oligonucleotide microarrays: System establishment and expression profiling in human tissues. Genome Res. 2004;14:2486–2494. doi: 10.1101/gr.2845604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lena AM, et al. miR-203 represses “stemness” by repressing DeltaNp63. Cell Death Differ. 2008;15:1187–1195. doi: 10.1038/cdd.2008.69. [DOI] [PubMed] [Google Scholar]
- 38.Yi R, Poy MN, Stoffel M, Fuchs E. A skin microRNA promotes differentiation by repressing “stemness.”. Nature. 2008;452:225–229. doi: 10.1038/nature06642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rupp RA, Snider L, Weintraub H. Xenopus embryos regulate the nuclear localization of XMyoD. Genes Dev. 1994;8:1311–1323. doi: 10.1101/gad.8.11.1311. [DOI] [PubMed] [Google Scholar]
- 40.Dorsky RI, et al. Maternal and embryonic expression of zebrafish lef1. Mech Dev. 1999;86:147–150. doi: 10.1016/s0925-4773(99)00101-x. [DOI] [PubMed] [Google Scholar]
- 41.Chen PY, et al. The developmental miRNA profiles of zebrafish as determined by small RNA cloning. Genes Dev. 2005;19:1288–1293. doi: 10.1101/gad.1310605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Griffiths-Jones S. The microRNA registry. Nucleic Acids Res. 2004;32:D109–D111. doi: 10.1093/nar/gkh023. (database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.